Assessment of Circulating Nucleic Acids in Cancer: From Current Status to Future Perspectives and Potential Clinical Applications
Abstract
Simple Summary
Abstract
1. Introduction
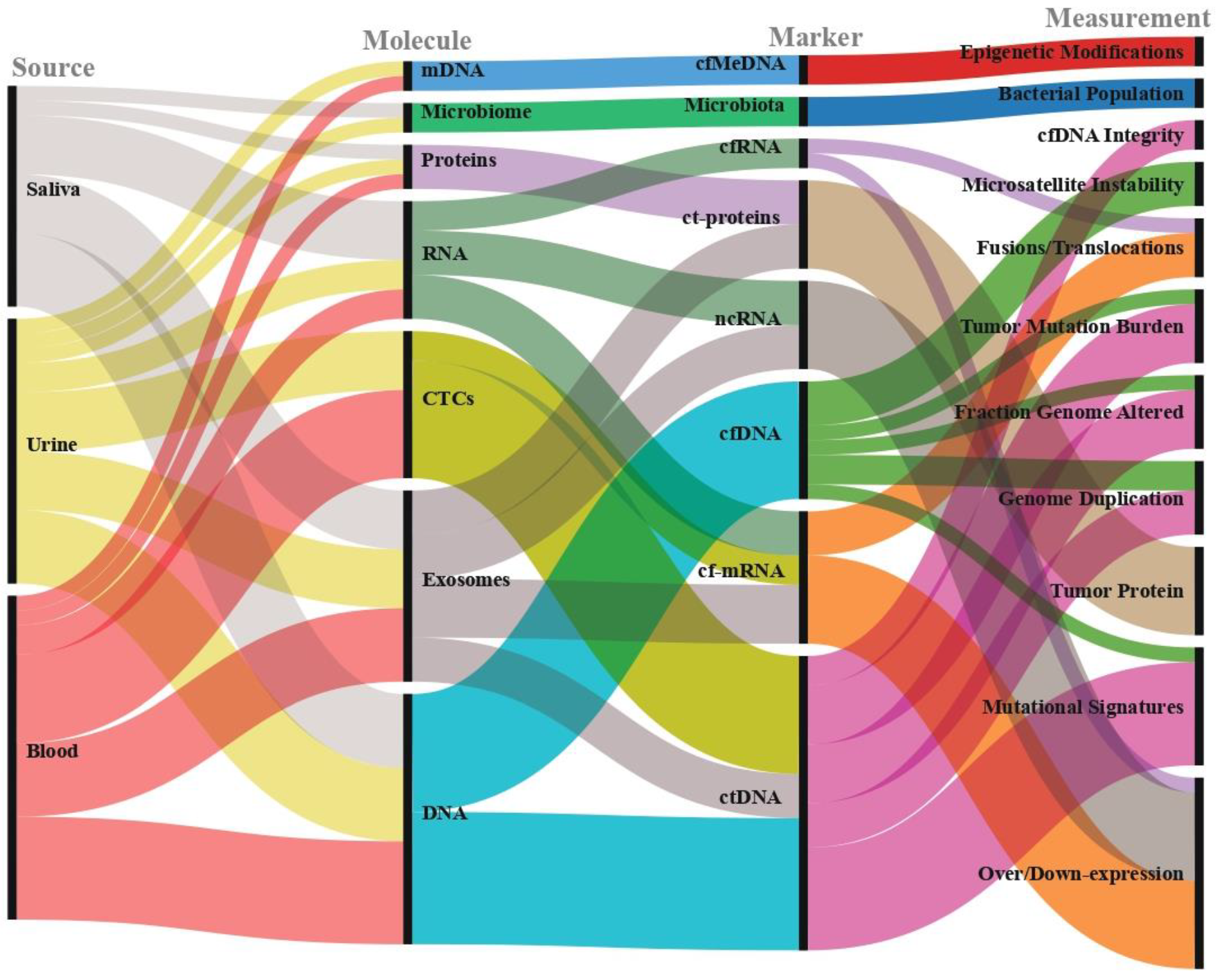
2. Pre-Analytical Management of Samples
2.1. Sample Types
2.2. Collection Tubes
2.3. Centrifugation and Long-Term Storage
3. cfNAs Extraction
4. cfNAs Quality Control
5. cfNAs Technical Applications
5.1. Mutation Detecation
5.2. Other Genetic Alterations
5.3. The Cell-Free RNA Transcriptome
5.4. Technologies for cfDNA Methylation Assessment
6. Current Challenges in ctDNA Detection and Strategies for Its Optimization
7. Current and Potential Applications of ctDNA Assessment
7.1. Assessment of Actionable Mutations for Therapeutic Purposes and Detection of Primary and Secondary Resistance to Systemic Therapy
7.2. Early Diagnosis
7.3. Early Detection of Recurrence/Relapse
7.4. ctDNA as An Independent Prognostic Factor
8. Theranostic Applications of cfRNA Assessment
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical Utility of Circulating Non-Coding RNAs—An Update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid Biopsy Enters the Clinic-Implementation Issues and Future Challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. ctDNA and CTCs in Liquid Biopsy - Current Status and Where We Need to Progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Aucamp, J.; Pretorius, P.J. Cell-Free DNA: Preanalytical Variables. Clin. Chim. Acta 2015, 450, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Grölz, D.; Hauch, S.; Schlumpberger, M.; Guenther, K.; Voss, T.; Sprenger-Haussels, M.; Oelmüller, U. Liquid Biopsy Preservation Solutions for Standardized Pre-Analytical Workflows-Venous Whole Blood and Plasma. Curr. Pathobiol. Rep. 2018, 6, 275–286. [Google Scholar] [CrossRef]
- Pantel, K.; Terstappen, L.W.; Baggiani, B.; Krahn, T.; Schlange, T. Abstract 1826: IMI’s CANCER-ID: Status of Liquid Biopsy Standardization. Cancer Res. 2016, 76, 1826. [Google Scholar] [CrossRef]
- Parpart-Li, S.; Bartlett, B.; Popoli, M.; Adleff, V.; Tucker, L.; Steinberg, R.; Georgiadis, A.; Phallen, J.; Brahmer, J.; Azad, N.; et al. The Effect of Preservative and Temperature on the Analysis of Circulating Tumor DNA. Clin. Cancer Res. 2017, 23, 2471–2477. [Google Scholar] [CrossRef]
- van Dessel, L.F.; Beije, N.; Helmijr, J.C.A.; Vitale, S.R.; Kraan, J.; Look, M.P.; de Wit, R.; Sleijfer, S.; Jansen, M.P.H.M.; Martens, J.W.M.; et al. Application of Circulating Tumor DNA in Prospective Clinical Oncology Trials-Standardization of Preanalytical Conditions. Mol. Oncol. 2017, 11, 295–304. [Google Scholar] [CrossRef]
- Diefenbach, R.J.; Lee, J.H.; Kefford, R.F.; Rizos, H. Evaluation of Commercial Kits for Purification of Circulating Free DNA. Cancer Genet. 2018, 228–229, 21–27. [Google Scholar] [CrossRef]
- Pérez-Barrios, C.; Nieto-Alcolado, I.; Torrente, M.; Jiménez-Sánchez, C.; Calvo, V.; Gutierrez-Sanz, L.; Palka, M.; Donoso-Navarro, E.; Provencio, M.; Romero, A. Comparison of Methods for Circulating Cell-Free DNA Isolation Using Blood from Cancer Patients: Impact on Biomarker Testing. Transl. Lung Cancer Res. 2016, 5, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Sorber, L.; Zwaenepoel, K.; Deschoolmeester, V.; Roeyen, G.; Lardon, F.; Rolfo, C.; Pauwels, P. A Comparison of Cell-Free DNA Isolation Kits: Isolation and Quantification of Cell-Free DNA in Plasma. J. Mol. Diagn. 2017, 19, 162–168. [Google Scholar] [CrossRef] [PubMed]
- van Dessel, L.F.; Vitale, S.R.; Helmijr, J.C.A.; Wilting, S.M.; van der Vlugt-Daane, M.; Oomen-de Hoop, E.; Sleijfer, S.; Martens, J.W.M.; Jansen, M.P.H.M.; Lolkema, M.P. High-Throughput Isolation of Circulating Tumor DNA: A Comparison of Automated Platforms. Mol. Oncol. 2019, 13, 392–402. [Google Scholar] [CrossRef]
- Warton, K.; Graham, L.-J.; Yuwono, N.; Samimi, G. Comparison of 4 Commercial Kits for the Extraction of Circulating DNA from Plasma. Cancer Genet. 2018, 228–229, 143–150. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and Comparison of in Vitro Degradation Kinetics of DNA in Serum, Urine and Saliva: A Qualitative Study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef]
- Di Meo, A.; Bartlett, J.; Cheng, Y.; Pasic, M.D.; Yousef, G.M. Liquid Biopsy: A Step Forward towards Precision Medicine in Urologic Malignancies. Mol. Cancer 2017, 16, 80. [Google Scholar] [CrossRef]
- Ding, S.; Song, X.; Geng, X.; Liu, L.; Ma, H.; Wang, X.; Wei, L.; Xie, L.; Song, X. Saliva-Derived cfDNA Is Applicable for EGFR Mutation Detection but Not for Quantitation Analysis in Non-Small Cell Lung Cancer. Thorac Cancer 2019, 10, 1973–1983. [Google Scholar] [CrossRef]
- Gerber, T.; Taschner-Mandl, S.; Saloberger-Sindhöringer, L.; Popitsch, N.; Heitzer, E.; Witt, V.; Geyeregger, R.; Hutter, C.; Schwentner, R.; Ambros, I.M.; et al. Assessment of Pre-Analytical Sample Handling Conditions for Comprehensive Liquid Biopsy Analysis. J. Mol. Diagn. 2020, 22, 1070–1086. [Google Scholar] [CrossRef]
- Fernando, M.R.; Norton, S.E.; Luna, K.K.; Lechner, J.M.; Qin, J. Stabilization of Cell-Free RNA in Blood Samples Using a New Collection Device. Clin. Biochem. 2012, 45, 1497–1502. [Google Scholar] [CrossRef]
- Das, K.; Fernando, M.R.; Basiaga, S.; Wigginton, S.M.; Williams, T. Effects of a Novel Cell Stabilizing Reagent on DNA Amplification by PCR as Compared to Traditional Stabilizing Reagents. Acta Histochem. 2014, 116, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Cruickshank-Quinn, C.; Zheng, L.K.; Quinn, K.; Bowler, R.; Reisdorph, R.; Reisdorph, N. Impact of Blood Collection Tubes and Sample Handling Time on Serum and Plasma Metabolome and Lipidome. Metabolites 2018, 8, 88. [Google Scholar] [CrossRef] [PubMed]
- Alidousty, C.; Brandes, D.; Heydt, C.; Wagener, S.; Wittersheim, M.; Schäfer, S.C.; Holz, B.; Merkelbach-Bruse, S.; Büttner, R.; Fassunke, J.; et al. Comparison of Blood Collection Tubes from Three Different Manufacturers for the Collection of Cell-Free DNA for Liquid Biopsy Mutation Testing. J. Mol. Diagn. 2017, 19, 801–804. [Google Scholar] [CrossRef]
- Toro, P.V.; Erlanger, B.; Beaver, J.A.; Cochran, R.L.; VanDenBerg, D.A.; Yakim, E.; Cravero, K.; Chu, D.; Zabransky, D.J.; Wong, H.Y.; et al. Comparison of Cell Stabilizing Blood Collection Tubes for Circulating Plasma Tumor DNA. Clin. Biochem. 2015, 48, 993–998. [Google Scholar] [CrossRef]
- Schmidt, B.; Reinicke, D.; Reindl, I.; Bork, I.; Wollschläger, B.; Lambrecht, N.; Fleischhacker, M. Liquid Biopsy-Performance of the PAXgene® Blood ccfDNA Tubes for the Isolation and Characterization of Cell-Free Plasma DNA from Tumor Patients. Clin. Chim. Acta 2017, 469, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Ward Gahlawat, A.; Lenhardt, J.; Witte, T.; Keitel, D.; Kaufhold, A.; Maass, K.K.; Pajtler, K.W.; Sohn, C.; Schott, S. Evaluation of Storage Tubes for Combined Analysis of Circulating Nucleic Acids in Liquid Biopsies. Int. J. Mol. Sci. 2019, 20, 704. [Google Scholar] [CrossRef] [PubMed]
- Van Paemel, R.; De Koker, A.; Caggiano, C.; Morlion, A.; Mestdagh, P.; De Wilde, B.; Vandesompele, J.; De Preter, K. Genome-Wide Study of the Effect of Blood Collection Tubes on the Cell-Free DNA Methylome. bioRxiv 2020. [Google Scholar] [CrossRef]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Somer-Shely, T.; Ryan, W.L. A Novel Approach to Stabilize Fetal Cell-Free DNA Fraction in Maternal Blood Samples for Extended Period of Time. PLoS ONE 2018, 13, e0208508. [Google Scholar] [CrossRef]
- Poole, J.C.; Wu, S.-F.; Lu, T.T.; Vibat, C.R.T.; Pham, A.; Samuelsz, E.; Patel, M.; Chen, J.; Daher, T.; Singh, V.M.; et al. Analytical Validation of the Target Selector ctDNA Platform Featuring Single Copy Detection Sensitivity for Clinically Actionable EGFR, BRAF, and KRAS Mutations. PLoS ONE 2019, 14, e0223112. [Google Scholar] [CrossRef]
- Martínez-Fernández, M.; Paramio, J.M.; Dueñas, M. RNA Detection in Urine: From RNA Extraction to Good Normalizer Molecules. J. Mol. Diagn. 2016, 18, 15–22. [Google Scholar] [CrossRef]
- Murugesan, K.; Hogan, C.A.; Palmer, Z.; Reeve, B.; Theron, G.; Andama, A.; Somoskovi, A.; Steadman, A.; Madan, D.; Andrews, J.; et al. Investigation of Preanalytical Variables Impacting Pathogen Cell-Free DNA in Blood and Urine. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Li, P.; Ning, J.; Luo, X.; Du, H.; Zhang, Q.; Zhou, G.; Du, Q.; Ou, Z.; Wang, L.; Wang, Y. New Method to Preserve the Original Proportion and Integrity of Urinary Cell-Free DNA. J. Clin. Lab. Anal. 2019, 33, e22668. [Google Scholar] [CrossRef]
- Langevin, S.M.; Kuhnell, D.; Biesiada, J.; Zhang, X.; Medvedovic, M.; Talaska, G.G.; Burns, K.A.; Kasper, S. Comparability of the Small RNA Secretome across Human Biofluids Concomitantly Collected from Healthy Adults. PLoS ONE 2020, 15, e0229976. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Moin, S.F.; Khan, R.S.; Agwan, M.A.S.; Alwadaani, A.H.; Zafar, M.S. Human Salivary Protein Extraction from RNAPro·SALTM, Pure·SALTM, and Passive Drooling Method. Eur. J. Dent. 2017, 11, 385–389. [Google Scholar] [CrossRef]
- Trigg, R.M.; Martinson, L.J.; Parpart-Li, S.; Shaw, J.A. Factors That Influence Quality and Yield of Circulating-Free DNA: A Systematic Review of the Methodology Literature. Heliyon 2018, 4, e00699. [Google Scholar] [CrossRef] [PubMed]
- Page, K.; Powles, T.; Slade, M.J.; DE Bella, M.T.; Walker, R.A.; Coombes, R.C.; Shaw, J.A. The Importance of Careful Blood Processing in Isolation of Cell-Free DNA. Ann. N. Y. Acad. Sci. 2006, 1075, 313–317. [Google Scholar] [CrossRef]
- El Messaoudi, S.; Rolet, F.; Mouliere, F.; Thierry, A.R. Circulating Cell Free DNA: Preanalytical Considerations. Clin. Chim. Acta 2013, 424, 222–230. [Google Scholar] [CrossRef]
- Sozzi, G.; Roz, L.; Conte, D.; Mariani, L.; Andriani, F.; Verderio, P.; Pastorino, U. Effects of Prolonged Storage of Whole Plasma or Isolated Plasma DNA on the Results of Circulating DNA Quantification Assays. J. Natl. Cancer Inst. 2005, 97, 1848–1850. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, R.; Judicone, C.; Poncelet, P.; Robert, S.; Arnaud, L.; Sampol, J.; Dignat-George, F. Impact of Pre-Analytical Parameters on the Measurement of Circulating Microparticles: Towards Standardization of Protocol. J. Thromb. Haemost. 2012, 10, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhu, Z.-Z.; Lu, Y.; Liu, F.; Zhang, W.; Huang, G.; Zhu, G.; Jiang, B. A Modified Extraction Method of Circulating Free DNA for Epidermal Growth Factor Receptor Mutation Analysis. Yonsei Med. J. 2012, 53, 132–137. [Google Scholar] [CrossRef]
- Oreskovic, A.; Brault, N.D.; Panpradist, N.; Lai, J.J.; Lutz, B.R. Analytical Comparison of Methods for Extraction of Short Cell-Free DNA from Urine. J. Mol. Diagn. 2019, 21, 1067–1078. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid Biopsies Come of Age: Towards Implementation of Circulating Tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Lee, H.; Park, C.; Na, W.; Park, K.H.; Shin, S. Precision Cell-Free DNA Extraction for Liquid Biopsy by Integrated Microfluidics. NPJ Precision Oncol. 2020, 4, 1–10. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. Comparison of Methods for the Isolation of Cell-Free DNA from Cell Culture Supernatant. Tumour Biol. 2020, 42, 1010428320916314. [Google Scholar] [CrossRef] [PubMed]
- Lampignano, R.; Neumann, M.H.D.; Weber, S.; Kloten, V.; Herdean, A.; Voss, T.; Groelz, D.; Babayan, A.; Tibbesma, M.; Schlumpberger, M.; et al. Multicenter Evaluation of Circulating Cell-Free DNA Extraction and Downstream Analyses for the Development of Standardized (Pre)analytical Work Flows. Clin. Chem. 2020, 66, 149–160. [Google Scholar] [CrossRef] [PubMed]
- van der Leest, P.; Boonstra, P.A.; Elst, A.T.; van Kempen, L.C.; Tibbesma, M.; Koopmans, J.; Miedema, A.; Tamminga, M.; Groen, H.J.M.; Reyners, A.K.L.; et al. Comparison of Circulating Cell-Free DNA Extraction Methods for Downstream Analysis in Cancer Patients. Cancers 2020, 12, 1222. [Google Scholar] [CrossRef]
- Johansson, G.; Andersson, D.; Filges, S.; Li, J.; Muth, A.; Godfrey, T.E.; Ståhlberg, A. Considerations and Quality Controls When Analyzing Cell-Free Tumor DNA. Biomol. Detect Quantif 2019, 17, 100078. [Google Scholar] [CrossRef]
- Streleckiene, G.; Reid, H.M.; Arnold, N.; Bauerschlag, D.; Forster, M. Quantifying Cell Free DNA in Urine: Comparison between Commercial Kits, Impact of Gender and Inter-Individual Variation. Biotechniques 2018, 64, 225–230. [Google Scholar] [CrossRef]
- Volik, S.; Alcaide, M.; Morin, R.D.; Collins, C. Cell-Free DNA (cfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies. Mol. Cancer Res. 2016, 14, 898–908. [Google Scholar] [CrossRef]
- Alcaide, M.; Rushton, C.; Morin, R.D. Ultrasensitive Detection of Circulating Tumor DNA in Lymphoma via Targeted Hybridization Capture and Deep Sequencing of Barcoded Libraries. Methods Mol. Biol. 2019, 1956, 383–435. [Google Scholar] [CrossRef]
- Gorgannezhad, L.; Umer, M.; Islam, M.N.; Nguyen, N.-T.; Shiddiky, M.J.A. Circulating Tumor DNA and Liquid Biopsy: Opportunities, Challenges, and Recent Advances in Detection Technologies. Lab. Chip. 2018, 18, 1174–1196. [Google Scholar] [CrossRef]
- Janku, F.; Vibat, C.R.T.; Kosco, K.; Holley, V.R.; Cabrilo, G.; Meric-Bernstam, F.; Stepanek, V.M.; Lin, P.P.; Leppin, L.; Hassaine, L.; et al. BRAF V600E Mutations in Urine and Plasma Cell-Free DNA from Patients with Erdheim-Chester Disease. Oncotarget 2014, 5, 3607–3610. [Google Scholar] [CrossRef]
- Vendrell, J.A.; Mau-Them, F.T.; Béganton, B.; Godreuil, S.; Coopman, P.; Solassol, J. Circulating Cell Free Tumor DNA Detection as a Routine Tool forLung Cancer Patient Management. Int. J. Mol. Sci. 2017, 18, 264. [Google Scholar] [CrossRef]
- Bos, M.K.; Nasserinejad, K.; Jansen, M.P.H.M.; Angus, L.; Atmodimedjo, P.N.; de Jonge, E.; Dinjens, W.N.M.; van Schaik, R.H.N.; Del Re, M.; Dubbink, H.J.; et al. Comparison of Variant Allele Frequency and Number of Mutant Molecules as Units of Measurement for Circulating Tumor DNA. Mol. Oncol. 2021, 15, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, V.; Rutkowski, A.J.; Meuser, E.; Hedley Carr, T.; Harrington, E.A.; Carl Barrett, J. Optimisation of Robust Singleplex and Multiplex Droplet Digital PCR Assays for High Confidence Mutation Detection in Circulating Tumour DNA. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Hu, S.; Zhang, L.; Xin, J.; Sun, C.; Wang, L.; Ding, K.; Wang, B. Tumor Circulome in the Liquid Biopsies for Cancer Diagnosis and Prognosis. Theranostics 2020, 10, 4544–4556. [Google Scholar] [CrossRef] [PubMed]
- Remon, J.; García-Campelo, R.; de Álava, E.; Vera, R.; Rodríguez-Peralto, J.L.; Rodríguez-Lescure, Á.; Bellosillo, B.; Garrido, P.; Rojo, F.; Álvarez-Alegret, R. Liquid Biopsy in Oncology: A Consensus Statement of the Spanish Society of Pathology and the Spanish Society of Medical Oncology. Clin. Transl. Oncol. 2020, 22, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, M.; Dawson, S.-J.; Tsui, D.W.Y.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.-F.; Kingsbury, Z.; Wong, A.S.C.; et al. Non-Invasive Analysis of Acquired Resistance to Cancer Therapy by Sequencing of Plasma DNA. Nature 2013, 497, 108–112. [Google Scholar] [CrossRef]
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and Analytical Validation of FoundationOne Liquid CDx, a Novel 324-Gene cfDNA-Based Comprehensive Genomic Profiling Assay for Cancers of Solid Tumor Origin. PLoS ONE 2020, 15, e0237802. [Google Scholar] [CrossRef] [PubMed]
- Lanman, R.B.; Mortimer, S.A.; Zill, O.A.; Sebisanovic, D.; Lopez, R.; Blau, S.; Collisson, E.A.; Divers, S.G.; Hoon, D.S.B.; Kopetz, E.S.; et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS ONE 2015, 10, e0140712. [Google Scholar] [CrossRef]
- Ho, G.Y.F.; Wang, T.; Kwok, H.-H.; Rasul, R.; Peila, R.; Guzman, M.; Ip, M.S.M.; Lam, D.C.L. Longitudinal Multi-Gene Panel Assessment of Circulating Tumor DNA Revealed Tumor Burden and Molecular Characteristics along Treatment Course of Non-Small Cell Lung Cancer. Transl. Lung Cancer Res. 2020, 9, 1873–1884. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.J.; Gauthier, M.-E.A.; Chan, C.-L.; Thompson, T.J.; De Sousa, S.M.C.; Puttick, C.; Grady, J.P.; Gayevskiy, V.; Tao, J.; Ying, K.; et al. Development and Validation of a Targeted Gene Sequencing Panel for Application to Disparate Cancers. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef]
- Cimmino, F.; Lasorsa, V.A.; Vetrella, S.; Iolascon, A.; Capasso, M. A Targeted Gene Panel for Circulating Tumor DNA Sequencing in Neuroblastoma. Front. Oncol. 2020, 10, 596191. [Google Scholar] [CrossRef] [PubMed]
- Gobbini, E.; Swalduz, A.; Levra, M.G.; Ortiz-Cuaran, S.; Toffart, A.-C.; Pérol, M.; Moro-Sibilot, D.; Saintigny, P. Implementing ctDNA Analysis in the Clinic: Challenges and Opportunities in Non-Small Cell Lung Cancer. Cancers 2020, 12, 3112. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-C.; Du, M.; Wang, L. Bioinformatics Analysis for Circulating Cell-Free DNA in Cancer. Cancers 2019, 11, 805. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.; Kristiansson, H.; Kubista, M.; Ståhlberg, A. Ultrasensitive Circulating Tumor DNA Analysis Enables Precision Medicine: Experimental Workflow Considerations. Expert Rev. Mol. Diagn. 2021, 1–12. [Google Scholar] [CrossRef]
- Gale, D.; Lawson, A.R.J.; Howarth, K.; Madi, M.; Durham, B.; Smalley, S.; Calaway, J.; Blais, S.; Jones, G.; Clark, J.; et al. Development of a Highly Sensitive Liquid Biopsy Platform to Detect Clinically-Relevant Cancer Mutations at Low Allele Fractions in Cell-Free DNA. PLoS ONE 2018, 13, e0194630. [Google Scholar] [CrossRef]
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and Quantification of Rare Mutations with Massively Parallel Sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535. [Google Scholar] [CrossRef]
- Ståhlberg, A.; Krzyzanowski, P.M.; Egyud, M.; Filges, S.; Stein, L.; Godfrey, T.E. Simple Multiplexed PCR-Based Barcoding of DNA for Ultrasensitive Mutation Detection by next-Generation Sequencing. Nat. Protoc. 2017, 12, 664–682. [Google Scholar] [CrossRef]
- Gregory, M.T.; Bertout, J.A.; Ericson, N.G.; Taylor, S.D.; Mukherjee, R.; Robins, H.S.; Drescher, C.W.; Bielas, J.H. Targeted Single Molecule Mutation Detection with Massively Parallel Sequencing. Nucleic Acids Res. 2016, 44, e22. [Google Scholar] [CrossRef][Green Version]
- Schmitt, M.W.; Kennedy, S.R.; Salk, J.J.; Fox, E.J.; Hiatt, J.B.; Loeb, L.A. Detection of Ultra-Rare Mutations by next-Generation Sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 14508–14513. [Google Scholar] [CrossRef]
- Cantsilieris, S.; Stessman, H.A.; Shendure, J.; Eichler, E.E. Targeted Capture and High-Throughput Sequencing Using Molecular Inversion Probes (MIPs). Methods Mol. Biol. 2017, 1492, 95–106. [Google Scholar] [CrossRef]
- Hong, L.Z.; Hong, S.; Wong, H.T.; Aw, P.P.K.; Cheng, Y.; Wilm, A.; de Sessions, P.F.; Lim, S.G.; Nagarajan, N.; Hibberd, M.L.; et al. BAsE-Seq: A Method for Obtaining Long Viral Haplotypes from Short Sequence Reads. Genome Biol. 2014, 15, 1–14. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An Ultrasensitive Method for Quantitating Circulating Tumor DNA with Broad Patient Coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Elazezy, M.; Joosse, S.A. Techniques of Using Circulating Tumor DNA as a Liquid Biopsy Component in Cancer Management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chng, K.R.; Boey, E.J.H.; Ng, A.H.Q.; Wilm, A.; Nagarajan, N. INC-Seq: Accurate Single Molecule Reads Using Nanopore Sequencing. Gigascience 2016, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Rodda, A.E.; Parker, B.J.; Spencer, A.; Corrie, S.R. Extending Circulating Tumor DNA Analysis to Ultralow Abundance Mutations: Techniques and Challenges. ACS Sens. 2018, 3, 540–560. [Google Scholar] [CrossRef] [PubMed]
- Mosko, M.J.; Nakorchevsky, A.A.; Flores, E.; Metzler, H.; Ehrich, M.; van den Boom, D.J.; Sherwood, J.L.; Nygren, A.O.H. Ultrasensitive Detection of Multiplexed Somatic Mutations Using MALDI-TOF Mass Spectrometry. J. Mol. Diagn. 2016, 18, 23–31. [Google Scholar] [CrossRef]
- Kamps-Hughes, N.; McUsic, A.; Kurihara, L.; Harkins, T.T.; Pal, P.; Ray, C.; Ionescu-Zanetti, C. ERASE-Seq: Leveraging Replicate Measurements to Enhance Ultralow Frequency Variant Detection in NGS Data. PLoS ONE 2018, 13, e0195272. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical Relevance of Blood-Based ctDNA Analysis: Mutation Detection and beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef]
- Ruhen, O.; Mirzai, B.; Clark, M.E.; Nguyen, B.; Salomon, C.; Erber, W.; Meehan, K. Comparison of Circulating Tumour DNA and Extracellular Vesicle DNA by Low-Pass Whole-Genome Sequencing Reveals Molecular Drivers of Disease in a Breast Cancer Patient. Biomedicines 2020, 9, 14. [Google Scholar] [CrossRef]
- Dietz, S.; Christopoulos, P.; Yuan, Z.; Angeles, A.K.; Gu, L.; Volckmar, A.-L.; Ogrodnik, S.J.; Janke, F.; Fratte, C.D.; Zemojtel, T.; et al. Longitudinal Therapy Monitoring of ALK-Positive Lung Cancer by Combined Copy Number and Targeted Mutation Profiling of Cell-Free DNA. EBioMedicine 2020, 62. [Google Scholar] [CrossRef] [PubMed]
- Kinde, I.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. FAST-SeqS: A Simple and Efficient Method for the Detection of Aneuploidy by Massively Parallel Sequencing. PLoS ONE 2012, 7, e41162. [Google Scholar] [CrossRef]
- Belic, J.; Koch, M.; Ulz, P.; Auer, M.; Gerhalter, T.; Mohan, S.; Fischereder, K.; Petru, E.; Bauernhofer, T.; Geigl, J.B.; et al. Rapid Identification of Plasma DNA Samples with Increased ctDNA Levels by a Modified FAST-SeqS Approach. Clin. Chem. 2015, 61, 838–849. [Google Scholar] [CrossRef]
- Kirkizlar, E.; Zimmermann, B.; Constantin, T.; Swenerton, R.; Hoang, B.; Wayham, N.; Babiarz, J.E.; Demko, Z.; Pelham, R.J.; Kareht, S.; et al. Detection of Clonal and Subclonal Copy-Number Variants in Cell-Free DNA from Patients with Breast Cancer Using a Massively Multiplexed PCR Methodology. Transl. Oncol. 2015, 8, 407–416. [Google Scholar] [CrossRef]
- Douville, C.; Cohen, J.D.; Ptak, J.; Popoli, M.; Schaefer, J.; Silliman, N.; Dobbyn, L.; Schoen, R.E.; Tie, J.; Gibbs, P.; et al. Assessing Aneuploidy with Repetitive Element Sequencing. Proc. Natl. Acad. Sci. USA 2020, 117, 4858–4863. [Google Scholar] [CrossRef]
- Kato, S.; Okamura, R.; Baumgartner, J.M.; Patel, H.; Leichman, L.; Kelly, K.; Sicklick, J.K.; Fanta, P.T.; Lippman, S.M.; Kurzrock, R. Analysis of Circulating Tumor DNA and Clinical Correlates in Patients with Esophageal, Gastroesophageal Junction, and Gastric Adenocarcinoma. Clin. Cancer Res. 2018, 24, 6248–6256. [Google Scholar] [CrossRef]
- Clifton, K.; Rich, T.A.; Parseghian, C.; Raymond, V.M.; Dasari, A.; Pereira, A.A.L.; Willis, J.; Loree, J.M.; Bauer, T.M.; Chae, Y.K.; et al. Identification of Actionable Fusions as an Anti-EGFR Resistance Mechanism Using a Circulating Tumor DNA Assay. JCO Precision Oncol. 2019, 1–15. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Rooney, M.; Nagy, R.J.; Lin, J.J.; Chin, E.; Ferris, L.A.; Ackil, J.; Lennerz, J.K.; Lanman, R.B.; Gainor, J.F.; et al. Molecular Analysis of Plasma From Patients With ROS1-Positive NSCLC. J. Thorac. Oncol. 2019, 14, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Poehlein, C.H.; Marton, M.J.; Laterza, O.F.; Levitan, D. Measuring Tumor Mutational Burden (TMB) in Plasma from mCRPC Patients Using Two Commercial NGS Assays. Sci. Rep. 2019, 9, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Duan, J.; Cai, S.; Han, M.; Dong, H.; Zhao, J.; Zhu, B.; Wang, S.; Zhuo, M.; Sun, J.; et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients with Non-Small Cell Lung Cancer with Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019, 5, 696–702. [Google Scholar] [CrossRef]
- Willis, J.; Lefterova, M.I.; Artyomenko, A.; Kasi, P.M.; Nakamura, Y.; Mody, K.; Catenacci, D.V.T.; Fakih, M.; Barbacioru, C.; Zhao, J.; et al. Validation of Microsatellite Instability Detection Using a Comprehensive Plasma-Based Genotyping Panel. Clin. Cancer Res. 2019, 25, 7035–7045. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Mercado, M.; Manterola, L.; Larrea, E.; Goicoechea, I.; Arestin, M.; Armesto, M.; Otaegui, D.; Lawrie, C.H. The Circulating Transcriptome as a Source of Non-Invasive Cancer Biomarkers: Concepts and Controversies of Non-Coding and Coding RNA in Body Fluids. J. Cell. Mol. Med. 2015, 19, 2307–2323. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Li, X.; Zhang, F.; Zhang, C.; Guan, Q.; Cao, X.; Zhu, W.; Zhang, X.; Cheng, Y.; Ou, K.; et al. Circulating lncRNAs Associated with Occurrence of Colorectal Cancer Progression. Am. J. Cancer Res. 2015, 5, 2258–2265. [Google Scholar]
- Chapin, S.C.; Appleyard, D.C.; Pregibon, D.C.; Doyle, P.S. Rapid microRNA Profiling on Encoded Gel Microparticles. Angew. Chem. Int. Ed Engl. 2011, 50, 2289–2293. [Google Scholar] [CrossRef]
- Moldovan, L.; Batte, K.E.; Trgovcich, J.; Wisler, J.; Marsh, C.B.; Piper, M. Methodological Challenges in Utilizing miRNAs as Circulating Biomarkers. J. Cell. Mol. Med. 2014, 18, 371–390. [Google Scholar] [CrossRef] [PubMed]
- Shore, S.; Henderson, J.M.; Lebedev, A.; Salcedo, M.P.; Zon, G.; McCaffrey, A.P.; Paul, N.; Hogrefe, R.I. Small RNA Library Preparation Method for Next-Generation Sequencing Using Chemical Modifications to Prevent Adapter Dimer Formation. PLoS ONE 2016, 11, e0167009. [Google Scholar] [CrossRef]
- Kashi, K.; Henderson, L.; Bonetti, A.; Carninci, P. Discovery and Functional Analysis of lncRNAs: Methodologies to Investigate an Uncharacterized Transcriptome. Biochim. Biophys. Acta 2016, 1859, 3–15. [Google Scholar] [CrossRef]
- Bustin, S.; Dhillon, H.S.; Kirvell, S.; Greenwood, C.; Parker, M.; Shipley, G.L.; Nolan, T. Variability of the Reverse Transcription Step: Practical Implications. Clin. Chem. 2015, 61, 202–212. [Google Scholar] [CrossRef]
- Koh, W.; Pan, W.; Gawad, C.; Fan, H.C.; Kerchner, G.A.; Wyss-Coray, T.; Blumenfeld, Y.J.; El-Sayed, Y.Y.; Quake, S.R. Noninvasive in Vivo Monitoring of Tissue-Specific Global Gene Expression in Humans. Proc. Natl. Acad. Sci. USA 2014, 111, 7361–7366. [Google Scholar] [CrossRef]
- Ibarra, A.; Zhuang, J.; Zhao, Y.; Salathia, N.S.; Huang, V.; Acosta, A.D.; Aballi, J.; Toden, S.; Karns, A.P.; Purnajo, I.; et al. Non-Invasive Characterization of Human Bone Marrow Stimulation and Reconstitution by Cell-Free Messenger RNA Sequencing. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Song, C.-X.; Yin, S.; Ma, L.; Wheeler, A.; Chen, Y.; Zhang, Y.; Liu, B.; Xiong, J.; Zhang, W.; Hu, J.; et al. 5-Hydroxymethylcytosine Signatures in Cell-Free DNA Provide Information about Tumor Types and Stages. Cell Res. 2017, 27, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Rand, A.C.; Jain, M.; Eizenga, J.M.; Musselman-Brown, A.; Olsen, H.E.; Akeson, M.; Paten, B. Mapping DNA Methylation with High-Throughput Nanopore Sequencing. Nat. Methods 2017, 14, 411–413. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Kokubun, S.; Itoi, E.; Roach, H.I. Improved Quantification of DNA Methylation Using Methylation-Sensitive Restriction Enzymes and Real-Time PCR. Epigenetics 2007, 2, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Tycko, B.; Liu, T.-M.; Lin, J.-J.L.; Huang, T.H.-M. Methods in DNA Methylation Profiling. Epigenomics 2009, 1, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Bonora, G.; Rubbi, L.; Morselli, M.; Ma, F.; Chronis, C.; Plath, K.; Pellegrini, M. DNA Methylation Estimation Using Methylation-Sensitive Restriction Enzyme Bisulfite Sequencing (MREBS). PLoS ONE 2019, 14, e0214368. [Google Scholar] [CrossRef] [PubMed]
- Darst, R.P.; Pardo, C.E.; Ai, L.; Brown, K.D.; Kladde, M.P. Bisulfite Sequencing of DNA. Curr. Protoc. Mol. Biol. 2010, 91, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Olova, N.; Krueger, F.; Andrews, S.; Oxley, D.; Berrens, R.V.; Branco, M.R.; Reik, W. Comparison of Whole-Genome Bisulfite Sequencing Library Preparation Strategies Identifies Sources of Biases Affecting DNA Methylation Data. Genome Biol. 2018, 19, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Skvortsova, K.; Zotenko, E.; Luu, P.-L.; Gould, C.M.; Nair, S.S.; Clark, S.J.; Stirzaker, C. Comprehensive Evaluation of Genome-Wide 5-Hydroxymethylcytosine Profiling Approaches in Human DNA. Epigenetics Chromatin 2017, 10, 1–20. [Google Scholar] [CrossRef]
- Holmes, E.E.; Jung, M.; Meller, S.; Leisse, A.; Sailer, V.; Zech, J.; Mengdehl, M.; Garbe, L.-A.; Uhl, B.; Kristiansen, G.; et al. Performance Evaluation of Kits for Bisulfite-Conversion of DNA from Tissues, Cell Lines, FFPE Tissues, Aspirates, Lavages, Effusions, Plasma, Serum, and Urine. PLoS ONE 2014, 9, e93933. [Google Scholar] [CrossRef]
- Cheuk, I.W.Y.; Shin, V.Y.; Kwong, A. Detection of Methylated Circulating DNA as Noninvasive Biomarkers for Breast Cancer Diagnosis. J. Breast Cancer 2017, 20, 12–19. [Google Scholar] [CrossRef]
- Delpu, Y.; Cordelier, P.; Cho, W.C.; Torrisani, J. DNA Methylation and Cancer Diagnosis. Int. J. Mol. Sci. 2013, 14, 15029–15058. [Google Scholar] [CrossRef]
- Singh, A.; Gupta, S.; Badarukhiya, J.A.; Sachan, M. Detection of Aberrant Methylation of HOXA9 and HIC1 through Multiplex MethyLight Assay in Serum DNA for the Early Detection of Epithelial Ovarian Cancer. Int. J. Cancer 2020, 147, 1740–1752. [Google Scholar] [CrossRef] [PubMed]
- Eads, C.A.; Danenberg, K.D.; Kawakami, K.; Saltz, L.B.; Blake, C.; Shibata, D.; Danenberg, P.V.; Laird, P.W. MethyLight: A High-Throughput Assay to Measure DNA Methylation. Nucleic Acids Res. 2000, 28, E32. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Davies, J.J.; Wittig, D.; Oakeley, E.J.; Haase, M.; Lam, W.L.; Schübeler, D. Chromosome-Wide and Promoter-Specific Analyses Identify Sites of Differential DNA Methylation in Normal and Transformed Human Cells. Nat. Genet. 2005, 37, 853–862. [Google Scholar] [CrossRef]
- Down, T.A.; Rakyan, V.K.; Turner, D.J.; Flicek, P.; Li, H.; Kulesha, E.; Gräf, S.; Johnson, N.; Herrero, J.; Tomazou, E.M.; et al. A Bayesian Deconvolution Strategy for Immunoprecipitation-Based DNA Methylome Analysis. Nat. Biotechnol. 2008, 26, 779–785. [Google Scholar] [CrossRef]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive Tumour Detection and Classification Using Plasma Cell-Free DNA Methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.Y.; Burgener, J.M.; Bratman, S.V.; De Carvalho, D.D. Preparation of cfMeDIP-Seq Libraries for Methylome Profiling of Plasma Cell-Free DNA. Nat. Protoc. 2019, 14, 2749–2780. [Google Scholar] [CrossRef]
- Galardi, F.; Luca, F.D.; Romagnoli, D.; Biagioni, C.; Moretti, E.; Biganzoli, L.; Leo, A.D.; Migliaccio, I.; Malorni, L.; Benelli, M. Cell-Free DNA-Methylation-Based Methods and Applications in Oncology. Biomolecules 2020, 10, 1677. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Broche, J.; Lungu, C.; Bashtrykov, P. Biotechnological Applications of MBD Domain Proteins for DNA Methylation Analysis. J. Mol. Biol. 2019. [Google Scholar] [CrossRef]
- Hoppers, A.; Williams, L.; Ponnaluri, V.K.C.; Sexton, B.; Saleh, L.; Campbell, M.; Marks, K.; Samaranayake, M.; Ettwiller, L.; Guan, S.; et al. Enzymatic Methyl-Seq: Next Generation Methylomes. J. Biomol. Tech. 2020, 31, S15. [Google Scholar]
- Liu, Y.; Siejka-Zielińska, P.; Velikova, G.; Bi, Y.; Yuan, F.; Tomkova, M.; Bai, C.; Chen, L.; Schuster-Böckler, B.; Song, C.-X. Bisulfite-Free Direct Detection of 5-Methylcytosine and 5-Hydroxymethylcytosine at Base Resolution. Nat. Biotechnol. 2019, 37, 424–429. [Google Scholar] [CrossRef]
- Lam, D.; Luu, P.-L.; Song, J.Z.; Qu, W.; Risbridger, G.P.; Lawrence, M.G.; Lu, J.; Trau, M.; Korbie, D.; Clark, S.J.; et al. Comprehensive Evaluation of Targeted Multiplex Bisulphite PCR Sequencing for Validation of DNA Methylation Biomarker Panels. Clin. Epigenetics 2020, 12, 1–16. [Google Scholar] [CrossRef]
- Erger, F.; Nörling, D.; Borchert, D.; Leenen, E.; Habbig, S.; Wiesener, M.S.; Bartram, M.P.; Wenzel, A.; Becker, C.; Toliat, M.R.; et al. cfNOMe — A Single Assay for Comprehensive Epigenetic Analyses of Cell-Free DNA. Genome Med. 2020, 12, 1–14. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.B.; Siu, L.L.; et al. How Liquid Biopsies Can Change Clinical Practice in Oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef] [PubMed]
- Nawroz, H.; Koch, W.; Anker, P.; Stroun, M.; Sidransky, D. Microsatellite Alterations in Serum DNA of Head and Neck Cancer Patients. Nat. Med. 1996, 2, 1035–1037. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Bardelli, A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Sartore-Bianchi, A.; Nagy, R.J.; Raghav, K.; Odegaard, J.I.; Lanman, R.B.; Trusolino, L.; Marsoni, S.; Siena, S.; Bardelli, A. Plasma HER2 (ERBB2) Copy Number Predicts Response to HER2-Targeted Therapy in Metastatic Colorectal Cancer. Clin. Cancer Res. 2019, 25, 3046–3053. [Google Scholar] [CrossRef]
- Aravanis, A.M.; Lee, M.; Klausner, R.D. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017, 168, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced Detection of Circulating Tumor DNA by Fragment Size Analysis. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef]
- Sobhani, N.; Generali, D.; Zanconati, F.; Bortul, M.; Scaggiante, B. Cell-Free DNA Integrity for the Monitoring of Breast Cancer: Future Perspectives? World J. Clin. Oncol. 2018, 9, 26–32. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-Wide Cell-Free DNA Fragmentation in Patients with Cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Qiu, J.; Xu, J.; Zhang, K.; Gu, W.; Nie, L.; Wang, G.; Luo, Y. Refining Cancer Management Using Integrated Liquid Biopsy. Theranostics 2020, 10, 2374–2384. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor–Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Toppmeyer, D.L.; Press, M.F. Testing Considerations for Phosphatidylinositol-3-Kinase Catalytic Subunit Alpha as an Emerging Biomarker in Advanced Breast Cancer. Cancer Med. 2020, 9, 6463–6472. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Guardant360® CDx. FDA. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf20/P200010A.pdf (accessed on 15 February 2021).
- US Food and Drug Administration. FoundationOne® Liquid CDx (F1 Liquid CDx).FDA. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf20/P200016A.pdf (accessed on 25 February 2021).
- Zheng, D.; Ye, X.; Zhang, M.Z.; Sun, Y.; Wang, J.Y.; Ni, J.; Zhang, H.P.; Zhang, L.; Luo, J.; Zhang, J.; et al. Plasma EGFR T790M ctDNA Status Is Associated with Clinical Outcome in Advanced NSCLC Patients with Acquired EGFR-TKI Resistance. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Vidal, J.; Muinelo, L.; Dalmases, A.; Jones, F.; Edelstein, D.; Iglesias, M.; Orrillo, M.; Abalo, A.; Rodríguez, C.; Brozos, E.; et al. Plasma ctDNA RAS Mutation Analysis for the Diagnosis and Treatment Monitoring of Metastatic Colorectal Cancer Patients. Ann. Oncol. 2017, 28, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- Calapre, L.; Warburton, L.; Millward, M.; Ziman, M.; Gray, E.S. Circulating Tumour DNA (ctDNA) as a Liquid Biopsy for Melanoma. Cancer Lett. 2017, 404, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-Y.; Xie, N.; Tian, C.; Yang, X.; Liu, L.; Li, J.; Xiao, H.; Wu, H.; Lu, J.; Gao, J.; et al. Identifying Circulating Tumor DNA Mutation Profiles in Metastatic Breast Cancer Patients with Multiline Resistance. EBioMedicine 2018, 32, 111–118. [Google Scholar] [CrossRef]
- De Santo, I.; McCartney, A.; Migliaccio, I.; Di Leo, A.; Malorni, L. The Emerging Role of ESR1 Mutations in Luminal Breast Cancer as a Prognostic and Predictive Biomarker of Response to Endocrine Therapy. Cancers 2019, 11, 1894. [Google Scholar] [CrossRef]
- Khagi, Y.; Goodman, A.M.; Daniels, G.A.; Patel, S.P.; Sacco, A.G.; Randall, J.M.; Bazhenova, L.A.; Kurzrock, R. Hypermutated Circulating Tumor DNA: Correlation with Response to Checkpoint Inhibitor-Based Immunotherapy. Clin. Cancer Res. 2017, 23, 5729–5736. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating Tumor DNA as an Early Marker of Therapeutic Response in Patients with Metastatic Colorectal Cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Kruger, S.; Heinemann, V.; Ross, C.; Diehl, F.; Nagel, D.; Ormanns, S.; Liebmann, S.; Prinz-Bravin, I.; Westphalen, C.B.; Haas, M.; et al. Repeated mutKRAS ctDNA Measurements Represent a Novel and Promising Tool for Early Response Prediction and Therapy Monitoring in Advanced Pancreatic Cancer. Ann. Oncol. 2018, 29, 2348–2355. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Guan, Y.; Yi, Z.; Chang, L.; Li, Q.; Chen, S.; Zhu, W.; Guan, X.; Li, C.; Qian, H.; et al. Assessing Tumor Heterogeneity Using ctDNA to Predict and Monitor Therapeutic Response in Metastatic Breast Cancer. Int. J. Cancer 2020, 146, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive Human Cell-Type Methylation Atlas Reveals Origins of Circulating Cell-Free DNA in Health and Disease. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. A Decade of Exploring the Cancer Epigenome — Biological and Translational Implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V. CCGA Consortium Sensitive and Specific Multi-Cancer Detection and Localization Using Methylation Signatures in Cell-Free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Nuzzo, P.V.; Berchuck, J.E.; Korthauer, K.; Spisak, S.; Nassar, A.H.; Abou Alaiwi, S.; Chakravarthy, A.; Shen, S.Y.; Bakouny, Z.; Boccardo, F.; et al. Detection of Renal Cell Carcinoma Using Plasma and Urine Cell-Free DNA Methylomes. Nat. Med. 2020, 26, 1041–1043. [Google Scholar] [CrossRef]
- Nassiri, F.; Chakravarthy, A.; Feng, S.; Shen, S.Y.; Nejad, R.; Zuccato, J.A.; Voisin, M.R.; Patil, V.; Horbinski, C.; Aldape, K.; et al. Detection and Discrimination of Intracranial Tumors Using Plasma Cell-Free DNA Methylomes. Nat. Med. 2020, 26, 1044–1047. [Google Scholar] [CrossRef]
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-Invasive Early Detection of Cancer Four Years before Conventional Diagnosis Using a Blood Test. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Swigart, L.B.; Wu, H.-T.; Hirst, G.L.; Yau, C.; Wolf, D.M.; Tin, A.; Salari, R.; Shchegrova, S.; Pawar, H.; et al. Circulating Tumor DNA in Neoadjuvant-Treated Breast Cancer Reflects Response and Survival. Ann. Oncol. 2021, 32, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Radovich, M.; Jiang, G.; Hancock, B.A.; Chitambar, C.; Nanda, R.; Falkson, C.; Lynce, F.C.; Gallagher, C.; Isaacs, C.; Blaya, M.; et al. Association of Circulating Tumor DNA and Circulating Tumor Cells After Neoadjuvant Chemotherapy With Disease Recurrence in Patients With Triple-Negative Breast Cancer: Preplanned Secondary Analysis of the BRE12-158 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1410–1415. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Wang, Y.; Christie, M.; Simons, K.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; McKendrick, J.; et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murillas, I.; Chopra, N.; Comino-Méndez, I.; Beaney, M.; Tovey, H.; Cutts, R.J.; Swift, C.; Kriplani, D.; Afentakis, M.; Hrebien, S.; et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019, 5, 1473–1478. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Swanton, C. Early Stage NSCLC - Challenges to Implementing ctDNA-Based Screening and MRD Detection. Nat. Rev. Clin. Oncol. 2018, 15, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, A.; Díez-González, L.; García-Olmo, D.C.; Templeton, A.J.; Vera-Badillo, F.; José Escribano, M.; Serrano-Heras, G.; Corrales-Sánchez, V.; Seruga, B.; Andrés-Pretel, F.; et al. Circulating DNA and Survival in Solid Tumors. Cancer Epidemiol. Biomarkers Prev. 2016, 25, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Zaporozhchenko, I.A.; Ponomaryova, A.A.; Rykova, E.Y.; Laktionov, P.P. The Potential of Circulating Cell-Free RNA as a Cancer Biomarker: Challenges and Opportunities. Expert Rev. Mol. Diagn. 2018, 18, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.M.H. Circulating microRNAs as Biomarkers in Cancer Diagnosis. Life Sci. 2020, 248, 117473. [Google Scholar] [CrossRef]
- Tong, Y.; Zhao, Z.; Liu, B.; Bao, A.; Zheng, H.; Gu, J.; McGrath, M.; Xia, Y.; Tan, B.; Song, C.; et al. 5’/ 3’ Imbalance Strategy to Detect ALK Fusion Genes in Circulating Tumor RNA from Patients with Non-Small Cell Lung Cancer. J. Exp. Clin. Cancer Res. 2018, 37, 68. [Google Scholar] [CrossRef]
- Mellert, H.S.; Alexander, K.E.; Jackson, L.P.; Pestano, G.A. A Blood-Based Test for the Detection of ROS1 and RET Fusion Transcripts from Circulating Ribonucleic Acid Using Digital Polymerase Chain Reaction. J. Vis. Exp. 2018. [Google Scholar] [CrossRef]
- Guo, R.; Offin, M.; RoseBrannon, A.; Chang, J.; Chow, A.; Delasos, L.; Girshman, J.; Wilkins, O.; McCarthy, C.G.; Makhnin, A.; et al. MET Exon14–altered Lung Cancers and MET Inhibitor Resistance. Clin. Cancer Res. 2021, 27, 799–806. [Google Scholar] [CrossRef]
| Specimen | Collection Tubes | Storage Time and Temperature | Type of cfNAs | Ref. |
|---|---|---|---|---|
| Blood | cell-free DNA BCT (Streck, Omaha, NE, USA) | Up to 7 days 18 to 25 °C | cfDNA | [22] |
| RNA Complete BCT (Streck, Omaha, NE, USA) | Up to 7 days 18 to 25 °C | cfRNA, exosomes | [23] | |
| PAXgene Blood ccfDNA (PreAnalytiX GmbH, Hombrechtikon, Switzerland) | Up to 7 days at RT (15–25 °C). Up to 24 h at 35 °C | cfDNA | [24,25] | |
| Cell-Free DNA Collection (Roche Diagnostics, GmbH, Mannheim, Germany) | Up to 7 days at RT | cfDNA | [23] | |
| Norgen Biotek cfDNA Preservative (Norgen Biotek, Corp., Thorold, ON, Canada) | cfDNA 30 days at RT and for up to 8 days at 37 °C. cfRNA for 30 days at RT. | cfDNA, cfRNA | [26] | |
| ImproGene Cell Free DNA (Improve, Instruments Co., Ltd., Guangzhou, China) | 7–14 days under 4–30 °C | cfDNA | * | |
| Biomatrica LBgard Blood (Biomatrica, Inc., San Diego, CA, USA) | Up to 7 days under 4–25 °C. Up to 24 h at 37 °C | cfDNA | [27] | |
| Blood Stasis TM 21-ccfDNA (Mabio Genomics, Inc., Gaithersburg, MD, USA) | Up to 3 days at RT | cfDNA | [6] | |
| NICE® Check cfDNA (EONE-Diagnomics, Genome Center, Incheon, Korea) | NA | cfDNA | * | |
| Blood Exo DNA ProTeck (ProTeck, CFGenome LLC, Denver, CO, USA) | 4, 22 and 30 °C for 21, 28 and 7 days, | cfDNA | [28] | |
| CEE-Sure TM BCT (Biocept, San Diego, CA, USA) | cfDNA up to 8 days at under 6–37 °C. cfRNA for 30 days at RT. CTCs for 14 days at RT | cfDNA | [29] | |
| Urine | Norgen Biotek Urine Preservative (Norgen Biotek, Corp., Thorold, ON, Canada) | 2 years at RT | cfRNA, microRNA, DNA, RNA, proteins | [30] |
| Cell-Free DNA Urine Preserve (Streck; Omaha, NE, USA) | Up to 7 days when stored at 6 to 37 °C. | cfDNA | [31] | |
| Urine collection (Human UPSBio Inc., Atlanta, GA, USA) | Up to 7 days at RT | cfDNA | [32] | |
| Urine Conditioning Buffer UCB (Zymo Research, Irvine, CA, USA) | 1 months at RT | DNA, RNA, cfDNA | * | |
| Saliva | Saliva Exosome Collection and Preservation Kit (Norgen, Biotek, Corp., Thorold, ON, Canada) | 2 years at RT | Exosomes, RNA | [33] |
| Pure SAL TM (Oasis Diagnostics, Vancouver, WA, USA) | NA | cfDNA, cfRNA, exosomes, proteins | [34] | |
| RNA ProSAL TM (Oasis Diagnostics, Vancouver, WA, USA) | NA | RNA, cfDNA, cfRNA, exosomes | [34] |
| Analysis | Analyte | Approaches | Methods | |
|---|---|---|---|---|
| Mutations: point mutation, indels, amplifications, CNVs, deletions, translocations | cfNAs | Single-molecule | NGS | INC-Seq Nanopore; Pacific BioSciences PacBio |
| Single-gene | PCR | qPCR; ARMS-PCR; COLD-PCR; bi-AP; MAP; ddPCR; BEAMing | ||
| Gene panels | NGS | TAm-Seq; Safe-SeqS; SiMSen-Seq; CypherSeq; DuplexSeq; smMIPs; BAsE-Seq; CAPP-Seq; mFAST-SeqS; mmPCR-NGS;RealSeqS; bTMB assay | ||
| Genome | NGS | WGS; WES; low-pass sequencing | ||
| Other | spectroscopy | MALDI; SERS; UltraSEEK | ||
| Transcriptome quantification | miRNA, cf-mRNA, lncRNAs, ncRNAs | Few transcripts | PCR | qRT-PCR |
| Multi-transcripts | Hybridization | Microarray | ||
| 68 target miRNAs | Flow cytometry | FireFly | ||
| exome | NGS | Whole-transcriptome RNA-sequencing | ||
| Epigenetic modification | cfMeDNA | Single-molecule | NGS | Nanopore |
| Specific CpG site | PCR | Sodium bisulfite; MSP; qMSP; EpiTect MethyLight PCR; SYBR Green-based qMSP | ||
| All CpG sites | NGS | MethylCap; cfMeDIP-seq; EM-Seq; TAPS; cfNOMe | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cirmena, G.; Dameri, M.; Ravera, F.; Fregatti, P.; Ballestrero, A.; Zoppoli, G. Assessment of Circulating Nucleic Acids in Cancer: From Current Status to Future Perspectives and Potential Clinical Applications. Cancers 2021, 13, 3460. https://doi.org/10.3390/cancers13143460
Cirmena G, Dameri M, Ravera F, Fregatti P, Ballestrero A, Zoppoli G. Assessment of Circulating Nucleic Acids in Cancer: From Current Status to Future Perspectives and Potential Clinical Applications. Cancers. 2021; 13(14):3460. https://doi.org/10.3390/cancers13143460
Chicago/Turabian StyleCirmena, Gabriella, Martina Dameri, Francesco Ravera, Piero Fregatti, Alberto Ballestrero, and Gabriele Zoppoli. 2021. "Assessment of Circulating Nucleic Acids in Cancer: From Current Status to Future Perspectives and Potential Clinical Applications" Cancers 13, no. 14: 3460. https://doi.org/10.3390/cancers13143460
APA StyleCirmena, G., Dameri, M., Ravera, F., Fregatti, P., Ballestrero, A., & Zoppoli, G. (2021). Assessment of Circulating Nucleic Acids in Cancer: From Current Status to Future Perspectives and Potential Clinical Applications. Cancers, 13(14), 3460. https://doi.org/10.3390/cancers13143460






