Simple Summary
An important advance in the diagnostic and surveillance toolbox for oncologists is circulating tumor DNA (ctDNA). This technology can detect microscopic levels of cancer tissue before, during, or after treatment. Various groups from across the globe have published their experiences with the use of ctDNA to either guide therapy or monitor outcomes. The use of ctDNA likely cannot supplant the need for tissue biopsies, but it can complement other diagnostic and therapeutic monitoring mechanisms.
Abstract
With the addition of molecular testing to the oncologist’s diagnostic toolbox, patients have benefitted from the successes of gene- and immune-directed therapies. These therapies are often most effective when administered to the subset of malignancies harboring the target identified by molecular testing. An important advance in the application of molecular testing is the liquid biopsy, wherein circulating tumor DNA (ctDNA) is analyzed for point mutations, copy number alterations, and amplifications by polymerase chain reaction (PCR) and/or next-generation sequencing (NGS). The advantages of evaluating ctDNA over tissue DNA include (i) ctDNA requires only a tube of blood, rather than an invasive biopsy, (ii) ctDNA can plausibly reflect DNA shedding from multiple metastatic sites while tissue DNA reflects only the piece of tissue biopsied, and (iii) dynamic changes in ctDNA during therapy can be easily followed with repeat blood draws. Tissue biopsies allow comprehensive assessment of DNA, RNA, and protein expression in the tumor and its microenvironment as well as functional assays; however, tumor tissue acquisition is costly with a risk of complications. Herein, we review the ways in which ctDNA assessment can be leveraged to understand the dynamic changes of molecular landscape in cancers.
1. Introduction
A liquid biopsy is a minimally invasive technique for measuring diagnostically significant tumor-derived markers in body fluids. Although any liquid can be biopsied (e.g., blood, urine, ascites, and cerebrospinal fluid), herein we will be referring to blood biopsies when we speak of liquid biopsies. The types of components that can be interrogated in a liquid biopsy include circulating tumor cells, circulating extracellular nucleic acids (cell-free DNA (cfDNA) and its neoplastic fraction—circulating tumor DNA (ctDNA)), as well as extracellular vesicles (such as exosomes), and a variety of glycoproteins. We will be focused on ctDNA and cfDNA. cfDNA is a broad term that refers to DNA which is freely circulating in the blood but is not necessarily of tumor origin [1]; ctDNA is fragmented DNA in the bloodstream that is of tumor origin and is not associated with cells.
The use of next-generation sequencing (NGS) of ctDNA from a blood biopsy has gone, in the last decade, from the unimaginable to the routine. NGS of ctDNA has provided insights into potential genomic-derived treatment options such as identifying novel targets as well as predicting responses to treatments (Figure 1) [2].
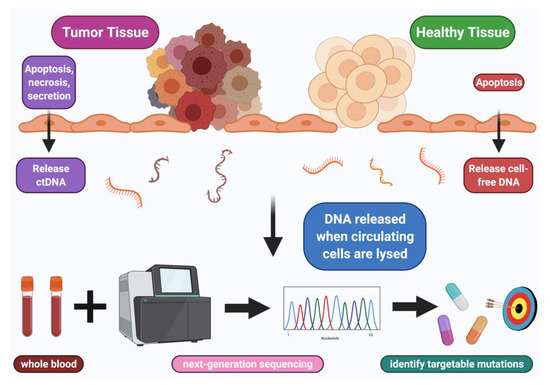
Figure 1.
ctDNA employed to identify targetable mutations. Blood sample collected and next-generation sequencing performed on shed ctDNA from tumor, which can identify genomic alterations, some of which may be pharmacologically tractable.
Liquid biopsies can also be used to evaluate microsatellite stability/instability (MSI-H) and high tumor mutational burden (TMB-H), both of which are critical parameters for predicting immune checkpoint blockade response [3,4,5]. Further, ctDNA can be exploited to monitor response and predict resistance in some tumors [6,7,8,9,10].
The half-life of ctDNA ranges from 30 min to two hours. Changes in ctDNA can be used to monitor tumors dynamically [11]. Both the concentration of ctDNA and the number of somatic alterations found within a sample have been implicated in some studies as a surrogate for tumor stage and size as well as tumor aggressiveness [12,13,14].
The implementation of diagnostics using ctDNA has been leveraged as a companion diagnostic test, e.g., for detecting EGFR inhibitor sensitive mutations for the use of erlotinib in non-small cell lung cancer [15]. Even so, ctDNA may provide important risk stratification data [16].
A challenge for the utility of ctDNA is that clonal hematopoiesis of indeterminate potential (CHIP) may confound results; in other words, some presumptive ctDNA mutants may be derived from aberrations in blood cells, particularly those that accompany aging, rather than abnormalities in the tumor, yet mutational burden from CH is low and can be excluded by sequencing healthy control tissue [17].
Herein, we will examine the multiple potential uses of liquid biopsy with NGS of ctDNA in oncology:
- ○
- early diagnosis of cancer;
- ○
- ctDNA as a prognostic variable;
- ○
- measurement of residual disease;
- ○
- discerning molecular alterations that can inform therapeutic decision-making; and
- ○
- monitoring response, resistance, and burden/aggressiveness of disease.
2. Comparison of CTCs, ctDNA, and Tissue DNA
Circulating tumor cells (CTCs), ctDNA, and tissue DNA (tDNA) are all potentially exploitable for providing insight and data about tumor genomes (Table 1). Acquisition of CTCs and ctDNA are both noninvasive, requiring only a venipuncture, and are considered to be liquid biopsies. In contrast, tissue DNA requires an invasive biopsy.

Table 1.
Comparison of CTCs, circulating tumor DNA (ctDNA), and tissue DNA (tDNA).
CTCs are tumor cells that are shed from growing and dying tumors that require isolation; thus, they technically require specialized equipment. However, CTCs can be a rich source of information about the genomic, transcriptomic, and proteomic content of the tumor; if grown in culture, CTCs can also provide functional assays. In contrast, ctDNA, while being easier to isolate that CTCs, cannot be cultured and the information obtainable from ctDNA is generally restricted to genomic analysis [18]. In this regard, blood-derived CTCs and tissue samples share similarities, as both can be isolated, cultured, and provide genomic, transcriptomic, and proteomic tumor data.
In a blood sample (~10 cc), there will be tens to hundreds of ctDNA fragments for testing, whereas there will likely only be a handful of CTCs.
One limitation of tissue DNA is that it is obtained from a discrete piece of tumor tissue; thus, tissue DNA cannot reflect heterogeneity amongst metastatic sites and is more difficult to be followed serially [19]. CTCs and ctDNA are, however, shed from multiple metastatic sites and therefore better reflect tissue heterogeneity than a tissue biopsy. On the other hand, the requirement for extremely sensitive techniques for genomic interrogation of CTCs and ctDNA means that tissue assays often yield greater numbers of positive genomic alterations, and tissue NGS assays are generally more comprehensive than those applied to ctDNA.
A unique advantage to CTCs and ctDNA is their amenability to longitudinal follow up with a simple blood test in order to predict therapeutic response and resistance [19].
3. Liquid Biopsy and Dynamics of Normal Versus Tumor Cell-Free DNA (cfDNA)
Elevated levels of cfDNA were found in patients with cancers but can be detected during pregnancy and in patients with history of organ transplant [20]. Generally, the blood concentration of cfDNA can vary from 0–5 to >1000 ng/mL in cancer patients and between 0 and 100 ng/mL in otherwise healthy patients [21,22]. The large range of cfDNA and ctDNA found in patients with cancer is in part due to the fact that various tumor types can have wide variations in ctDNA shedding and that the amount of ctDNA can reflect tumor burden. Patients with brain, kidney, and thyroid cancers have been found to have lower levels of cfDNA than those patients with pancreatic, colorectal, ovarian, breast, gastroesophageal, and melanoma [13,23]. Additionally, premalignant and early-stage cancers generally have lower levels of cfDNA compared to patients with advanced disease [21].
Not all of the cfDNA in the bloodstream of cancer patients is ctDNA, and it is important to recognize what fraction of cfDNA is actually from a cancer. It is believed that ~0.1–89% of cfDNA is made up of ctDNA and that the ratio may increase as a cancer progresses [13,24,25]. The sizes of cfDNA are estimated to be between 40 and 200 base pairs [26,27,28]. If wrapped in chromatin, the DNA in these vesicles can make up to 2 million base pairs [29]. These fragments are believed to be part of tumor metabolism and growth; fragments from necrotic tumor tissue can be over 10,000 kilobases [30].
The amount of cfDNA found within the bloodstream is dependent on the balance of release and clearance of cfDNA. Clearance can occur within the primary tumor tissue, within the blood, or within various filtration organs: spleen, liver, and lymph nodes [31]. Elevated levels of cfDNA in patients with cancer is believed to be in part because of lack of clearance and subsequent accumulation. Within the bloodstream, degradation of cfDNA is performed in large part by circulating enzymes: deoxyribonuclease (DNAse) I, plasma factor VII-activating protease, and factor H [32,33]. Within the spleen and liver, Kupffer cells and macrophages have been implicated in removing cfDNA and nucleosomes from circulation [34]. The presence of tumor in patients with cancer may be in part the reason for higher levels of cfDNA detected and also in part due to inability to clear these fragments within these various mechanisms.
4. How ctDNA Enters and Leaves the Circulation
It is unclear exactly how ctDNA enters the blood stream; however, it is postulated that when primary tumor cells or metastatic cells die via apoptosis or necrosis, DNA fragments may be released into the bloodstream [22,35]. The amount of ctDNA that can be found within the blood stream is heavily dependent on the overall tumor biology and burden. The half-life of ctDNA is estimated to be between 30 min and two hours; ctDNA is rapidly degraded by bloodstream DNases [31].
5. Technological Methods for cfDNA Extraction and Sequencing
Various techniques to detect ctDNA are available all with varying advantages and limitations (Table 2). These techniques include droplet digital polymerase chain reaction (ddPCR), beads, emulsion, amplification, and magnetics (BEAMing), tagged-amplicon deep sequencing (TAm-Seq), cancer personalized profiling by deep sequencing (CAPP-Seq), whole exome sequencing (WES), and whole genome sequencing (WGS).

Table 2.
Examples of techniques to detect ctDNA.
ddPCR can identify potentially rare mutations, calculate copy number variants, as well as inform on miRNA [36]. This method also allows for detection of very low levels of genomic material, 0.01–1.0% [37]. The most notable limitation of this method of ctDNA detection, however; is that only characterized sequences can be screened via this method.
The use of BEAMing allows for the assessment of characterized alterations (e.g., SNVs, indels, and amplifications) and combines PCR with flow cytometry [43]. This allows for the detection of alterations at exceedingly low levels—0.01%—with marked concordance to tissue testing of 91.8% [38].
The CAPP-Seq technique utilizes large genomic libraries combined with individual patient sample sequence signatures to identify alterations within ctDNA. This method combines statistical assessment of well-characterized tumor alterations with DNA oligonucleotides to identify patient specific alterations [39]. This method allows for the identification of various genomic alterations such as insertions/deletions, single nucleotide variants, rearrangements, and copy variants. A limitation of CAPP-Seq includes the inability to identify fusions, in contrast to ddPCR, TAm-Seq, WES, and WGS [39].
The TAm-Seq technique allows for highly sensitive and specific analysis ~97% along with the ability to detect low levels of ctDNA, 2%. This method uses primers to tag and identify the desired genomic sequence. The limitation with this technique is that the sequence needs to be characterized to be included in the analysis [40].
Whole exome sequencing allows for comprehensive analysis and characterization of potentially all tumor mutations. In doing so, the sensitivity may be lower than other modalities because it includes all exomic alterations. The limitations of WES relate to error rate and sensitivity [41,44].
WGS includes the entire tumor genome to discern characterized/deleterious alterations as well as many uncharacterized genomic events (variants of uncertain significance (VUSs) and is mainly used for CNAs [42].
6. Clinical Laboratory Improvement Amendments (CLIA) Grade Commercially Available ctDNA Assays
There are several CLIA-grade commercially available ctDNA assays that clinicians can order to potentially inform treatment decisions for patients. One of the most widely available of these tests is the Guardant360 CDx from Guardant Health, which was first accessible in 2014. The Guardant360 assay includes 73 genes commonly altered in cancers and can identify single-nucleotide variants (SNVs), insertions/deletions (indels), fusions, and copy number alterations (CNAs) (https://www.therapyselect.de/sites/default/files/downloads/guardant360/guardant360_specification-sheet_en.pdf, accessed date: 10 January 2021). This Guardant360 assay requires two 10 cc tubes of whole blood and is reported to have results in 7 calendar days after receipt of the samples.
In 2018, Foundation Medicine released their ctDNA assay called FoundationOne Liquid, which now includes 311 genes implicated in cancers (https://assets.ctfassets.net/w98cd481qyp0/wVEm7VtICYR0sT5C1VbU7/55b945602d7dc78f42b3306ca1caa451/FoundationOne_Liquid_CDx_Technical_Specifications.pdf, accessed date: 10 January 2021). The FoundationOne Liquid assay included base substitutions, indels, rearrangements, copy number alterations, and MSI-H status. The FoundationOne Liquid assay requires two 8.5 cc tubes of whole blood and reports to have results within less than two weeks after receipt of the samples.
Also in 2018, Tempus introduced Tempus xF, a ctDNA assay, which includes 105 genes implicated in cancers. The Tempus xF assay included SNVs, indels, rearrangements/fusions, CNAs, and MSI-H status (https://www.tempus.com/wp-content/uploads/2020/02/xF-Validation_013020-2.pdf, accessed date: 10 January 2021). The Tempus xF assay requires two 8 cc tubes of whole blood and reports to have results in nine to 14 days after receipt of the samples.
7. Food and Drug Administration (FDA) Approvals for ctDNA Tests
In August 2020, the FDA approved the use of FoundationOne Liquid CDx test from Foundation Medicine, Inc. as a companion diagnostic test for patients with ovarian cancer to identify mutations in BRCA1/2 for the use of rucaparib, for patients with metastatic hormone-resistant prostate cancer with mutations in BRCA1/2 and ATM for the use of olaparib, for patients with metastatic hormone-resistant prostate cancer with mutations in BRCA1/2 for the use of rucaparib, for patients with non-small cell lung cancer (NSCLC) with ALK rearrangement for the use of alectinib, for patients with NSCLC with EGFR exon 19 deletions and EGFR exon 21 L858R alterations for the use of gefinitinb, erlotinib, and osimertinib, and for patients with breast cancer with mutations in PIK3CA C420R, E542K, E545A, E545D [1635G > T only], E545G, E545K, Q546E, Q546R, H1047L, H1047R, and H1047Y for the use of alpelisib [45,46].
Guardant360 CDx by Guardant Health Inc. was also approved in 2020 to identify EGFR exon 19 deletions, L858R, and T790M mutations in patients with NSCLC for the use of Osimertinib [47].
The Therascreen PIK3CA RGQ PCR Kit was approved in 2019 to detect 11 mutations in the PIK3CA gene in patients with metastatic breast cancer for the use of alpelisib [48].
Additionally, Cobas EGFR Mutation Test v2 was also approved in 2016 to identify EGFR L858R mutations in patients with NSCLC for the use of erlotinib [49].
8. Clinical Uses of ctDNA
8.1. ctDNA for Early Diagnosis of Cancer
Early cancer detection could transform outcomes by detecting lethal tumors at a time when the malignancies are curable, and treatment invokes less morbidity. However, the technical, biological, and clinical hurdles to developing an effective pan-cancer screening test for early cancer are substantial.
Liquid biopsies with NGS of ctDNA are an attractive tool, but the very small amounts of ctDNA in early disease is still a major technical challenge, as is the issue that non-cancerous normal tissue may have somatic mutations indistinguishable from those in cancer, but as mentioned above, CH mutations can be filtered out by using healthy tissue control samples. Still, Cohen et al. developed a noninvasive blood test, called CancerSEEK, that detected eight common cancer types through assessment of circulating proteins and mutations in cfDNA. In a study of 1005 patients previously diagnosed with non-metastatic cancer and 850 healthy control individuals, CancerSEEK detected cancer with a 99% specificity and a sensitivity of 69% to 98% (depending on type of malignancy) [50].
Another methodology that has recently been exploited is assessing ctDNA methylation patterns, noting that increased methylation of tumor suppressor genes can be seen as an early inciting event in the carcinogenesis of various tumors, such as hepatocellular and colorectal carcinomas [51]. A prospective case–control study evaluated the performance of pan-cancer targeted methylation analysis of cfDNA. With 6689 participants (2482 cancers (>50 cancer types), 4207 healthy), specificity was 99.3% and stage I–III sensitivity was 43.9% in all cancer types [52]. Other unique technologies aimed at early cancer detection continue to be explored.
8.2. ctDNA as a Prognostic Variable
To date, multiple studies have analyzed the utility of ctDNA to be able to assess disease-free survival (DFS) and overall survival (OS) (Table 3) [53,54,55,56,57,58,59,60,61,62,63,64,65,66,67]. Factors that predict poorer outcomes include concordance between tissue and liquid ctDNA alterations (shown for both TP53 and KRAS mutations) [68,69], higher percent ctDNA (perhaps reflecting higher tumor burden, and higher number of ctDNA alterations [14].

Table 3.
Examples of studies investigating clinical applications of ctDNA.
Multiple studies have shown that ctDNA can be an important prognostic factor. For instance, in triple-negative breast cancer patients who had received or were receiving neoadjuvant chemotherapy, the detection of ctDNA was associated with a significantly worse DFS (p = 0.027) [53]. Additionally, at the last post-chemotherapy pre-surgery time point, detection of ctDNA was strongly associated with shorter DFS (p = 0.013) and OS (p = 0.006) [53]. In patients receiving adjuvant chemotherapy for locally advanced rectal cancer, 122 (77%) of 159 patients had pre-surgical detectable ctDNA and after surgery only 12 of 140 (8.6%) with negative ctDNA (hazard ratio (HR) 12, p < 0.001) experienced recurrence [54]. Further, post-op ctDNA detection predicted recurrence regardless of adjuvant chemotherapy (chemotherapy: HR 10, p < 0.001; no chemotherapy: HR 16, p < 0.001) and ctDNA detection predicted higher recurrence rate among patients with a pathological complete response (HR 14, p = 0.014) or with pathologic node-positive disease (HR 11, p < 0.001) [54]. A cohort study of patients with local advanced anal squamous cell cancer found that, in 33 patients, ctDNA detection after chemoradiation was associated with shorter DFS (p < 0.0001) [55]. Additionally, this study reported that ctDNA was associated with stage (64% in stage II and 100% in stage III; p = 0.008) and baseline ctDNA levels were higher in pathological node positive (median 85 copies/mL, range = 8–9333) than pathological node negative disease (median 32 copies/mL, range = 3–1350) p = 0.03 [55]. In another study, this one in pancreatic cancer, higher levels of total %ctDNA were an independent prognostic factor for worse survival (hazard ratio, 4.35; 95% confidence interval, 1.85–10.24 (multivariate, p = 0.001)) [63].
8.3. ctDNA to Measure Residual Disease
The ability of ctDNA to track tumor-specific mutations and to detect occult cancer lend themselves naturally to assessment of minimal residual disease. Further, the ease of plasma sampling permits ctDNA levels to be serially followed in order to longitudinally trend mutation status and frequently assess dynamic changes in levels of ctDNA, as reflected by percent ctDNA (or variant allele fraction (VAF).
Multiple studies are now beginning to confirm clinical utility of ctDNA in evaluating minimal residual disease [73]. For instance, declines in circulating allele fractions of relevant mutations have been associated with clinical outcomes in melanoma, colorectal cancer, breast and ovarian cancer, and EGFR-positive lung cancer [74,75,76,77,78]. As examples, in a study that monitored patients with colorectal cancer pre- and post-surgery, pretreatment ctDNA was detected in 93.4% (100/107) of patients; post-operative ctDNA status was assessed in 107 patients, of whom, 13% (14/107) were minimal residual disease-positive. Of the positive patients, 42.9% (6/14) eventually relapsed while only 8.6% (8/93) of the negative patients relapsed (HR: 10; 95% CI: 3.3–30; p < 0.001). In multivariate analysis, ctDNA status was the most significant prognostic factor associated with relapse-free survival (HR: 28.8, 95% CI: 3.5–234.1; p < 0.001) [79]. Similarly, in patients undergoing surgery for peritoneal metastases, high levels of pre-operative ctDNA and new postoperative ctDNA alterations in the context of preoperative alterations predicted worse outcomes [59]. Applications may include using ctDNA to determine escalation or de-escalation of adjuvant therapy.
8.4. Discerning ctDNA Molecular Alterations That Can Inform Decision Making
Multiple studies demonstrate the important use of ctDNA interrogation for prosecuting treatment. In fact, as mentioned above, the FDA has approved several ctDNA tests as companion diagnostics [45,46,47]: detection of BRCA1/2 alterations for the use of the poly (ADP-ribose) polymerase (PARP) inhibitor rucaparib in ovarian cancer; BRCA1/2 and ATM mutations for the use of the PARP inhibitor olaparib in prostate cancer; ALK and EGFR alterations to be treated with the ALK inhibitor alectinib or the EGFR inhibitors gefitinib, erlotinib, and osimertinib in NSCLC; and a variety of PIK3CA alterations to be treated with the PIK3CA inhibitor alpelisib in breast cancer.
Numerous other studies support the utility of ctDNA for genomic characterization aimed at assisting therapeutic choice. For instance, one study in patients with advanced breast cancer found that 68% (42/62) of patients had ≥1 characterized/pathogenic ctDNA alteration (non-VUS) [57]. A similar study in patients with advanced and resected esophageal, gastroesophageal junction, and gastric adenocarcinoma found that 76% (42/55) of patients had a ctDNA alteration, with 69% (38/55) having ≥1 characterized/deleterious (non-VUS) [58]. In gynecologic cancers, therapy matched to ctDNA alterations (n = 33 patients) was independently associated with improved survival (HR: 0.34, p = 0.007) compared to unmatched therapy (n = 28 patients) in multivariate analysis [60]. In a study focused on EGFR amplification, such amplifications were detected in cfDNA in a significant subset of pan-cancer patients—8.5% of 28,584. Most patients had coexisting alterations. Importantly, responses were observed in five of nine patients who received EGFR inhibitors, including patients who showed ctDNA EGFR amplifications, but no amplifications in the tissue DNA [64]. Taken together, it is apparent that ctDNA molecular alterations play a vital 79 Burden/Aggressiveness of Disease
Resistant ctDNA alterations that may emerge months before changes in scans are noted and can inform an understanding of mechanisms of resistance in colorectal, lung, and breast cancers, as examples [80,81,82]. For instance, ctDNA was used to identify early resistance mutations in patients with HER2-amplified breast cancer; PI3K/mTOR pathway alterations were the major cause of resistance [83]. This information may be exploitable with the addition of another targeted therapeutic [84]. Using ctDNA in a longitudinal fashion could allow for concomitant or sequential targeting of multiple gene mutations in real-time. This strategy and the ability of ctDNA to offer this information prior to imaging and without the need for additional tissue biopsies may be part of the holy grail of getting the right drug to the right patient at the right time. Based on criteria established by the OncoKB database, and other evidential reports, studies have shown that over one-quarter of cancers harbored level 1 actionable targets in their ctDNA [85]. The ability to find these mutations early in the treatment course could potentially alter the trajectory of recognizing mutation acquisition, thus enhancing patient outcomes.
Furthermore, ctDNA can be an early marker of response. For instance, drug-induced tumor apoptosis may occur for EGFR-targeted therapy in lung cancer within days of initial dosing, and daily sampling of ctDNA may facilitate early assessment of patient response within the first week of treatment with EGFR inhibitors [10]. Similarly, ctDNA has been used to predict response to treatment before radiographic response in colorectal cancer [75]. This measurable entity portends survival even in the setting of neoadjuvant therapy of breast cancer [86].
Similarly, early plasma ctDNA changes predicted response to first-line pembrolizumab in in patients with lung cancer [70]. Finally, genome-wide sequencing of cfDNA identified copy number alterations that could be used for monitoring early response (or resistance) to immunotherapy in cancer patients [72,87].
Multiple publications also show that both %ctDNA (VAF) and number of alterations in ctDNA predict a poor prognosis, possibly because they reflect tumor burden and/or aggressiveness [14].
9. The Issue of Concordance between ctDNA and Tissue DNA
Several studies have examined the concordance in molecular alterations between tissue and ctDNA samples. In general, concordance is variable ranging from ~50% to over 95% [69,88]. The literature suggests that the results from liquid biopsies and from tissue biopsies, vis a vis NGS, are highly reproducible [89]. Therefore, biological differences most likely account for discrepant ctDNA and tissue DNA NGS results.
The biologic attributes that underlie differences between tissue and ctDNA results include (i) shedding of DNA into the bloodstream may be limited from some sites, (ii) ctDNA can be suppressed by treatment, and (iii) tissue DNA tests the genomics in a small sample of tissue, whereas ctDNA may reflect shed DNA from multiple metastatic sites. Both tissue and ctDNA may be confounded by germline alterations and by clonal hematopoiesis of indeterminate potential, though ctDNA may be more vulnerable to such confounders.
Interestingly, studies now show that concordance between ctDNA and tissue DNA alterations, at least for TP53 and for KRAS, is associated with worse outcomes [68,69].
10. Conclusions and Future Directions
Tumors release ctDNA into the bloodstream. The amount of ctDNA discernable, as reflected by percent of DNA VAF and the number of ctDNA alterations, may be an indicator of tumor burden and/or aggressiveness, with higher numbers predicting worse prognosis.
Blood-derived ctDNA may provide crucial molecular information as a complement to the tumor biopsy for the following reasons: (i) some cancer tissue is not easily or safely accessible for biopsy; (ii) even if accessible, tumor biopsies can be complex and expensive procedures with morbidity; (iii) over time, the tissue that was biopsied may become less representative of the tumor, since malignancies undergo genomic evolution; (iv) genomic aberrations discerned in a tissue biopsy reflect the content of the small tissue sample, while ctDNA NGS abnormalities may reflect the heterogeneous alterations found in shed DNA from many metastatic sites; and (v) dynamic changes in ctDNA can occur and reflect response or resistance to treatment. Furthermore, evaluating ctDNA pre- or post-surgery may serve as a predictive tool for recurrence risk. Finally, ctDNA may be exploitable for early detection of lethal cancers when they are still curable and/or do not require drastic, life-altering interventions.
There are also disadvantages to ctDNA as compared to tissue DNA assessment: (i) ctDNA is found in only small amounts in the circulation, making it difficult to detect alterations, and (ii) ctDNA carrying tumor-specific alterations may represent only a small fraction of the total genomic alterations in the tumor, since not all cancer-derived DNA may be shed into the blood. Therefore, variability in concordance rates between blood-derived ctDNA samples and tissue samples can be caused by spatial and temporal variables, as well as by dynamic changes driven by therapy and disease evolution; and (iii) ctDNA is more liable to be confounded by alterations of clonal hematopoiesis of indeterminate potential, and perhaps also by germline alterations.
Taken together, the literature indicates that assessment of blood-derived ctDNA is a powerful and transformative technology which can inform genomic decision making for gene- and immune-targeted therapy, can predict prognosis, and can be followed serially to assess response, resistance, and residual disease.
Author Contributions
Conceptualization, J.J.A., F.J. and R.K.; writing—original draft preparation, J.J.A. and R.K. writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
R.K. is funded in part by the Joan and Irwin Jacobs Fund and NIH P30 CA023100. Figure created with biorender.com, accessed date: 10 January 2021.
Conflicts of Interest
J.J.A. has no conflicts of interest. F.J. has the following conflicts of interest: serves in an advisory role for Asana, Baush Health, Cardiff Oncology, Deciphera, Guardant Health, Ideaya, IFM Therapeutics, Immunomet, Illumina, Jazz Pharmaceuticals, Novartis, PureTech Health, Sotio, and Synlogic; has stocks in Cardiff Oncology and his institution receives funding from Agios, Asana, Astellas, Astex, Bayer, Bicara, BioMed Valley Discoveries, Bioxcel, Bristol-Myers Squibb, Deciphera, FujiFilm Pharma, Genentech, Ideaya, JS Innopharm, Eli Lilly, Merck, Novartis, Novellus, Plexxikon, Proximagen, Sanofi, Sotio, SpringBank Pharmaceuticals, SQZ Biotechnologies, Synlogic, Synthorx, and Symphogen. R.K. has the following conflicts of interest: Stock and Other Equity Interests (IDbyDNA, CureMatch, Inc., and Soluventis); Consulting or Advisory Role (Gaido, LOXO, X-Biotech, Actuate Therapeutics, Roche, NeoMed, and Soluventis); Speaker’s fee (Roche); Research Funding (Incyte, Genentech, Merck Serono, Pfizer, Sequenom, Foundation Medicine, Guardant Health, Grifols, Konica Minolta, and OmniSeq [All institutional]); Board Member and co-Founder (CureMatch, Inc.); Board Member CureMetrix.
References
- Husain, H.; Velculescu, V.E. Cancer DNA in the Circulation: The Liquid Biopsy. JAMA 2017, 318, 1272–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, N.C.; Kingston, B.; Kilburn, L.S.; Kernaghan, S.; Wardley, A.M.; Macpherson, I.R.; Baird, R.D.; Roylance, R.; Stephens, P.; Oikonomidou, O.; et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): A multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020, 21, 1296–1308. [Google Scholar] [CrossRef]
- Khagi, Y.; Goodman, A.M.; Daniels, G.A.; Patel, S.P.; Sacco, A.G.; Randall, J.M.; Bazhenova, L.A.; Kurzrock, R. Hypermutated Circulating Tumor DNA: Correlation with Response to Checkpoint Inhibitor-Based Immunotherapy. Clin. Cancer Res. 2017, 23, 5729–5736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willis, J.; Lefterova, M.I.; Artyomenko, A.; Kasi, P.M.; Nakamura, Y.; Mody, K.; Catenacci, D.V.T.; Fakih, M.; Barbacioru, C.; Zhao, J.; et al. Validation of Microsatellite Instability Detection Using a Comprehensive Plasma-Based Genotyping Panel. Clin. Cancer Res. 2019, 25, 7035–7045. [Google Scholar] [CrossRef] [PubMed]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Reece, M.; Saluja, H.; Hollington, P.; Karapetis, C.S.; Vatandoust, S.; Young, G.P.; Symonds, E.L. The Use of Circulating Tumor DNA to Monitor and Predict Response to Treatment in Colorectal Cancer. Front. Genet. 2019, 10, 1118. [Google Scholar] [CrossRef] [Green Version]
- Osumi, H.; Shinozaki, E.; Yamaguchi, K.; Zembutsu, H. Early change in circulating tumor DNA as a potential predictor of response to chemotherapy in patients with metastatic colorectal cancer. Sci. Rep. 2019, 9, 17358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.S.; Eun, J.W.; Choi, J.H.; Woo, H.G.; Cho, H.J.; Ahn, H.R.; Suh, C.W.; Baek, G.O.; Cho, S.W.; Cheong, J.Y. MLH1 single-nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma. Sci. Rep. 2020, 10, 17862. [Google Scholar] [CrossRef]
- Sharbatoghli, M.; Vafaei, S.; Aboulkheyr Es, H.; Asadi-Lari, M.; Totonchi, M.; Madjd, Z. Prediction of the treatment response in ovarian cancer: A ctDNA approach. J. Ovarian Res. 2020, 13, 124. [Google Scholar] [CrossRef]
- Husain, H.; Melnikova, V.O.; Kosco, K.; Woodward, B.; More, S.; Pingle, S.C.; Weihe, E.; Park, B.H.; Tewari, M.; Erlander, M.G.; et al. Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine. Clin. Cancer Res. 2017, 23, 4716–4723. [Google Scholar] [CrossRef] [Green Version]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef]
- Thierry, A.R.; Mouliere, F.; Gongora, C.; Ollier, J.; Robert, B.; Ychou, M.; Del Rio, M.; Molina, F. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010, 38, 6159–6175. [Google Scholar] [CrossRef] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef] [Green Version]
- Vu, P.; Khagi, Y.; Riviere, P.; Goodman, A.; Kurzrock, R. Total Number of Alterations in Liquid Biopsies Is an Independent Predictor of Survival in Patients with Advanced Cancers. JCO Precis. Oncol. 2020, 4. [Google Scholar] [CrossRef]
- Ossandon, M.R.; Agrawal, L.; Bernhard, E.J.; Conley, B.A.; Dey, S.M.; Divi, R.L.; Guan, P.; Lively, T.G.; McKee, T.C.; Sorg, B.S.; et al. Circulating Tumor DNA Assays in Clinical Cancer Research. J. Natl. Cancer Inst. 2018, 110, 929–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Saw, R.P.; Thompson, J.F.; Lo, S.; Spillane, A.J.; Shannon, K.F.; Stretch, J.R.; Howle, J.; Menzies, A.M.; Carlino, M.S.; et al. Pre-operative ctDNA predicts survival in high-risk stage III cutaneous melanoma patients. Ann. Oncol. 2019, 30, 815–822. [Google Scholar] [CrossRef]
- Okamura, R.; Piccioni, D.E.; Boichard, A.; Lee, S.; Jimenez, R.E.; Sicklick, J.K.; Kato, S.; Kurzrock, R. High Prevalence of Clonal Hematopoiesis-type Genomic Abnormalities in Cell-free DNA in Invasive Gliomas After Treatment. Int. J. Cancer 2021, 148, 2839–2847. [Google Scholar] [CrossRef]
- Qin, Z.; Ljubimov, V.A.; Zhou, C.; Tong, Y.; Liang, J. Cell-free circulating tumor DNA in cancer. Chin. J. Cancer 2016, 35, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, C.R.; Zhou, L.; El-Deiry, W.S. Circulating Tumor Cells Versus Circulating Tumor DNA in Colorectal Cancer: Pros and Cons. Curr. Colorectal. Cancer Rep. 2016, 12, 151–161. [Google Scholar] [CrossRef] [Green Version]
- Sharon, E.; Shi, H.; Kharbanda, S.; Koh, W.; Martin, L.R.; Khush, K.K.; Valantine, H.; Pritchard, J.K.; De Vlaminck, I. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS Comput. Biol. 2017, 13, e1005629. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef] [Green Version]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Zill, O.A.; Banks, K.C.; Fairclough, S.R.; Mortimer, S.A.; Vowles, J.V.; Mokhtari, R.; Gandara, D.R.; Mack, P.C.; Odegaard, J.I.; Nagy, R.J.; et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin. Cancer Res. 2018, 24, 3528–3538. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.Q.; Raleigh, J.M.; Callahan, J.; Vergara, I.A.; Ftouni, S.; Hatzimihalis, A.; Colebatch, A.J.; Li, J.; Semple, T.; Doig, K.; et al. Circulating Tumor DNA Analysis and Functional Imaging Provide Complementary Approaches for Comprehensive Disease Monitoring in Metastatic Melanoma. JCO Precis. Oncol. 2017, 1, 1–14. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S.J.; Pogrebniak, K.; Rueda, O.M.; Provenzano, E.; Grant, J.; Chin, S.F.; Tsui, D.W.Y.; Marass, F.; Gale, D.; et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 2015, 6, 8760. [Google Scholar] [CrossRef] [Green Version]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Nakahira, K.; Guo, X.; Choi, A.M.; Gu, Z. Very Short Mitochondrial DNA Fragments and Heteroplasmy in Human Plasma. Sci. Rep. 2016, 6, 36097. [Google Scholar] [CrossRef] [Green Version]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De, S.; et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muhanna, N.; Di Grappa, M.A.; Chan, H.H.L.; Khan, T.; Jin, C.S.; Zheng, Y.; Irish, J.C.; Bratman, S.V. Cell-Free DNA Kinetics in a Pre-Clinical Model of Head and Neck Cancer. Sci. Rep. 2017, 7, 16723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, F.; Kulasingam, V.; Diamandis, E.P.; Hoon, D.S.; Kinzler, K.; Pantel, K.; Alix-Panabieres, C. Circulating Tumor DNA as a Cancer Biomarker: Fact or Fiction? Clin. Chem. 2016, 62, 1054–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephan, F.; Marsman, G.; Bakker, L.M.; Bulder, I.; Stavenuiter, F.; Aarden, L.A.; Zeerleder, S. Cooperation of factor VII-activating protease and serum DNase I in the release of nucleosomes from necrotic cells. Arthritis Rheumatol. 2014, 66, 686–693. [Google Scholar] [CrossRef]
- Martin, M.; Leffler, J.; Smolag, K.I.; Mytych, J.; Bjork, A.; Chaves, L.D.; Alexander, J.J.; Quigg, R.J.; Blom, A.M. Factor H uptake regulates intracellular C3 activation during apoptosis and decreases the inflammatory potential of nucleosomes. Cell Death Differ. 2016, 23, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Du Clos, T.W.; Volzer, M.A.; Hahn, F.F.; Xiao, R.; Mold, C.; Searles, R.P. Chromatin clearance in C57Bl/10 mice: Interaction with heparan sulphate proteoglycans and receptors on Kupffer cells. Clin. Exp. Immunol. 1999, 117, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Stroun, M.; Lyautey, J.; Lederrey, C.; Olson-Sand, A.; Anker, P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin. Chim. Acta 2001, 313, 139–142. [Google Scholar] [CrossRef]
- Zhang, B.O.; Xu, C.W.; Shao, Y.; Wang, H.T.; Wu, Y.F.; Song, Y.Y.; Li, X.B.; Zhang, Z.; Wang, W.J.; Li, L.Q.; et al. Comparison of droplet digital PCR and conventional quantitative PCR for measuring EGFR gene mutation. Exp. Ther. Med. 2015, 9, 1383–1388. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Jing, C.; Wu, J.; Ni, J.; Sha, H.; Xu, X.; Du, Y.; Lou, R.; Dong, S.; Feng, J. Circulating tumor DNA detection: A potential tool for colorectal cancer management. Oncol. Lett. 2019, 17, 1409–1416. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Foncillas, J.; Alba, E.; Aranda, E.; Diaz-Rubio, E.; Lopez-Lopez, R.; Tabernero, J.; Vivancos, A. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: An expert taskforce review. Ann. Oncol. 2017, 28, 2943–2949. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra168. [Google Scholar] [CrossRef] [PubMed]
- Giroux Leprieur, E.; Helias-Rodzewicz, Z.; Takam Kamga, P.; Costantini, A.; Julie, C.; Corjon, A.; Dumenil, C.; Dumoulin, J.; Giraud, V.; Labrune, S.; et al. Sequential ctDNA whole-exome sequencing in advanced lung adenocarcinoma with initial durable tumor response on immune checkpoint inhibitor and late progression. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Imperial, R.; Nazer, M.; Ahmed, Z.; Kam, A.E.; Pluard, T.J.; Bahaj, W.; Levy, M.; Kuzel, T.M.; Hayden, D.M.; Pappas, S.G.; et al. Matched Whole-Genome Sequencing (WGS) and Whole-Exome Sequencing (WES) of Tumor Tissue with Circulating Tumor DNA (ctDNA) Analysis: Complementary Modalities in Clinical Practice. Cancers 2019, 11, 1399. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Foncillas, J.; Tabernero, J.; Elez, E.; Aranda, E.; Benavides, M.; Camps, C.; Jantus-Lewintre, E.; Lopez, R.; Muinelo-Romay, L.; Montagut, C.; et al. Prospective multicenter real-world RAS mutation comparison between OncoBEAM-based liquid biopsy and tissue analysis in metastatic colorectal cancer. Br. J. Cancer 2018, 119, 1464–1470. [Google Scholar] [CrossRef]
- LB057—Whole Exome Sequencing of Tumor Tissue and Circulating Tumor DNA Ingastrointestinal Stromal Tumors (GIST). Available online: https://www.abstractsonline.com/pp8/#!/9325/presentation/4552 (accessed on 10 January 2021).
- U.S. Food & Drug. FDA Approves Liquid Biopsy NGS Companion Diagnostic Test for Multiple Cancers and Biomarkers. Available online: https://www.fda.gov/drugs/fda-approves-liquid-biopsy-ngs-companion-diagnostic-test-multiple-cancers-and-biomarkers (accessed on 10 January 2021).
- U.S. Food & Drug. Oncology (Cancer)/Hematologic Malignancies Approval Notifications. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/hematologyoncology-cancer-approvals-safety-notifications (accessed on 10 January 2021).
- U.S. Food & Drug. Guardant360 CDx—P200010. Available online: https://www.fda.gov/medical-devices/recently-approved-devices/guardant360-cdx-p200010 (accessed on 10 January 2021).
- U.S. Food & Drug. The Therascreen PIK3CA RGQ PCR Kit-P190001 and P190004. Available online: https://www.fda.gov/medical-devices/recently-approved-devices/therascreen-pik3ca-rgq-pcr-kit-p190001-and-p190004 (accessed on 10 January 2021).
- U.S. Food & Drug. Cobas EGFR Mutation Test v2. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/cobas-egfr-mutation-test-v2 (accessed on 10 January 2021).
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K.; et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; Consortium, C. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Cavallone, L.; Aguilar, A.; Aldamry, M.; Lafleur, J.; Brousse, S.; Lan, C.; Alirezaie, N.; Bareke, E.; Majewski, J.; Pelmus, M.; et al. Circulating tumor DNA (ctDNA) during and after neoadjuvant chemotherapy and prior to surgery is a powerful prognostic factor in triple-negative breast cancer (TNBC). J. Clin. Oncol. 2019, 37, 594. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.; Wang, Y.; Li, L.; Kinde, I.; Elsaleh, H.; Wong, R.; Kosmider, S.; Yip, D.; Lee, M.; et al. The potential of circulating tumor DNA (ctDNA) to guide adjuvant chemotherapy decision making in locally advanced rectal cancer (LARC). J. Clin. Oncol. 2017, 35, 3521. [Google Scholar] [CrossRef]
- Cabel, L.; Jeannot, E.; Bieche, I.; Vacher, S.; Callens, C.; Bazire, L.; Morel, A.; Bernard-Tessier, A.; Chemlali, W.; Schnitzler, A.; et al. Prognostic Impact of Residual HPV ctDNA Detection after Chemoradiotherapy for Anal Squamous Cell Carcinoma. Clin. Cancer Res. 2018, 24, 5767–5771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, I.S.; Kato, S.; Fanta, P.T.; Leichman, L.; Okamura, R.; Raymond, V.M.; Lanman, R.B.; Lippman, S.M.; Kurzrock, R. Genomic Profiling of Blood-Derived Circulating Tumor DNA from Patients with Colorectal Cancer: Implications for Response and Resistance to Targeted Therapeutics. Mol. Cancer Ther. 2019, 18, 1852–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shatsky, R.; Parker, B.A.; Bui, N.Q.; Helsten, T.; Schwab, R.B.; Boles, S.G.; Kurzrock, R. Next-Generation Sequencing of Tissue and Circulating Tumor DNA: The UC San Diego Moores Center for Personalized Cancer Therapy Experience with Breast Malignancies. Mol. Cancer Ther. 2019, 18, 1001–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, S.; Okamura, R.; Baumgartner, J.M.; Patel, H.; Leichman, L.; Kelly, K.; Sicklick, J.K.; Fanta, P.T.; Lippman, S.M.; Kurzrock, R. Analysis of Circulating Tumor DNA and Clinical Correlates in Patients with Esophageal, Gastroesophageal Junction, and Gastric Adenocarcinoma. Clin. Cancer Res. 2018, 24, 6248–6256. [Google Scholar] [CrossRef] [Green Version]
- Baumgartner, J.M.; Raymond, V.M.; Lanman, R.B.; Tran, L.; Kelly, K.J.; Lowy, A.M.; Kurzrock, R. Preoperative Circulating Tumor DNA in Patients with Peritoneal Carcinomatosis is an Independent Predictor of Progression-Free Survival. Ann. Surg. Oncol. 2018, 25, 2400–2408. [Google Scholar] [CrossRef] [PubMed]
- Charo, L.M.; Eskander, R.N.; Okamura, R.; Patel, S.P.; Nikanjam, M.; Lanman, R.B.; Piccioni, D.E.; Kato, S.; McHale, M.T.; Kurzrock, R. Clinical implications of plasma circulating tumor DNA in gynecologic cancer patients. Mol. Oncol. 2021, 15, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Okamura, R.; Kurzrock, R.; Mallory, R.J.; Fanta, P.T.; Burgoyne, A.M.; Clary, B.M.; Kato, S.; Sicklick, J.K. Comprehensive genomic landscape and precision therapeutic approach in biliary tract cancers. Int. J. Cancer 2021, 148, 702–712. [Google Scholar] [CrossRef]
- Schwaederle, M.C.; Patel, S.P.; Husain, H.; Ikeda, M.; Lanman, R.B.; Banks, K.C.; Talasaz, A.; Bazhenova, L.; Kurzrock, R. Utility of Genomic Assessment of Blood-Derived Circulating Tumor DNA (ctDNA) in Patients with Advanced Lung Adenocarcinoma. Clin. Cancer Res. 2017, 23, 5101–5111. [Google Scholar] [CrossRef] [Green Version]
- Patel, H.; Okamura, R.; Fanta, P.; Patel, C.; Lanman, R.B.; Raymond, V.M.; Kato, S.; Kurzrock, R. Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J. Hematol. Oncol. 2019, 12, 130. [Google Scholar] [CrossRef] [Green Version]
- Kato, S.; Okamura, R.; Mareboina, M.; Lee, S.; Goodman, A.; Patel, S.P.; Fanta, P.T.; Schwab, R.B.; Vu, P.; Raymond, V.M.; et al. Revisiting Epidermal Growth Factor Receptor (EGFR) Amplification as a Target for Anti-EGFR Therapy: Analysis of Cell-Free Circulating Tumor DNA in Patients with Advanced Malignancies. JCO Precis. Oncol. 2019, 3. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, S.; Schwaederle, M.; Mohindra, M.; Fontes Jardim, D.L.; Kurzrock, R. MET alterations detected in blood-derived circulating tumor DNA correlate with bone metastases and poor prognosis. J. Hematol. Oncol. 2018, 11, 76. [Google Scholar] [CrossRef] [Green Version]
- Li, B.T.; Janku, F.; Jung, B.; Hou, C.; Madwani, K.; Alden, R.; Razavi, P.; Reis-Filho, J.S.; Shen, R.; Isbell, J.M.; et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: Results from the Actionable Genome Consortium. Ann. Oncol. 2019, 30, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Janku, F.; Zhang, S.; Waters, J.; Liu, L.; Huang, H.J.; Subbiah, V.; Hong, D.S.; Karp, D.D.; Fu, S.; Cai, X.; et al. Development and Validation of an Ultradeep Next-Generation Sequencing Assay for Testing of Plasma Cell-Free DNA from Patients with Advanced Cancer. Clin. Cancer Res. 2017, 23, 5648–5656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, S.; Okamura, R.; Kato, S.; Soussi, T.; Kurzrock, R. Survival Implications of the Relationship between Tissue versus Circulating Tumor DNA TP53 Mutations-A Perspective from a Real-World Precision Medicine Cohort. Mol. Cancer Ther. 2020, 19, 2612–2620. [Google Scholar] [CrossRef] [PubMed]
- Mardinian, K.; Okamura, R.; Kato, S.; Kurzrock, R. Temporal and spatial effects and survival outcomes associated with concordance between tissue and blood KRAS alterations in the pan-cancer setting. Int. J. Cancer 2020, 146, 566–576. [Google Scholar] [CrossRef]
- Ricciuti, B.; Jones, G.; Severgnini, M.; Alessi, J.V.; Recondo, G.; Lawrence, M.; Forshew, T.; Lydon, C.; Nishino, M.; Cheng, M.; et al. Early plasma circulating tumor DNA (ctDNA) changes predict response to first-line pembrolizumab-based therapy in non-small cell lung cancer (NSCLC). J. Immunother. Cancer 2021, 9. [Google Scholar] [CrossRef]
- Janku, F.; Huang, H.J.; Fujii, T.; Shelton, D.N.; Madwani, K.; Fu, S.; Tsimberidou, A.M.; Piha-Paul, S.A.; Wheler, J.J.; Zinner, R.G.; et al. Multiplex KRASG12/G13 mutation testing of unamplified cell-free DNA from the plasma of patients with advanced cancers using droplet digital polymerase chain reaction. Ann. Oncol. 2017, 28, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.J.; Goodman, A.M.; Kato, S.; Ellison, C.K.; Daniels, G.A.; Kim, L.; Nakashe, P.; McCarthy, E.; Mazloom, A.R.; McLennan, G.; et al. Genome-Wide Sequencing of Cell-Free DNA Identifies Copy-Number Alterations That Can Be Used for Monitoring Response to Immunotherapy in Cancer Patients. Mol. Cancer Ther. 2019, 18, 448–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, Y.K.; Oh, M.S. Detection of Minimal Residual Disease Using ctDNA in Lung Cancer: Current Evidence and Future Directions. J. Thorac. Oncol. 2019, 14, 16–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, S.J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef] [Green Version]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Parkinson, C.A.; Gale, D.; Piskorz, A.M.; Biggs, H.; Hodgkin, C.; Addley, H.; Freeman, S.; Moyle, P.; Sala, E.; Sayal, K.; et al. Exploratory Analysis of TP53 Mutations in Circulating Tumour DNA as Biomarkers of Treatment Response for Patients with Relapsed High-Grade Serous Ovarian Carcinoma: A Retrospective Study. PLoS Med. 2016, 13, e1002198. [Google Scholar] [CrossRef]
- Lipson, E.J.; Velculescu, V.E.; Pritchard, T.S.; Sausen, M.; Pardoll, D.M.; Topalian, S.L.; Diaz, L.A., Jr. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. J. Immunother. Cancer 2014, 2, 42. [Google Scholar] [CrossRef] [Green Version]
- Mok, T.; Wu, Y.L.; Lee, J.S.; Yu, C.J.; Sriuranpong, V.; Sandoval-Tan, J.; Ladrera, G.; Thongprasert, S.; Srimuninnimit, V.; Liao, M.; et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015, 21, 3196–3203. [Google Scholar] [CrossRef] [Green Version]
- Anandappa, G.; Starling, N.; Begum, R.; Bryant, A.; Sharma, S.; Renner, D.; Aresu, M.; Peckitt, C.; Sethi, H.; Feber, A.; et al. Minimal residual disease (MRD) detection with circulating tumor DNA (ctDNA) from personalized assays in stage II-III colorectal cancer patients in a U.K. multicenter prospective study (TRACC). J. Clin. Oncol. 2021, 39, 102. [Google Scholar] [CrossRef]
- Cao, H.; Liu, X.; Chen, Y.; Yang, P.; Huang, T.; Song, L.; Xu, R. Circulating Tumor DNA Is Capable of Monitoring the Therapeutic Response and Resistance in Advanced Colorectal Cancer Patients Undergoing Combined Target and Chemotherapy. Front. Oncol. 2020, 10, 466. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, T.; Yang, Z.; Zheng, Y.; Yu, R.; Wu, X.; Yan, J.; Shao, Y.W.; Shao, X.; Cao, W.; et al. Monitoring treatment efficacy and resistance in breast cancer patients via circulating tumor DNA genomic profiling. Mol. Genet. Genom. Med. 2020, 8, e1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz - Cuaran, S.; Mezquita, L.; Swalduz, A.; Aldea, M.; Mazieres, J.; Jovelet, C.; Lacroix, L.; Pradines, A.; Avrillon, V.; MahierAït Oukhatar, C.; et al. 1556P—Circulating tumour DNA (ctDNA) analysis depicts mechanisms of resistance and tumour response to BRAF inhibitors in BRAF-mutant non-small cell lung cancer (NSCLC). Ann. Oncol. 2019, 30, v641. [Google Scholar] [CrossRef]
- Razavi, P.; Dickler, M.N.; Shah, P.D.; Toy, W.; Brown, D.N.; Won, H.H.; Li, B.T.; Shen, R.; Vasan, N.; Modi, S.; et al. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat. Cancer 2020, 1, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Zhu, W.; Guan, Y.; Yang, L.; Xia, X.; Chen, S.; Li, Q.; Guan, X.; Yi, Z.; Qian, H.; et al. ctDNA dynamics: A novel indicator to track resistance in metastatic breast cancer treated with anti-HER2 therapy. Oncotarget 2016, 7, 66020–66031. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yao, Y.; Xu, Y.; Li, L.; Gong, Y.; Zhang, K.; Zhang, M.; Guan, Y.; Chang, L.; Xia, X.; et al. Pan-cancer circulating tumor DNA detection in over 10,000 Chinese patients. Nat. Commun. 2021, 12, 11. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Swigart, L.B.; Wu, H.T.; Hirst, G.L.; Yau, C.; Wolf, D.M.; Tin, A.; Salari, R.; Shchegrova, S.; Pawar, H.; et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann. Oncol. 2021, 32, 229–239. [Google Scholar] [CrossRef]
- Zhang, Q.; Luo, J.; Wu, S.; Si, H.; Gao, C.; Xu, W.; Abdullah, S.E.; Higgs, B.W.; Dennis, P.A.; van der Heijden, M.S.; et al. Prognostic and Predictive Impact of Circulating Tumor DNA in Patients with Advanced Cancers Treated with Immune Checkpoint Blockade. Cancer Discov. 2020, 10, 1842–1853. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.K.; Davis, A.A.; Jain, S.; Santa-Maria, C.; Flaum, L.; Beaubier, N.; Platanias, L.C.; Gradishar, W.; Giles, F.J.; Cristofanilli, M. Concordance of Genomic Alterations by Next-Generation Sequencing in Tumor Tissue versus Circulating Tumor DNA in Breast Cancer. Mol. Cancer Ther. 2017, 16, 1412–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.; Othman, T.; Chen, C.; Sandhu, J.; Ouyang, C.; Fakih, M. Guardant360 Circulating Tumor DNA Assay Is Concordant with FoundationOne Next-Generation Sequencing in Detecting Actionable Driver Mutations in Anti-EGFR Naive Metastatic Colorectal Cancer. Oncologist 2020, 25, 235–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).