Imaging Mass Spectrometry-Based Proteomic Analysis to Differentiate Melanocytic Nevi and Malignant Melanoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Sample Preparation Optimization
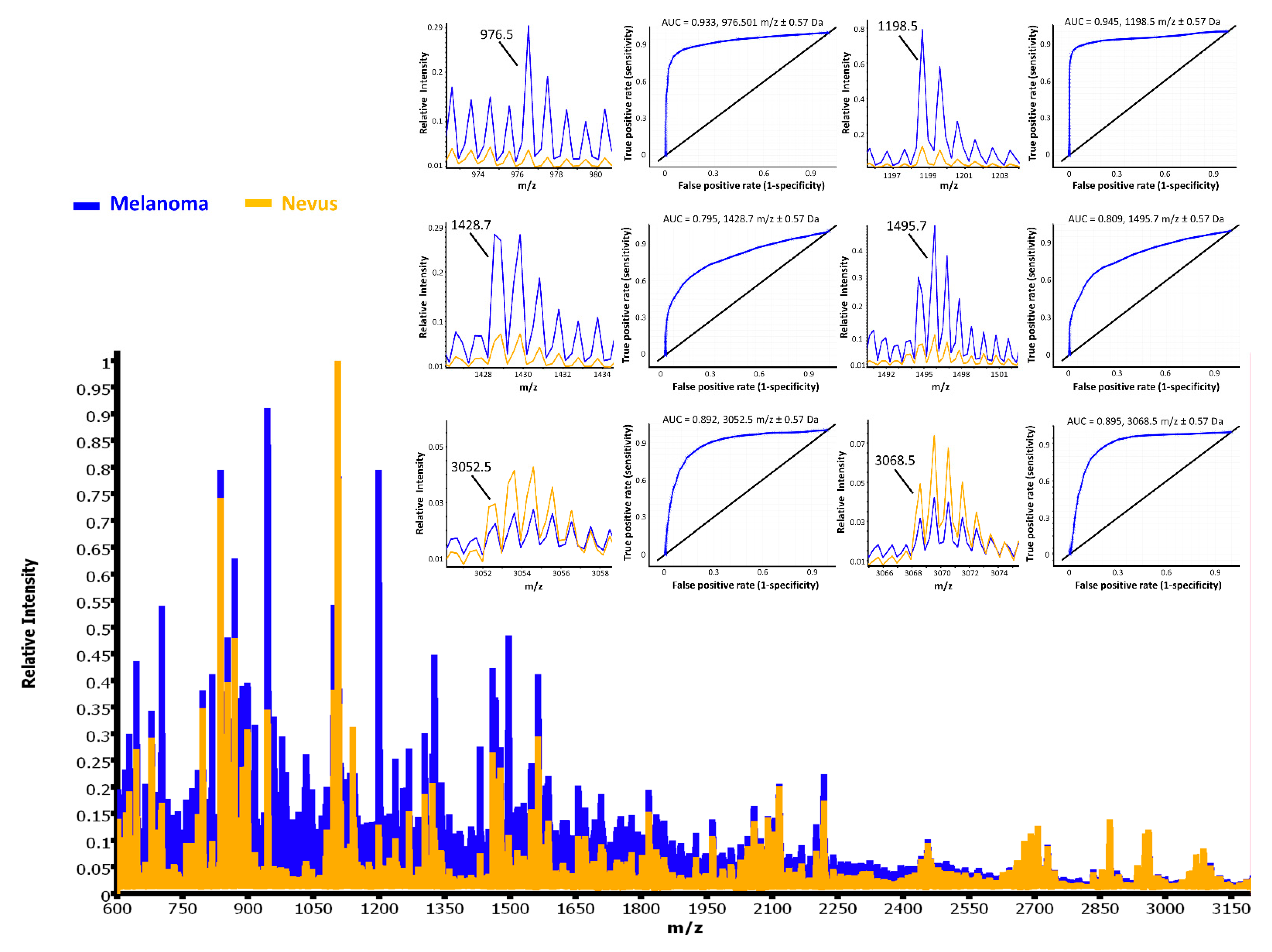
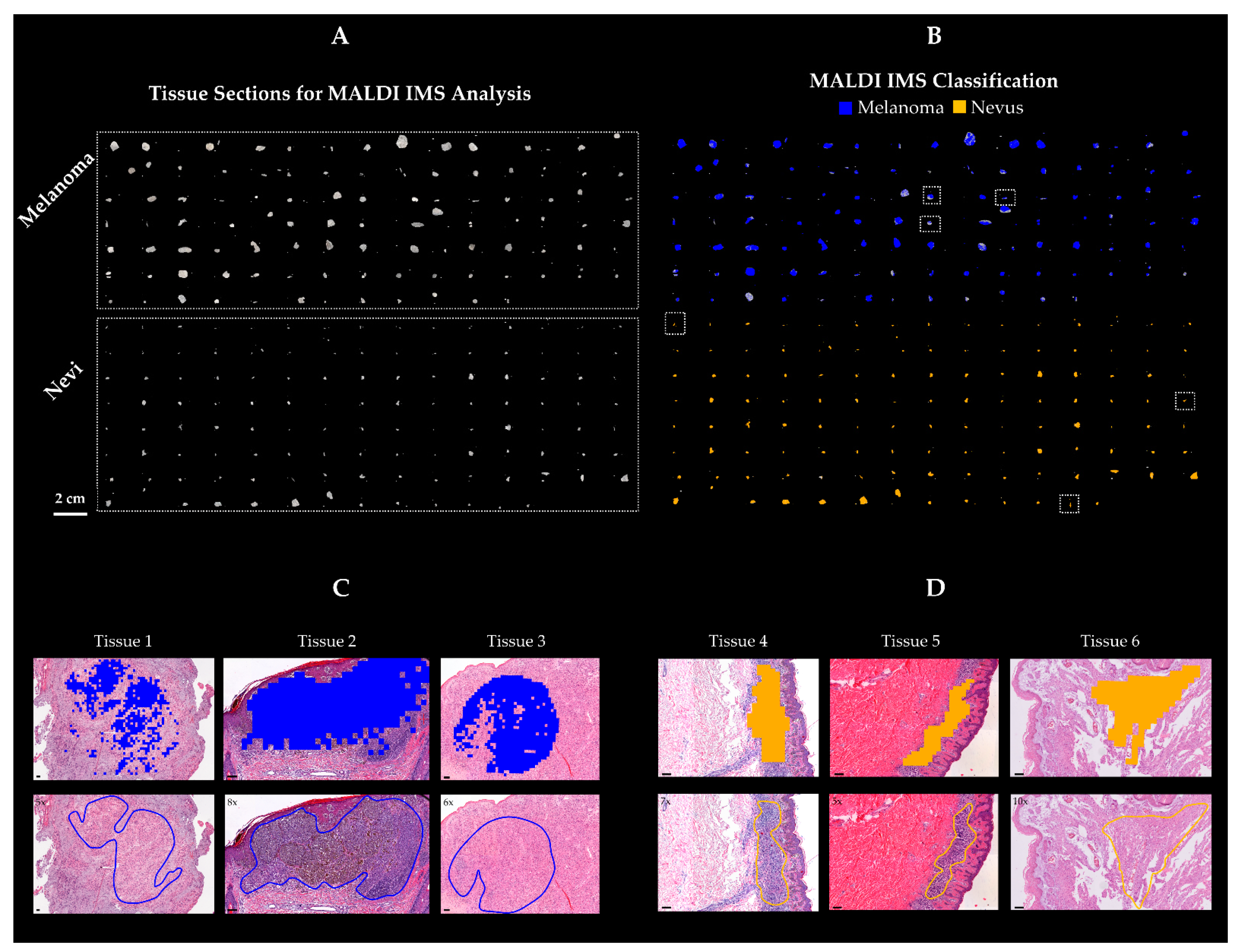
2.2. Statistical Analysis and Classification Study
2.3. Protein Identification
3. Discussion
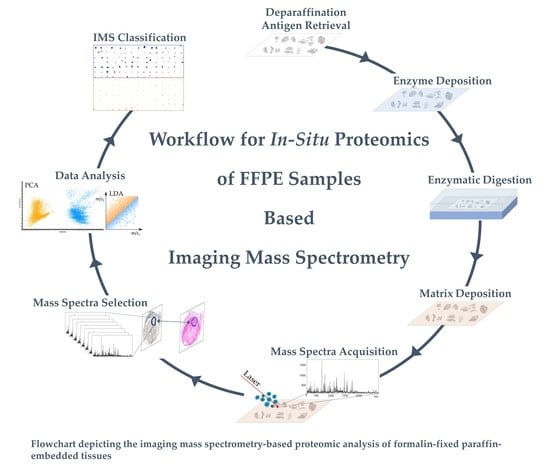
4. Materials and Methods
4.1. Tissue Collection and Preparation
4.2. On-Tissue Digestion and Matrix Application
4.3. MALDI Imaging Mass Spectrometry (IMS) Profiling
4.4. Data Analysis
4.5. Protein Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Bevona, C.; Goggins, W.; Quinn, T.; Fullerton, J.; Tsao, H. Cutaneous melanomas associated with nevi. Arch. Dermatol. 2003, 139, 1620–1624; discussion 1624. [Google Scholar] [CrossRef] [PubMed]
- Tsao, H.; Bevona, C.; Goggins, W.; Quinn, T. The transformation rate of moles (melanocytic nevi) into cutaneous melanoma: A population-based estimate. Arch. Dermatol. 2003, 139, 282–288. [Google Scholar] [CrossRef]
- Gruber, S.B.; Barnhill, R.L.; Stenn, K.S.; Roush, G.C. Nevomelanocytic proliferations in association with cutaneous malignant melanoma: A multivariate analysis. J. Am. Acad. Dermatol. 1989, 21, 773–780. [Google Scholar] [CrossRef]
- Farmer, E.R.; Gonin, R.; Hanna, M.P. Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Hum. Pathol. 1996, 27, 528–531. [Google Scholar] [CrossRef]
- Garola, R.; Singh, V. Utility of p16-Ki-67-HMB45 score in sorting benign from malignant Spitz tumors. Pathol. Res. Pract. 2019, 215, 152550. [Google Scholar] [CrossRef]
- Ritter, A.; Tronnier, M.; Vaske, B.; Mitteldorf, C. Reevaluation of established and new criteria in differential diagnosis of Spitz nevus and melanoma. Arch. Dermatol. Res. 2018, 310, 329–342. [Google Scholar] [CrossRef]
- See, S.H.C.; Finkelman, B.S.; Yeldandi, A.V. The diagnostic utility of PRAME and p16 in distinguishing nodal nevi from nodal metastatic melanoma. Pathol. Res. Pract. 2020, 216, 153105. [Google Scholar] [CrossRef]
- Gerami, P.; Li, G.; Pouryazdanparast, P.; Blondin, B.; Beilfuss, B.; Slenk, C.; Du, J.; Guitart, J.; Jewell, S.; Pestova, K. A highly specific and discriminatory FISH assay for distinguishing between benign and malignant melanocytic neoplasms. Am. J. Surg. Pathol. 2012, 36, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Cho-Vega, J.H.; Cao, T.; Ledon, J.; Moller, M.; Avisar, E.; Elgart, G.; Tan, J.H.; Fan, Y.S.; Grichnik, J.M. Diagnostic application of cyclin D1 fluorescent in situ hybridization for histologically undetermined early lesions of acral melanoma in situ: A case series. Ann. Diagn. Pathol. 2021, 50, 151681. [Google Scholar] [CrossRef]
- Pouryazdanparast, P.; Newman, M.; Mafee, M.; Haghighat, Z.; Guitart, J.; Gerami, P. Distinguishing epithelioid blue nevus from blue nevus-like cutaneous melanoma metastasis using fluorescence in situ hybridization. Am. J. Surg. Pathol. 2009, 33, 1396–1400. [Google Scholar] [CrossRef]
- Yanovich, G.; Agmon, H.; Harel, M.; Sonnenblick, A.; Peretz, T.; Geiger, T. Clinical Proteomics of Breast Cancer Reveals a Novel Layer of Breast Cancer Classification. Cancer Res. 2018, 78, 6001–6010. [Google Scholar] [CrossRef]
- Guo, T.; Li, L.; Zhong, Q.; Rupp, N.J.; Charmpi, K.; Wong, C.E.; Wagner, U.; Rueschoff, J.H.; Jochum, W.; Fankhauser, C.D.; et al. Multi-region proteome analysis quantifies spatial heterogeneity of prostate tissue biomarkers. Life Sci. Alliance 2018, 1. [Google Scholar] [CrossRef]
- Timms, J.F.; Hale, O.J.; Cramer, R. Advances in mass spectrometry-based cancer research and analysis: From cancer proteomics to clinical diagnostics. Expert Rev. Proteom. 2016, 13, 593–607. [Google Scholar] [CrossRef]
- Longuespee, R.; Casadonte, R.; Kriegsmann, M.; Pottier, C.; Picard de Muller, G.; Delvenne, P.; Kriegsmann, J.; De Pauw, E. MALDI mass spectrometry imaging: A cutting-edge tool for fundamental and clinical histopathology. Proteom. Clin. Appl. 2016, 10, 701–719. [Google Scholar] [CrossRef]
- Rauser, S.; Marquardt, C.; Balluff, B.; Deininger, S.O.; Albers, C.; Belau, E.; Hartmer, R.; Suckau, D.; Specht, K.; Ebert, M.P.; et al. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. J. Proteome Res. 2010, 9, 1854–1863. [Google Scholar] [CrossRef] [PubMed]
- Schwamborn, K.; Krieg, R.C.; Reska, M.; Jakse, G.; Knuechel, R.; Wellmann, A. Identifying prostate carcinoma by MALDI-Imaging. Int. J. Mol. Med. 2007, 20, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Kriegsmann, M.; Casadonte, R.; Kriegsmann, J.; Dienemann, H.; Schirmacher, P.; Hendrik Kobarg, J.; Schwamborn, K.; Stenzinger, A.; Warth, A.; Weichert, W. Reliable Entity Subtyping in Non-small Cell Lung Cancer by Matrix-assisted Laser Desorption/Ionization Imaging Mass Spectrometry on Formalin-fixed Paraffin-embedded Tissue Specimens. Mol. Cell. Proteom. 2016, 15, 3081–3089. [Google Scholar] [CrossRef]
- Kriegsmann, M.; Casadonte, R.; Maurer, N.; Stoehr, C.; Erlmeier, F.; Moch, H.; Junker, K.; Zgorzelski, C.; Weichert, W.; Schwamborn, K.; et al. Mass Spectrometry Imaging Differentiates Chromophobe Renal Cell Carcinoma and Renal Oncocytoma with High Accuracy. J. Cancer 2020, 11, 6081–6089. [Google Scholar] [CrossRef] [PubMed]
- Casadonte, R.; Kriegsmann, M.; Perren, A.; Baretton, G.; Deininger, S.O.; Kriegsmann, K.; Welsch, T.; Pilarsky, C.; Kriegsmann, J. Development of a Class Prediction Model to Discriminate Pancreatic Ductal Adenocarcinoma from Pancreatic Neuroendocrine Tumor by MALDI Mass Spectrometry Imaging. Proteom. Clin. Appl. 2019, 13, e1800046. [Google Scholar] [CrossRef] [PubMed]
- Van de Plas, R.; Yang, J.; Spraggins, J.; Caprioli, R.M. Image fusion of mass spectrometry and microscopy: A multimodality paradigm for molecular tissue mapping. Nat. Methods 2015, 12, 366–372. [Google Scholar] [CrossRef]
- Welinder, C.; Pawłowski, K.; Sugihara, Y.; Yakovleva, M.; Jönsson, G.; Ingvar, C.; Lundgren, L.; Baldetorp, B.; Olsson, H.; Rezeli, M.; et al. A protein deep sequencing evaluation of metastatic melanoma tissues. PLoS ONE 2015, 10, e0123661. [Google Scholar] [CrossRef]
- Azimi, A.; Kaufman, K.L.; Kim, J.; Ali, M.; Mann, G.J.; Fernandez-Penas, P. Proteomics: An emerging approach for the diagnosis and classification of cutaneous squamous cell carcinoma and its precursors. J. Dermatol. Sci. 2020, 99, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Matharoo-Ball, B.; Ratcliffe, L.; Lancashire, L.; Ugurel, S.; Miles, A.K.; Weston, D.J.; Rees, R.; Schadendorf, D.; Ball, G.; Creaser, C.S. Diagnostic biomarkers differentiating metastatic melanoma patients from healthy controls identified by an integrated MALDI-TOF mass spectrometry/bioinformatic approach. Proteom. Clin. Appl. 2007, 1, 605–620. [Google Scholar] [CrossRef]
- Mian, S.; Ugurel, S.; Parkinson, E.; Schlenzka, I.; Dryden, I.; Lancashire, L.; Ball, G.; Creaser, C.; Rees, R.; Schadendorf, D. Serum proteomic fingerprinting discriminates between clinical stages and predicts disease progression in melanoma patients. J. Clin. Oncol. 2005, 23, 5088–5093. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hardesty, W.M.; Kelley, M.C.; Mi, D.; Low, R.L.; Caprioli, R.M. Protein signatures for survival and recurrence in metastatic melanoma. J. Proteom. 2011, 74, 1002–1014. [Google Scholar] [CrossRef]
- Ly, A.; Longuespée, R.; Casadonte, R.; Wandernoth, P.; Schwamborn, K.; Bollwein, C.; Marsching, C.; Kriegsmann, K.; Hopf, C.; Weichert, W.; et al. Site-to-Site Reproducibility and Spatial Resolution in MALDI-MSI of Peptides from Formalin-Fixed Paraffin-Embedded Samples. Proteom. Clin. Appl. 2019, 13, e1800029. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Kerl, K.; Palmedo, G.; Wiesner, T.; Mentzel, T.; Rutten, A.; Scharer, L.; Paredes, B.; Hantschke, M.; Kutzner, H. A proposal for improving multicolor FISH sensitivity in the diagnosis of malignant melanoma using new combined criteria. Am. J. Dermatopathol. 2012, 34, 580–585. [Google Scholar] [CrossRef]
- Ebbelaar, C.F.; Jansen, A.M.L.; Bloem, L.T.; Blokx, W.A.M. Genome-wide copy number variations as molecular diagnostic tool for cutaneous intermediate melanocytic lesions: A systematic review and individual patient data meta-analysis. Virchows Arch. 2021. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.L.; Neishaboori, N.; Tetzlaff, M.T.; Chen, W.S.; Aung, P.P.; Curry, J.L.; Nagarajan, P.; Ivan, D.; Hwu, W.J.; Prieto, V.G.; et al. BAP-1 Expression Status by Immunohistochemistry in Cellular Blue Nevus and Blue Nevus-like Melanoma. Am. J. Dermatopathol. 2020, 42, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Beleaua, M.A.; Jung, I.; Braicu, C.; Milutin, D.; Gurzu, S. SOX11, SOX10 and MITF Gene Interaction: A Possible Diagnostic Tool in Malignant Melanoma. Life 2021, 11, 281. [Google Scholar] [CrossRef]
- Lozada, J.R.; Geyer, F.C.; Selenica, P.; Brown, D.; Alemar, B.; Merghoub, T.; Berger, M.F.; Busam, K.J.; Halpern, A.C.; Weigelt, B.; et al. Massively parallel sequencing analysis of benign melanocytic naevi. Histopathology 2019, 75, 29–38. [Google Scholar] [CrossRef]
- Lazova, R.; Seeley, E.H.; Keenan, M.; Gueorguieva, R.; Caprioli, R.M. Imaging mass spectrometry--a new and promising method to differentiate Spitz nevi from Spitzoid malignant melanomas. Am. J. Dermatopathol. 2012, 34, 82–90. [Google Scholar] [CrossRef]
- Hermann, J.; Noels, H.; Theelen, W.; Lellig, M.; Orth-Alampour, S.; Boor, P.; Jankowski, V.; Jankowski, J. Sample preparation of formalin-fixed paraffin-embedded tissue sections for MALDI-mass spectrometry imaging. Anal. Bioanal. Chem. 2020, 412, 1263–1275. [Google Scholar] [CrossRef]
- Vaysse, P.M.; Heeren, R.M.A.; Porta, T.; Balluff, B. Mass spectrometry imaging for clinical research—Latest developments, applications, and current limitations. Analyst 2017, 142, 2690–2712. [Google Scholar] [CrossRef] [PubMed]
- Korsching, E.; Packeisen, J.; Liedtke, C.; Hungermann, D.; Wülfing, P.; van Diest, P.J.; Brandt, B.; Boecker, W.; Buerger, H. The origin of vimentin expression in invasive breast cancer: Epithelial-mesenchymal transition, myoepithelial histogenesis or histogenesis from progenitor cells with bilinear differentiation potential? J. Pathol. 2005, 206, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Upton, M.P.; Hirohashi, S.; Tome, Y.; Miyazawa, N.; Suemasu, K.; Shimosato, Y. Expression of vimentin in surgically resected adenocarcinomas and large cell carcinomas of lung. Am. J. Surg. Pathol. 1986, 10, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Li, S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 2011, 68, 3033–3046. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, B.; Sun, B.; Wang, X.; Ban, X.; Sun, T.; Liu, Z.; Zhao, X. A novel function for vimentin: The potential biomarker for predicting melanoma hematogenous metastasis. J. Exp. Clin. Cancer Res. 2010, 29, 109. [Google Scholar] [CrossRef]
- Pawlak, G.; Helfman, D.M. Cytoskeletal changes in cell transformation and tumorigenesis. Curr. Opin. Genet. Dev. 2001, 11, 41–47. [Google Scholar] [CrossRef]
- Jasińska-Konior, K.; Wiecheć, O.; Sarna, M.; Panek, A.; Swakoń, J.; Michalik, M.; Urbańska, K.; Elas, M. Increased elasticity of melanoma cells after low-LET proton beam due to actin cytoskeleton rearrangements. Sci. Rep. 2019, 9, 7008. [Google Scholar] [CrossRef]
- Alomari, A.K.; Klump, V.; Neumeister, V.; Ariyan, S.; Narayan, D.; Lazova, R. Comparison of the expression of vimentin and actin in spitz nevi and spitzoid malignant melanomas. Am. J. Dermatopathol. 2015, 37, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Kashina, A.S. Regulation of actin isoforms in cellular and developmental processes. Semin. Cell Dev. Biol. 2020, 102, 113–121. [Google Scholar] [CrossRef]
- Viita, T.; Vartiainen, M.K. From Cytoskeleton to Gene Expression: Actin in the Nucleus. Handb. Exp. Pharmacol. 2017, 235, 311–329. [Google Scholar] [CrossRef]
- Viita, T.; Kyheroinen, S.; Prajapati, B.; Virtanen, J.; Frilander, M.J.; Varjosalo, M.; Vartiainen, M.K. Nuclear actin interactome analysis links actin to KAT14 histone acetyl transferase and mRNA splicing. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed]
- Kulis, M.; Esteller, M. DNA methylation and cancer. Adv. Genet. 2010, 70, 27–56. [Google Scholar] [CrossRef] [PubMed]
- Molden, R.C.; Bhanu, N.V.; LeRoy, G.; Arnaudo, A.M.; Garcia, B.A. Multi-faceted quantitative proteomics analysis of histone H2B isoforms and their modifications. Epigenetics Chromatin 2015, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, H.; Pessoa, G.C.; de Luna Vitorino, F.N.; Nsengimana, J.; Newton-Bishop, J.; Reis, E.M.; da Cunha, J.P.C.; Jasiulionis, M.G. Gene co-expression and histone modification signatures are associated with melanoma progression, epithelial-to-mesenchymal transition, and metastasis. Clin. Epigenetics 2020, 12, 127. [Google Scholar] [CrossRef]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Utility of histone H3K27me3 and H4K20me as diagnostic indicators of melanoma. Melanoma Res. 2020, 30, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Hurkmans, D.P.; Jensen, C.; Koolen, S.L.W.; Aerts, J.; Karsdal, M.A.; Mathijssen, R.H.J.; Willumsen, N. Blood-based extracellular matrix biomarkers are correlated with clinical outcome after PD-1 inhibition in patients with metastatic melanoma. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef]
- Osorio, A.; Milne, R.L.; Kuchenbaecker, K.; Vaclová, T.; Pita, G.; Alonso, R.; Peterlongo, P.; Blanco, I.; de la Hoya, M.; Duran, M.; et al. DNA glycosylases involved in base excision repair may be associated with cancer risk in BRCA1 and BRCA2 mutation carriers. PLoS Genet. 2014, 10, e1004256. [Google Scholar] [CrossRef] [PubMed]
- Casadonte, R.; Caprioli, R.M. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat. Protoc. 2011, 6, 1695–1709. [Google Scholar] [CrossRef] [PubMed]
- Boskamp, T.; Lachmund, D.; Casadonte, R.; Hauberg-Lotte, L.; Kobarg, J.H.; Kriegsmann, J.; Maass, P. Using the Chemical Noise Background in MALDI Mass Spectrometry Imaging for Mass Alignment and Calibration. Anal. Chem. 2020, 92, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Boskamp, T.; Casadonte, R.; Hauberg-Lotte, L.; Deininger, S.; Kriegsmann, J.; Maass, P. Cross-normalization of MALDI mass spectrometry imaging data improves site-to-site reproducibility. Anal. Chem. 2021. submitted for publication. [Google Scholar]


| Peptide Signature | Cohort | Tissue Type | n Patients | Correct Classification 1 | Incorrect Classification | Overall Accuracy |
|---|---|---|---|---|---|---|
| 187 Peptides | Training | Melanoma | 56 | 54 | 2 | 98% |
| Nevus | 57 | 57 | 0 | |||
| Testing | Melanoma | 45 | 41 | 4 | 93% | |
| Nevus | 45 | 43 | 2 | |||
| 18 Peptides | Training | Melanoma | 56 | 49 | 7 | 93% |
| Nevus | 57 | 56 | 1 | |||
| Testing | Melanoma | 45 | 39 | 6 | 92% | |
| Nevus | 45 | 44 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casadonte, R.; Kriegsmann, M.; Kriegsmann, K.; Hauk, I.; Meliß, R.R.; Müller, C.S.L.; Kriegsmann, J. Imaging Mass Spectrometry-Based Proteomic Analysis to Differentiate Melanocytic Nevi and Malignant Melanoma. Cancers 2021, 13, 3197. https://doi.org/10.3390/cancers13133197
Casadonte R, Kriegsmann M, Kriegsmann K, Hauk I, Meliß RR, Müller CSL, Kriegsmann J. Imaging Mass Spectrometry-Based Proteomic Analysis to Differentiate Melanocytic Nevi and Malignant Melanoma. Cancers. 2021; 13(13):3197. https://doi.org/10.3390/cancers13133197
Chicago/Turabian StyleCasadonte, Rita, Mark Kriegsmann, Katharina Kriegsmann, Isabella Hauk, Rolf R. Meliß, Cornelia S. L. Müller, and Jörg Kriegsmann. 2021. "Imaging Mass Spectrometry-Based Proteomic Analysis to Differentiate Melanocytic Nevi and Malignant Melanoma" Cancers 13, no. 13: 3197. https://doi.org/10.3390/cancers13133197
APA StyleCasadonte, R., Kriegsmann, M., Kriegsmann, K., Hauk, I., Meliß, R. R., Müller, C. S. L., & Kriegsmann, J. (2021). Imaging Mass Spectrometry-Based Proteomic Analysis to Differentiate Melanocytic Nevi and Malignant Melanoma. Cancers, 13(13), 3197. https://doi.org/10.3390/cancers13133197