Proteomics, Personalized Medicine and Cancer
Abstract
Simple Summary
Abstract
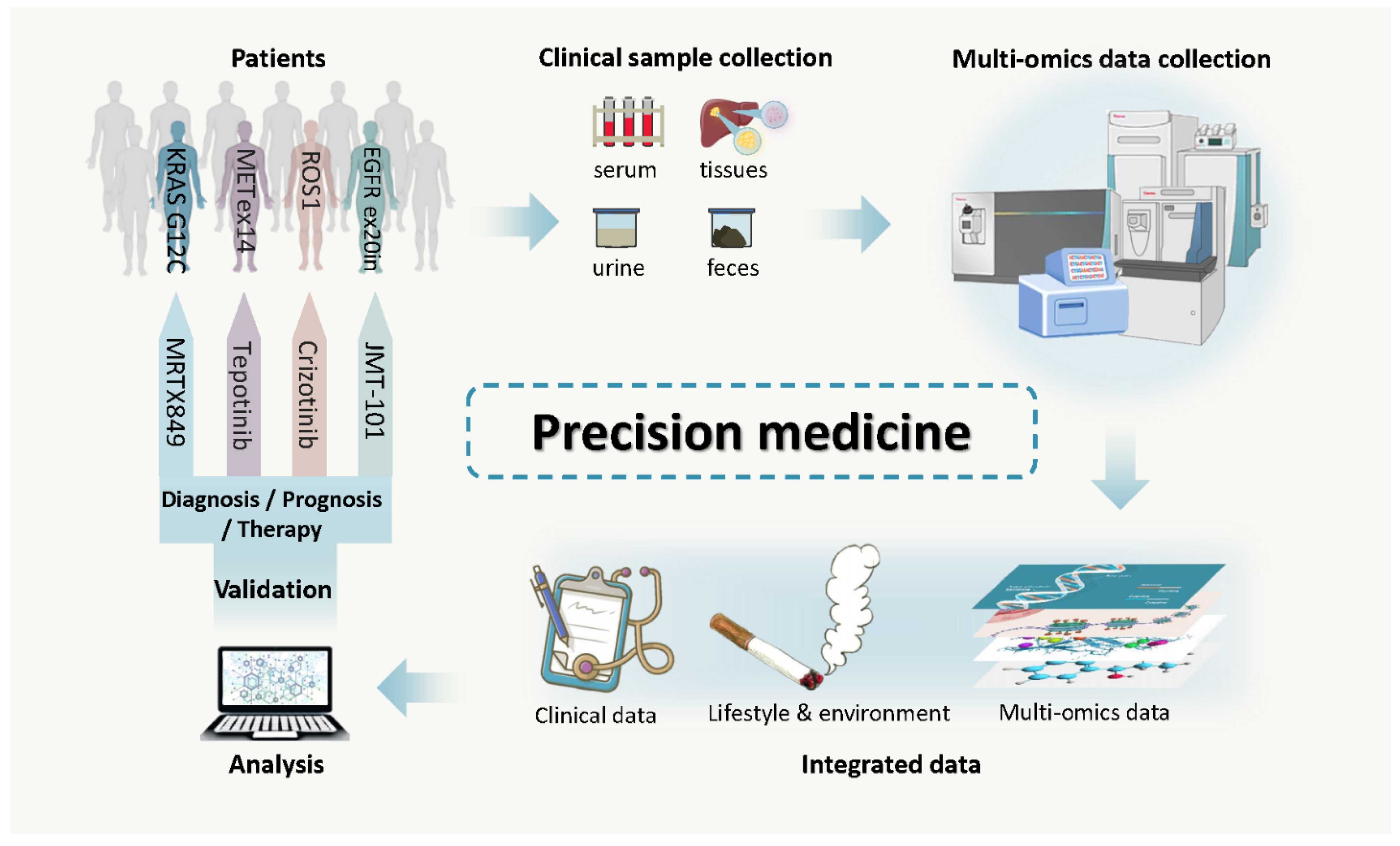
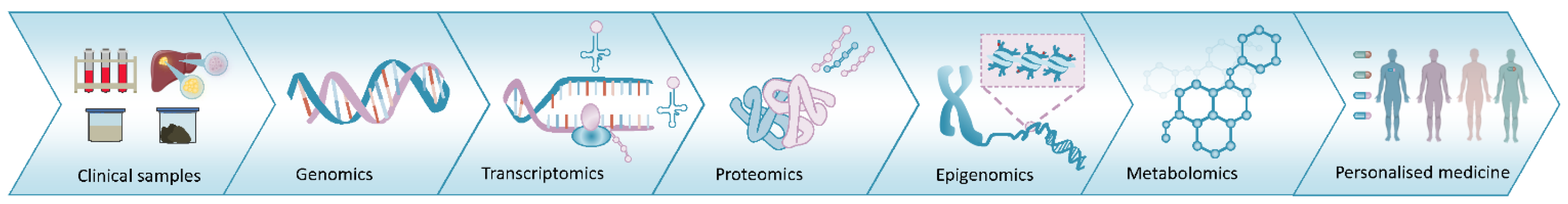
1. Introduction
2. Proteomics, the Current Status
2.1. iTRAQ and Other Labelling Strategies
2.2. Targeted Approaches
2.3. Label Free Approaches
2.4. Proteogenomics
2.5. Bioinformatics
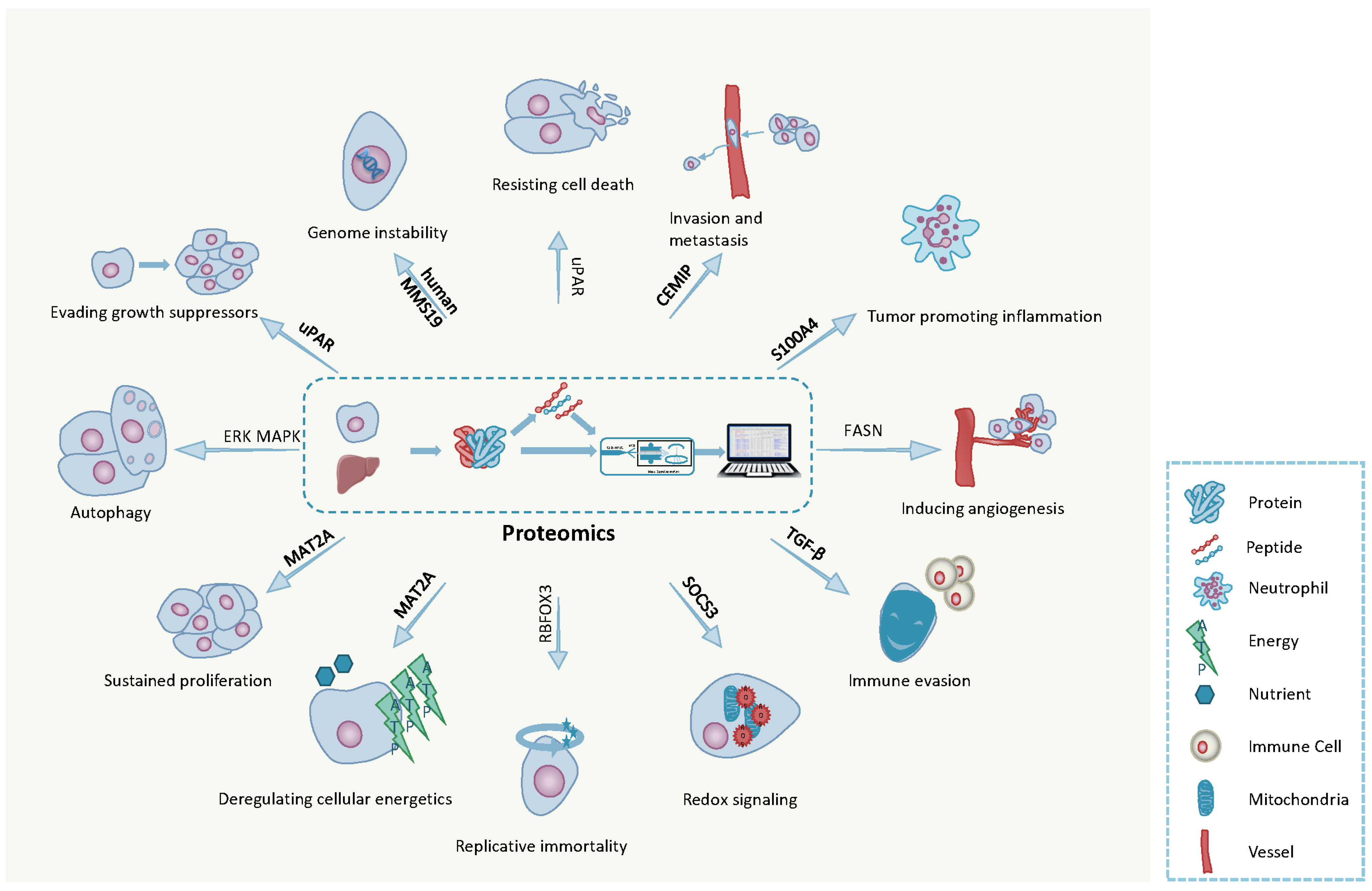
3. The Hallmarks of Cancer
3.1. Ten Hallmarks of Cancer
3.2. Emerging Hallmarks of Cancer
4. Cancer Biomarkers: Detection, Surveillance and Drug Efficacy
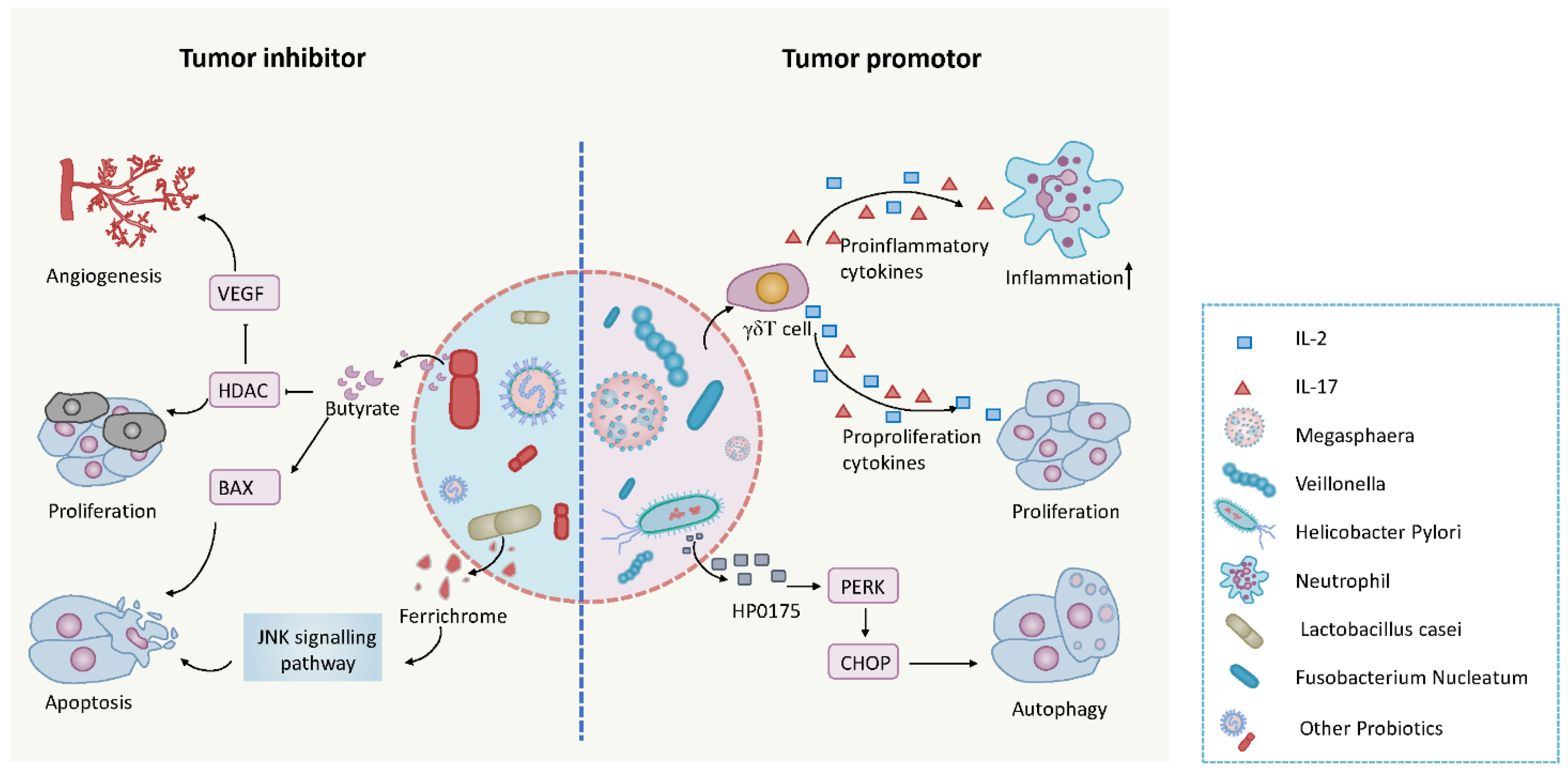
5. The Microbiome and Cancer
| Function | Microorganism | Type of Cancer | Mechanism | References |
|---|---|---|---|---|
| Cancer Therapy | Mycobacterium bovis BCG | Bladder | Stimulating the immune system and increasing the proinflammatory cytokines activation of cancer cells phagocytosis | [154,155,156,157,158] |
| Streptococcus pyogenes OK-432 | Lymphangioma Intraoral Ranula | Immune activation by increasing cytokine levels | [159,160,161,162] | |
| Clostridium novyi | Leiomyoma | Targeting and destroying tumor cells | [154,163,164,165] | |
| Salmonella Typhimurium VNP20009 | Melanoma, Pancreatic | Helping antitumor drugs target cancer | [166,167] | |
| Magnetococcus marinus | - | Targeted transport vector | [168,169] | |
| Bifidobacterium Longum | Colorectal | Enhancing the body’s immune function and regulating the expression of tumor-related genes and cytokines | [170,171,172,173,174] | |
| Listeria Monocytogenes LADD strain | Cervical, Oropharyngeal, Pancreatic, Lung and Mesothelioma | Targeted transport vector | [175,176,177] | |
| Escherichia Coli | - | Targeted transport vector | [178,179] | |
| Tumor Promoters | Gram-Negative Bacteria | Liver Colorectal | TLR2 and TLR4 mediated upregulation of Innate inflammation; Induction of IL-17/23 pathway cytokines | [180,181,182] |
| Helicobacter Pylori | Gastric | Inducing inflammation | [149,183,184] | |
| Clostridium species | HCC | Production of deoxycholic acid from bile and inducing inflammation | [185] | |
| Enterotoxigenic Bacteroides Fragilis | Colon | Inducing inflammation | [186] | |
| Fusobacterium | Colorectal | Inducing inflammation and protecting tumors from an immune cell attack | [187,188,189] | |
| Escherichia Coli | Colorectal | PKS inducing DNA breaks | [190] | |
| Chlamydia Pneumoniae | Lung | C. pneumoniae protein interfering with host cell behavior | [191] | |
| Chlamydia Trachomatis | Cervical, Ovarian | Promotes host cell DNA double-strand breaks, induces host cell genome instability and even transformation | [192] |
6. Future Directions/Perspectives
6.1. Top-Down MS
6.2. Differential Ion Mobility MS (DMS)
6.3. Imaging Mass Cytometry
6.4. Microarrays
6.5. Big Data, Artificial Intelligence (AI), Machine and Deep Learning
6.6. Single-Cell Proteomics
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2-DE | Two-dimensional gel electrophoresis |
| AI | Artificial Intelligence |
| AML | Acute myeloid leukemia |
| APOAII | Apolipoprotein-AII |
| APOLLO | Applied Proteogenomics Organizational Learning and Outcomes |
| ART | Antibiotic resistance |
| AS | Alternative splicing |
| AST | Antibiotic susceptibility testing |
| CAF | Cancer-associated fibroblasts |
| CASK | Calcium/calmodulin-dependent serine protein kinase |
| CCA | Cholangiocarcinoma |
| CD | Crohn’s disease |
| CDI | Clostridium difficile infection |
| CEMIP | Cell migration-inducing and hyaluronan-binding protein |
| CHAT | Cancer Hallmarks Analytics Tool |
| CPTAC | Clinical Proteomic Tumor Analysis Consortium |
| CRC | Colorectal cancer |
| DAVID | The Database for Annotation, Visualization and Integrated Discovery |
| DDA | Data dependent analysis |
| DIA | Data independent analysis |
| DMS | Differential ion mobility MS |
| EDRN | Early Detection Research Network |
| EV | Extracellular vesicles |
| FASN | Fatty acid synthase |
| FFPE | Formalin-fixed paraffin-embedded |
| FMT | Fecal microbiota transplantation |
| Fn | Fusobacterium nucleatum |
| GELFREE | Gel-eluted liquid fractionation entrapment electrophoresis |
| HCC | Hepatocellular carcinoma |
| HGSC | High-grade serous ovarian carcinoma |
| HoC | Hallmarks of Cancer |
| hPOP | Human Personal Omics Profiling |
| HPP | Human Proteomics Project |
| HPRL | Proteome Reference Library |
| hTERT | Human telomerase reverse transcriptase |
| HUPO | Human Protein Organisation |
| IARC | International Agency for Research on Cancer |
| IBD | Inflammatory bowel disease |
| ICP | Inductively coupled argon plasma |
| ICPC | International Cancer Proteogenome Consortium |
| IHC | Immunohistochemistry |
| IPA | Ingenuity pathway analysis |
| iTRAQ | Isobaric tagging for multiplexed relative and absolute protein quantitation |
| LOXL-4 | Lysyl oxidase-like 4 |
| LPS | Lipopolysaccharide |
| LRG | Leucine-rich α-2 glycoprotein |
| LUAD | Lung adenocarcinoma |
| MAT2A | Methionine adenosyltransferase 2a |
| mdCAT | Mass-defect-based carbonyl activated tag |
| MDD | Major depressive disorder |
| MRM | Multiple reaction monitoring |
| MudPIT | Multidimensional protein identification technology |
| NNMT | Nicotinamide N-methyltransferase |
| NSCLC | Non-small cell lung cancer |
| NTME | Nontumor microenvironment |
| OS | Overall survival |
| PCT | Pressure cycling technology |
| PDAC | Pancreatic ductal adenocarcinoma |
| PHOENIX | The Chinese Pilot Hub Of Encyclopedic Proteomix |
| PPTT | Plasmonic photothermal therapy |
| PRM | Parallel reaction monitoring |
| PTMs | Posttranslational modifications |
| QSOX1 | Quiescin sulfhydryl oxidase 1 |
| QTOF | Quadrupole time-of-flight |
| RA | Rheumatoid arthritis |
| RBFOX3 | RNA binding protein fox-1 homolog 3 |
| ROS | Reactive oxygen species |
| RPMAs | Reverse phase microarrays |
| RPPA | Reverse-phase protein microarray |
| RFS | Recurrence-free survival |
| SAPs | Single amino acid polymorphisms |
| SCFAs | short-chain fatty acids |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SERS | Surface-enhanced Raman scattering |
| SILAC | Stable isotope labeling by/with amino acids in cell culture |
| SRM | Selective reaction monitoring |
| suPAR | Soluble urokinase-type plasminogen activator receptor |
| SWATH-MS | Sequential window acquisition of all theoretical mass spectra |
| TCGA | The Cancer Genome Atlas |
| TME | Tumor microenvironment |
| TMT | Tandem mass tag |
| TNBC | Triple negative breast cancer |
| TOTT | The Oncology Think Tank |
| uPAR | Urokinase plasminogen activator receptor |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Mitsudomi, T.; Kosaka, T.; Endoh, H.; Horio, Y.; Hida, T.; Mori, S.; Hatooka, S.; Shinoda, M.; Takahashi, T.; Yatabe, Y. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J. Clin. Oncol. 2005, 23, 2513–2520. [Google Scholar] [CrossRef] [PubMed]
- Middleton, J.; Stover, D.; Hai, T. Chemotherapy-exacerbated breast cancer metastasis: A paradox explainable by dysregulated adaptive-response. Int. J. Mol. Sci. 2018, 19, 3333. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, G.; Pastoriza, J.; Wang, Y.; Harney, A.; Entenberg, D.; Pignatelli, J.; Sharma, V.; Xue, E.; Cheng, E.; D’Alfonso, T.; et al. Neoadjuvant chemotherapy induces breast cancer metastasis through a tmem-mediated mechanism. Sci. Transl. Med. 2017, 9, eaan0026. [Google Scholar] [CrossRef]
- Ming, H.; Li, B.; Zhou, L.; Goel, A.; Huang, C. Long non-coding rnas and cancer metastasis: Molecular basis and therapeutic implications. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188519. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhou, X.; Yang, J.; Liu, X.; Zhao, X.; Wang, Q.; Han, L.; Song, X.; Zhu, Z.; Tian, W.; et al. Ac1mmyr2 impairs high dose paclitaxel-induced tumor metastasis by targeting mir-21/cdk5 axis. Cancer Lett. 2015, 362, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Nice, E. The status of proteomics as we enter the 2020s: Towards personalised/precision medicine. Anal. Biochem. 2020, 113840. [Google Scholar] [CrossRef]
- Lander, E.; Linton, L.; Birren, B.; Nusbaum, C.; Zody, M.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Venter, J.; Adams, M.; Myers, E.; Li, P.; Mural, R.; Sutton, G.; Smith, H.; Yandell, M.; Evans, C.; Holt, R.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef]
- Adhikari, S.; Nice, E.; Deutsch, E.; Lane, L.; Omenn, G.; Pennington, S.; Paik, Y.; Overall, C.; Corrales, F.; Cristea, I.; et al. A high-stringency blueprint of the human proteome. Nat. Commun. 2020, 11, 5301. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Nice, E. The separation sciences, the front end to proteomics: An historical perspective. Biomed. Chromatogr. 2021, 35, e4995. [Google Scholar] [CrossRef]
- Omenn, G.; Lane, L.; Overall, C.; Cristea, I.; Corrales, F.; Lindskog, C.; Paik, Y.; Van Eyk, J.; Liu, S.; Pennington, S.; et al. Research on the human proteome reaches a major milestone: >90% of predicted human proteins now credibly detected, according to the hupo human proteome project. J. Proteome Res. 2020, 19, 4735–4746. [Google Scholar] [CrossRef] [PubMed]
- Kolch, W.; Mischak, H.; Pitt, A. The molecular make-up of a tumour: Proteomics in cancer research. Clin. Sci. 2005, 108, 369–383. [Google Scholar] [CrossRef]
- Ponomarenko, E.; Poverennaya, E.; Ilgisonis, E.; Pyatnitskiy, M.; Kopylov, A.; Zgoda, V.; Lisitsa, A.; Archakov, A. The size of the human proteome: The width and depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Wu, Y.; Han, T.; Zang, X.; Xiao, H.; Tang, Y.; Wu, R.; Fernández, F.; El-Sayed, M. Simultaneous time-dependent surface-enhanced raman spectroscopy, metabolomics, and proteomics reveal cancer cell death mechanisms associated with gold nanorod photothermal therapy. J. Am. Chem. Soc. 2016, 138, 15434–15442. [Google Scholar] [CrossRef]
- Wasinger, V.; Cordwell, S.; Cerpa-Poljak, A.; Yan, J.; Gooley, A.; Wilkins, M.; Duncan, M.; Harris, R.; Williams, K.; Humphery-Smith, I. Progress with gene-product mapping of the mollicutes: Mycoplasma genitalium. Electrophoresis 1995, 16, 1090–1094. [Google Scholar] [CrossRef]
- The Financial Times. Proteomics: Searching for the Real Stuff of Life. The Financial Times, 2021; 14. [Google Scholar]
- Al-Wajeeh, A.; Salhimi, S.; Al-Mansoub, M.; Khalid, I.; Harvey, T.; Latiff, A.; Ismail, M. Comparative proteomic analysis of different stages of breast cancer tissues using ultra high performance liquid chromatography tandem mass spectrometer. PLoS ONE 2020, 15, e0227404. [Google Scholar] [CrossRef]
- Nice, E.; Rothacker, J.; Weinstock, J.; Lim, L.; Catimel, B. Use of multidimensional separation protocols for the purification of trace components in complex biological samples for proteomics analysis. J. Chromatogr. A 2007, 1168, 190–210. [Google Scholar] [CrossRef]
- Duong, V.; Park, J.; Lee, H. Review of three-dimensional liquid chromatography platforms for bottom-up proteomics. Int. J. Mol. Sci. 2020, 21, 1524. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Jiang, B.; Zhao, B.; Zhang, L.; Zhang, Y. Recent advances in multidimensional separation for proteome analysis. Anal. Chem. 2019, 91, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Kota, U.; Stolowitz, M.L. Improving Proteome Coverage by Reducing Sample Complexity via Chromatography. Adv Exp Med Biol. 2016, 919, 83–143. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Poljak, A.; Ali, S.; Zhong, L.; Raftery, M.; Sachdev, P. Extending the depth of human plasma proteome coverage using simple fractionation techniques. J. Proteome Res. 2021, 20, 1261–1279. [Google Scholar] [CrossRef]
- Ahn, S.; Sharma, S.; Mohamedali, A.; Mahboob, S.; Redmond, W.; Pascovici, D.; Wu, J.; Zaw, T.; Adhikari, S.; Vaibhav, V.; et al. Potential early clinical stage colorectal cancer diagnosis using a proteomics blood test panel. Clin. Proteom. 2019, 16, 34. [Google Scholar] [CrossRef]
- Riley, N.; Hebert, A.; Coon, J. Proteomics moves into the fast lane. Cell Syst. 2016, 2, 142–143. [Google Scholar] [CrossRef] [PubMed]
- Borràs, E.; Sabidó, E. What is targeted proteomics? A concise revision of targeted acquisition and targeted data analysis in mass spectrometry. Proteomics 2017, 17, 1700180. [Google Scholar] [CrossRef]
- Kusebauch, U.; Campbell, D.; Deutsch, E.; Chu, C.; Spicer, D.; Brusniak, M.; Slagel, J.; Sun, Z.; Stevens, J.; Grimes, B.; et al. Human srmatlas: A resource of targeted assays to quantify the complete human proteome. Cell 2016, 166, 766–778. [Google Scholar] [CrossRef]
- Doerr, A. Dia mass spectrometry. Nat. Methods 2015, 12, 35. [Google Scholar] [CrossRef]
- Ludwig, C.; Gillet, L.; Rosenberger, G.; Amon, S.; Collins, B.; Aebersold, R. Data-independent acquisition-based swath-ms for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2018, 14, e8126. [Google Scholar] [CrossRef]
- Collins, B.; Hunter, C.; Liu, Y.; Schilling, B.; Rosenberger, G.; Bader, S.; Chan, D.; Gibson, B.; Gingras, A.; Held, J.; et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of swath-mass spectrometry. Nat. Commun. 2017, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Hood, B.; Bateman, N.; Conrads, T. Quantitative mass spectrometry by isotope dilution and multiple reaction monitoring (mrm). Methods Mol. Biol. 2017, 1606, 313–332. [Google Scholar] [CrossRef] [PubMed]
- Ronsein, G.; Pamir, N.; von Haller, P.; Kim, D.; Oda, M.; Jarvik, G.; Vaisar, T.; Heinecke, J. Parallel reaction monitoring (prm) and selected reaction monitoring (srm) exhibit comparable linearity, dynamic range and precision for targeted quantitative hdl proteomics. J. Proteom. 2015, 113, 388–399. [Google Scholar] [CrossRef]
- Bourmaud, A.; Gallien, S.; Domon, B. Parallel reaction monitoring using quadrupole-orbitrap mass spectrometer: Principle and applications. Proteomics 2016, 16, 2146–2159. [Google Scholar] [CrossRef] [PubMed]
- Gallien, S.; Bourmaud, A.; Kim, S.; Domon, B. Technical considerations for large-scale parallel reaction monitoring analysis. J. Proteom. 2014, 100, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Núñez, E.; Domont, G.; Nogueira, F. Itraq-based shotgun proteomics approach for relative protein quantification. Methods Mol. Biol. 2017, 1546, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Samuel, M.; Ang, C.; Keerthikumar, S.; Mathivanan, S. Label-based and label-free strategies for protein quantitation. Methods Mol. Biol. 2017, 1549, 31–43. [Google Scholar] [CrossRef]
- Gillet, L.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the ms/ms spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012, 11, O111-016717. [Google Scholar] [CrossRef]
- Whitman, J.; Lynch, K. Optimization and comparison of information-dependent acquisition (ida) to sequential window acquisition of all theoretical fragment ion spectra (swath) for high-resolution mass spectrometry in clinical toxicology. Clin. Chem. 2019, 65, 862–870. [Google Scholar] [CrossRef]
- Monroe, A.; Zhang, H.; Schunter, C.; Ravasi, T. Probing swath-ms as a tool for proteome level quantification in a nonmodel fish. Mol. Ecol. Resour. 2020, 20, 1647–1657. [Google Scholar] [CrossRef]
- Prudova, A.; Gocheva, V.; Auf dem Keller, U.; Eckhard, U.; Olson, O.; Akkari, L.; Butler, G.; Fortelny, N.; Lange, P.; Mark, J.; et al. Tails n-terminomics and proteomics show protein degradation dominates over proteolytic processing by cathepsins in pancreatic tumors. Cell Rep. 2016, 16, 1762–1773. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Howe, G.; Harrison, L.; Miller, A. A study of repeatability of dietary data over a seven-year period. Am. J. Epidemiol. 1989, 129, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Serada, S.; Fujimoto, M.; Ogata, A.; Terabe, F.; Hirano, T.; Iijima, H.; Shinzaki, S.; Nishikawa, T.; Ohkawara, T.; Iwahori, K.; et al. Itraq-based proteomic identification of leucine-rich alpha-2 glycoprotein as a novel inflammatory biomarker in autoimmune diseases. Ann. Rheum. Dis. 2010, 69, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, R.; Tang, Q.; Yu, Y.; Huang, Q.; Chen, Y.; Wang, G.; Wang, X. Leucine-rich alpha-2-glycoprotein-1 is up-regulated in colorectal cancer and is a tumor promoter. OncoTargets Ther. 2018, 11, 2745–2752. [Google Scholar] [CrossRef]
- Zhang, J.; Kim, S.; Li, L.; Kemp, C.; Jiang, C.; Lü, J. Proteomic and transcriptomic profiling of pten gene-knockout mouse model of prostate cancer. Prostate 2020, 80, 588–605. [Google Scholar] [CrossRef]
- Lin, P.; Yao, Z.; Sun, Y.; Li, W.; Liu, Y.; Liang, K.; Liu, Y.; Qin, J.; Hou, X.; Chen, L. Deciphering novel biomarkers of lymph node metastasis of thyroid papillary microcarcinoma using proteomic analysis of ultrasound-guided fine-needle aspiration biopsy samples. J. Proteom. 2019, 204, 103414. [Google Scholar] [CrossRef]
- Zhang, S.; Di, Y.; Yao, J.; Wang, Y.; Shu, H.; Yan, G.; Zhang, L.; Lu, H. Mass defect-based carbonyl activated tags (mdcats) for multiplex data-independent acquisition proteome quantification. Chem. Commun. 2021, 57, 737–740. [Google Scholar] [CrossRef]
- Kumar, V.; Ray, S.; Ghantasala, S.; Srivastava, S. An integrated quantitative proteomics workflow for cancer biomarker discovery and validation in plasma. Front. Oncol. 2020, 10, 543997. [Google Scholar] [CrossRef]
- Chen, X.; Wei, S.; Ji, Y.; Guo, X.; Yang, F. Quantitative proteomics using silac: Principles, applications, and developments. Proteomics 2015, 15, 3175–3192. [Google Scholar] [CrossRef]
- Shenoy, A.; Geiger, T. Super-silac: Current trends and future perspectives. Expert Rev. Proteom. 2015, 12, 13–19. [Google Scholar] [CrossRef]
- Cuomo, A.; Moretti, S.; Minucci, S.; Bonaldi, T. Silac-based proteomic analysis to dissect the "histone modification signature" of human breast cancer cells. Amino Acids 2011, 41, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Ahrné, E.; Baron, A.; Glatter, T.; Fava, L.; Santamaria, A.; Nigg, E.; Schmidt, A. Evaluation of data-dependent and -independent mass spectrometric workflows for sensitive quantification of proteins and phosphorylation sites. J. Proteome Res. 2014, 13, 5973–5988. [Google Scholar] [CrossRef] [PubMed]
- Whiteaker, J.; Zhao, L.; Yan, P.; Ivey, R.; Voytovich, U.; Moore, H.; Lin, C.; Paulovich, A. Peptide immunoaffinity enrichment and targeted mass spectrometry enables multiplex, quantitative pharmacodynamic studies of phospho-signaling. Mol. Cell. Proteom. 2015, 14, 2261–2273. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.; Froehlich, B.; Aguilar-Mahecha, A.; Aloyz, R.; Poetz, O.; Basik, M.; Batist, G.; Zahedi, R.; Borchers, C. Using two peptide isotopologues as internal standards for the streamlined quantification of low-abundance proteins by immuno-mrm and immuno-maldi. Anal. Chem. 2020, 92, 12407–12414. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Somasundaram, K. Targeted proteomics comes to the benchside and the bedside: Is it ready for us? BioEssays 2019, 41, e1800042. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Whiteaker, J.; Hoofnagle, A.; Baird, G.; Rodland, K.; Paulovich, A. Clinical potential of mass spectrometry-based proteogenomics. Nat. Rev. Clin. Oncol. 2019, 16, 256–268. [Google Scholar] [CrossRef]
- Do, M.; Kim, H.; Yeo, I.; Lee, J.; Park, I.; Ryu, H.; Kim, Y. Clinical application of multiple reaction monitoring-mass spectrometry to human epidermal growth factor receptor 2 measurements as a potential diagnostic tool for breast cancer therapy. Clin. Chem. 2020, 66, 1339–1348. [Google Scholar] [CrossRef]
- Ang, C.; Rothacker, J.; Patsiouras, H.; Gibbs, P.; Burgess, A.; Nice, E. Use of multiple reaction monitoring for multiplex analysis of colorectal cancer-associated proteins in human feces. Electrophoresis 2011, 32, 1926–1938. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Y.; Liu, W.; Xing, S.; Wang, D.; Chen, J.; Sun, L.; Mu, J.; Liu, W.; Xing, B.; et al. Identification of noninvasive diagnostic biomarkers for hepatocellular carcinoma by urinary proteomics. J. Proteom. 2020, 225, 103780. [Google Scholar] [CrossRef]
- Patel, V.; Thalassinos, K.; Slade, S.; Connolly, J.; Crombie, A.; Murrell, J.; Scrivens, J. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J. Proteome Res. 2009, 8, 3752–3759. [Google Scholar] [CrossRef]
- Tan, H.; Wang, N.; Zhang, C.; Chan, Y.; Yuen, M.; Feng, Y. Loxl4 fosters an immunosuppressive microenvironment during hepatocarcinogenesis. Hepatology 2020. [Google Scholar] [CrossRef]
- Guo, T.; Kouvonen, P.; Koh, C.; Gillet, L.; Wolski, W.; Röst, H.; Rosenberger, G.; Collins, B.; Blum, L.; Gillessen, S.; et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat. Med. 2015, 21, 407–413. [Google Scholar] [CrossRef]
- Powell, B.; Lazarev, A.; Carlson, G.; Ivanov, A.; Rozak, D. Pressure cycling technology in systems biology. Methods Mol. Biol. 2012, 881, 27–62. [Google Scholar] [CrossRef] [PubMed]
- Hallal, S.; Azimi, A.; Wei, H.; Ho, N.; Lee, M.; Sim, H.; Sy, J.; Shivalingam, B.; Buckland, M.; Alexander-Kaufman, K. A comprehensive proteomic swath-ms workflow for profiling blood extracellular vesicles: A new avenue for glioma tumour surveillance. Int. J. Mol. Sci. 2020, 21, 4754. [Google Scholar] [CrossRef] [PubMed]
- Sahni, S.; Krisp, C.; Molloy, M.; Nahm, C.; Maloney, S.; Gillson, J.; Gill, A.; Samra, J.; Mittal, A. Psmd11, ptprm and ptprb as novel biomarkers of pancreatic cancer progression. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129682. [Google Scholar] [CrossRef]
- Del Pilar Chantada-Vázquez, M.; López, A.; Vence, M.; Vázquez-Estévez, S.; Acea-Nebril, B.; Calatayud, D.; Jardiel, T.; Bravo, S.; Núñez, C. Proteomic investigation on bio-corona of au, ag and fe nanoparticles for the discovery of triple negative breast cancer serum protein biomarkers. J. Proteom. 2020, 212, 103581. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Yan, Y.; Caruso, F.; Nice, E. Emerging techniques in proteomics for probing nano-bio interactions. ACS Nano 2012, 6, 10438–10448. [Google Scholar] [CrossRef]
- Zhang, F.; Ge, W.; Ruan, G.; Cai, X.; Guo, T. Data-independent acquisition mass spectrometry-based proteomics and software tools: A glimpse in 2020. Proteomics 2020, 20, e1900276. [Google Scholar] [CrossRef]
- Midha, M.; Campbell, D.; Kapil, C.; Kusebauch, U.; Hoopmann, M.; Bader, S.; Moritz, R. Dialib-qc an assessment tool for spectral libraries in data-independent acquisition proteomics. Nat. Commun. 2020, 11, 5251. [Google Scholar] [CrossRef]
- Agus, D.; Jaffee, E.; Van Dang, C. Cancer moonshot 2.0. Lancet Oncol. 2021, 22, 164–165. [Google Scholar] [CrossRef]
- Rodriguez, H.; Pennington, S. Revolutionizing precision oncology through collaborative proteogenomics and data sharing. Cell 2018, 173, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Boehm, J.; Garnett, M.; Adams, D.; Francies, H.; Golub, T.; Hahn, W.; Iorio, F.; McFarland, J.; Parts, L.; Vazquez, F. Cancer research needs a better map. Nature 2021, 589, 514–516. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A. Proteogenomics: Concepts, applications and computational strategies. Nat. Methods 2014, 11, 1114–1125. [Google Scholar] [CrossRef]
- Gillette, M.; Satpathy, S.; Cao, S.; Dhanasekaran, S.; Vasaikar, S.; Krug, K.; Petralia, F.; Li, Y.; Liang, W.; Reva, B.; et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell 2020, 182, 200–225. [Google Scholar] [CrossRef] [PubMed]
- Petralia, F.; Tignor, N.; Reva, B.; Koptyra, M.; Chowdhury, S.; Rykunov, D.; Krek, A.; Ma, W.; Zhu, Y.; Ji, J.; et al. Integrated proteogenomic characterization across major histological types of pediatric brain cancer. Cell 2020, 183, 1962–1985. [Google Scholar] [CrossRef] [PubMed]
- Krug, K.; Jaehnig, E.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.; et al. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell 2020, 183, 1436–1456. [Google Scholar] [CrossRef]
- Edwards, N.; Oberti, M.; Thangudu, R.; Cai, S.; McGarvey, P.; Jacob, S.; Madhavan, S.; Ketchum, K. The cptac data portal: A resource for cancer proteomics research. J. Proteome Res. 2015, 14, 2707–2713. [Google Scholar] [CrossRef] [PubMed]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. Linkedomics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2017, 46, D956–D963. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, C.; Wang, X.; Zhai, L.; Ma, Y.; Mao, Y.; Qian, K.; Sun, C.; Liu, Z.; Jiang, S.; et al. Integrative proteomic characterization of human lung adenocarcinoma. Cell 2020, 182, 245–261. [Google Scholar] [CrossRef]
- Cristobal, A.; van den Toorn, H.; van de Wetering, M.; Clevers, H.; Heck, A.; Mohammed, S. Personalized proteome profiles of healthy and tumor human colon organoids reveal both individual diversity and basic features of colorectal cancer. Cell Rep. 2017, 18, 263–274. [Google Scholar] [CrossRef]
- Lindhorst, P.; Hummon, A. Proteomics of colorectal cancer: Tumors, organoids, and cell cultures-a minireview. Front. Mol. Biosci. 2020, 7, 604492. [Google Scholar] [CrossRef]
- Abe, Y.; Tada, A.; Isoyama, J.; Nagayama, S.; Yao, R.; Adachi, J.; Tomonaga, T. Improved phosphoproteomic analysis for phosphosignaling and active-kinome profiling in matrigel-embedded spheroids and patient-derived organoids. Sci. Rep. 2018, 8, 11401. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, G.; Zhang, L.; Wang, Q.; Li, H.; Han, Y.; Xie, L.; Yan, Z.; Li, Y.; An, Y.; et al. Comprehensive review of web servers and bioinformatics tools for cancer prognosis analysis. Front. Oncol. 2020, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Andrieux, G.; Ahmed, M.; Chakraborty, S. Integration of online omics-data resources for cancer research. Front. Genet. 2020, 11, 578345. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Cox, J. Perseus: A bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol. Biol. 2018, 1711, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.; Collisson, E.; Mills, G.; Shaw, K.; Ozenberger, B.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.; Ferluga, S.; Sharma, V.; Futschik, M.; Hilton, D.; Adams, C.; Lasonder, E.; Hanemann, C. Proteomic analysis discovers the differential expression of novel proteins and phosphoproteins in meningioma including nek9, hk2 and set and deregulation of rna metabolism. EBioMedicine 2019, 40, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Da, Z.; Gao, L.; Su, G.; Yao, J.; Fu, W.; Zhang, J.; Zhang, X.; Pei, Z.; Yue, P.; Bai, B.; et al. Bioinformatics combined with quantitative proteomics analyses and identification of potential biomarkers in cholangiocarcinoma. Cancer Cell Int. 2020, 20, 130. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Cantor, D.; Nice, E.; Baker, M. Recent findings from the human proteome project: Opening the mass spectrometry toolbox to advance cancer diagnosis, surveillance and treatment. Expert Rev. Proteom. 2015, 12, 279–293. [Google Scholar] [CrossRef]
- He, Y.; Mohamedali, A.; Huang, C.; Baker, M.; Nice, E. Oncoproteomics: Current status and future opportunities. Clin. Chim. Acta 2019, 495, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Kalev, P.; Hyer, M.; Gross, S.; Konteatis, Z.; Chen, C.; Fletcher, M.; Lein, M.; Aguado-Fraile, E.; Frank, V.; Barnett, A.; et al. Mat2a inhibition blocks the growth of mtap-deleted cancer cells by reducing prmt5-dependent mrna splicing and inducing DNA damage. Cancer Cell 2021, 39, 209–224. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, W.; Lu, W.; Chen, M.; Luo, M.; Zhang, C.; Li, Y.; Qin, G.; Shi, D.; Xiao, B.; et al. Rbfox3 promotes tumor growth and progression via htert signaling and predicts a poor prognosis in hepatocellular carcinoma. Theranostics 2017, 7, 3138–3154. [Google Scholar] [CrossRef] [PubMed]
- Stehling, O.; Vashisht, A.; Mascarenhas, J.; Jonsson, Z.; Sharma, T.; Netz, D.; Pierik, A.; Wohlschlegel, J.; Lill, R. Mms19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 2012, 337, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, G.; Hoshino, A.; Kenific, C.; Matei, I.; Steiner, L.; Freitas, D.; Kim, H.; Oxley, P.; Scandariato, I.; Casanova-Salas, I.; et al. Tumour exosomal cemip protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 2019, 21, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Bruning, U.; Morales-Rodriguez, F.; Kalucka, J.; Goveia, J.; Taverna, F.; Queiroz, K.; Dubois, C.; Cantelmo, A.; Chen, R.; Loroch, S.; et al. Impairment of angiogenesis by fatty acid synthase inhibition involves mtor malonylation. Cell Metab. 2018, 28, 866–880. [Google Scholar] [CrossRef]
- Ahn, S.; Mohamedali, A.; Pascovici, D.; Adhikari, S.; Sharma, S.; Nice, E.; Baker, M. Proteomics reveals cell-surface urokinase plasminogen activator receptor expression impacts most hallmarks of cancer. Proteomics 2019, 19, e1900026. [Google Scholar] [CrossRef]
- Yuan, K.; Lei, Y.; Chen, H.; Chen, Y.; Zhang, T.; Li, K.; Xie, N.; Wang, K.; Feng, X.; Pu, Q.; et al. Hbv-induced ros accumulation promotes hepatocarcinogenesis through snail-mediated epigenetic silencing of socs3. Cell Death Differ. 2016, 23, 616–627. [Google Scholar] [CrossRef]
- Xiao, H.; Jedrychowski, M.; Schweppe, D.; Huttlin, E.; Yu, Q.; Heppner, D.; Li, J.; Long, J.; Mills, E.; Szpyt, J.; et al. A quantitative tissue-specific landscape of protein redox regulation during aging. Cell 2020, 180, 968–983. [Google Scholar] [CrossRef]
- Bryant, K.; Stalnecker, C.; Zeitouni, D.; Klomp, J.; Peng, S.; Tikunov, A.; Gunda, V.; Pierobon, M.; Waters, A.; George, S.; et al. Combination of erk and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 2019, 25, 628–640. [Google Scholar] [CrossRef]
- Ambartsumian, N.; Klingelhöfer, J.; Grigorian, M. The multifaceted s100a4 protein in cancer and inflammation. Methods Mol. Biol. 2019, 1929, 339–365. [Google Scholar] [CrossRef] [PubMed]
- Ghahremanifard, P.; Chanda, A.; Bonni, S.; Bose, P. Tgf-β mediated immune evasion in cancer-spotlight on cancer-associated fibroblasts. Cancers 2020, 12, 3650. [Google Scholar] [CrossRef] [PubMed]
- Jafri, M.; Ansari, S.; Alqahtani, M.; Shay, J. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Gao, W.; Su, M.; Nice, E.C.; Zhang, W.; Lin, J.; Xie, N. Adaptive mechanisms of tumor therapy resistance driven by tumor microenvironment. Front. Cell Dev. Biol. 2021, 9, 357. [Google Scholar] [CrossRef]
- Chew, V.; Lai, L.; Pan, L.; Lim, C.; Li, J.; Ong, R.; Chua, C.; Leong, J.; Lim, K.; Toh, H.; et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc. Natl. Acad. Sci. USA 2017, 114, E5900–E5909. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.; Zhang, B.; McDermott, J.; Zhou, J.; Petyuk, V.; Chen, L.; Ray, D.; et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Eckert, M.; Coscia, F.; Chryplewicz, A.; Chang, J.; Hernandez, K.; Pan, S.; Tienda, S.; Nahotko, D.; Li, G.; Blaženović, I.; et al. Proteomics reveals nnmt as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019, 569, 723–728. [Google Scholar] [CrossRef]
- Alanazi, B.; Munje, C.; Rastogi, N.; Williamson, A.; Taylor, S.; Hole, P.; Hodges, M.; Doyle, M.; Baker, S.; Gilkes, A.; et al. Integrated nuclear proteomics and transcriptomics identifies s100a4 as a therapeutic target in acute myeloid leukemia. Leukemia 2020, 34, 427–440. [Google Scholar] [CrossRef]
- Holmström, K.; Finkel, T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 411–421. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, L.; Zhou, L.; Lei, Y.; Zhang, Y.; Huang, C. Redox signaling and unfolded protein response coordinate cell fate decisions under er stress. Redox Biol. 2019, 25, 101047. [Google Scholar] [CrossRef]
- Jia, M.; Qin, D.; Zhao, C.; Chai, L.; Yu, Z.; Wang, W.; Tong, L.; Lv, L.; Wang, Y.; Rehwinkel, J.; et al. Redox homeostasis maintained by gpx4 facilitates sting activation. Nat. Immunol. 2020, 21, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Helfinger, V.; Schröder, K. Redox control in cancer development and progression. Mol. Asp. Med. 2018, 63, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.-W. Cancer a redox disease: Cancer cells are universally disturbed in their electronic energy balance, an understanding that potentially revolutionises cancer therapy and prevention. J. Australas. Coll. Nutr. Environ. Med. 2013, 32, 12. [Google Scholar]
- Bachi, A.; Dalle-Donne, I.; Scaloni, A. Redox proteomics: Chemical principles, methodological approaches and biological/biomedical promises. Chem. Rev. 2013, 113, 596–698. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Vats, S.; Chia, A.; Tan, T.; Deng, S.; Ong, M.; Arfuso, F.; Yap, C.; Goh, B.; Sethi, G.; et al. Dual role of autophagy in hallmarks of cancer. Oncogene 2018, 37, 1142–1158. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T.; Komatsu, M. Autophagy in the liver: Functions in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 170–184. [Google Scholar] [CrossRef]
- Califf, R. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of pancreatic cancer: Global trends, etiology and risk factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef]
- Shen, J.; Person, M.; Zhu, J.; Abbruzzese, J.; Li, D. Protein expression profiles in pancreatic adenocarcinoma compared with normal pancreatic tissue and tissue affected by pancreatitis as detected by two-dimensional gel electrophoresis and mass spectrometry. Cancer Res. 2004, 64, 9018–9026. [Google Scholar] [CrossRef]
- Honda, K.; Okusaka, T.; Felix, K.; Nakamori, S.; Sata, N.; Nagai, H.; Ioka, T.; Tsuchida, A.; Shimahara, T.; Shimahara, M.; et al. Altered plasma apolipoprotein modifications in patients with pancreatic cancer: Protein characterization and multi-institutional validation. PLoS ONE 2012, 7, e46908. [Google Scholar] [CrossRef]
- Honda, K.; Kobayashi, M.; Okusaka, T.; Rinaudo, J.; Huang, Y.; Marsh, T.; Sanada, M.; Sasajima, Y.; Nakamori, S.; Shimahara, M.; et al. Plasma biomarker for detection of early stage pancreatic cancer and risk factors for pancreatic malignancy using antibodies for apolipoprotein-aii isoforms. Sci. Rep. 2015, 5, 15921. [Google Scholar] [CrossRef]
- Alves Martins, B.; de Bulhões, G.; Cavalcanti, I.; Martins, M.; de Oliveira, P.; Martins, A. Biomarkers in colorectal cancer: The role of translational proteomics research. Front. Oncol. 2019, 9, 1284. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Toiyama, Y.; Otake, K.; Ide, S.; Imaoka, H.; Okigami, M.; Okugawa, Y.; Fujikawa, H.; Saigusa, S.; Hiro, J.; et al. Successful identification of a predictive biomarker for lymph node metastasis in colorectal cancer using a proteomic approach. Oncotarget 2017, 8, 106935–106947. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wei, K.; Yu, H.; Jin, D.; Wang, G.; Yu, B. Prognostic value of ezrin in various cancers: A systematic review and updated meta-analysis. Sci. Rep. 2015, 5, 17903. [Google Scholar] [CrossRef] [PubMed]
- Aikawa, A.; Fujita, H.; Kosaka, T.; Minato, H.; Kiyokawa, E. Clinicopathological significance of heterogeneic ezrin expression in poorly differentiated clusters of colorectal cancers. Cancer Sci. 2019, 110, 2667–2675. [Google Scholar] [CrossRef] [PubMed]
- Pagliara, V.; Donadio, G.; De Tommasi, N.; Amodio, G.; Remondelli, P.; Moltedo, O.; Dal Piaz, F. Bioactive ent-kaurane diterpenes oridonin and irudonin prevent cancer cells migration by interacting with the actin cytoskeleton controller ezrin. Int. J. Mol. Sci. 2020, 21, 7186. [Google Scholar] [CrossRef]
- Sung, H.; Ahn, J.; Yoon, Y.; Na, S.; Choi, Y.; Kim, Y.; Lee, S.; Lee, E.; Cho, S.; Cho, J. Quiescin sulfhydryl oxidase 1 (qsox1) secreted by lung cancer cells promotes cancer metastasis. Int. J. Mol. Sci. 2018, 19, 3213. [Google Scholar] [CrossRef]
- Dietrich, S.; Oleś, M.; Lu, J.; Sellner, L.; Anders, S.; Velten, B.; Wu, B.; Hüllein, J.; da Silva Liberio, M.; Walther, T.; et al. Drug-perturbation-based stratification of blood cancer. J. Clin. Investig. 2018, 128, 427–445. [Google Scholar] [CrossRef]
- Li, B.; Jiang, J.; Assaraf, Y.; Xiao, H.; Chen, Z.; Huang, C. Surmounting cancer drug resistance: New insights from the perspective of n-methyladenosine rna modification. Drug Resist. Updates 2020, 53, 100720. [Google Scholar] [CrossRef]
- Talevi, A. Multi-target pharmacology: Possibilities and limitations of the "skeleton key approach" from a medicinal chemist perspective. Front. Pharmacol. 2015, 6, 205. [Google Scholar] [CrossRef]
- Coscia, F.; Lengyel, E.; Duraiswamy, J.; Ashcroft, B.; Bassani-Sternberg, M.; Wierer, M.; Johnson, A.; Wroblewski, K.; Montag, A.; Yamada, S.; et al. Multi-level proteomics identifies ct45 as a chemosensitivity mediator and immunotherapy target in ovarian cancer. Cell 2018, 175, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, J.; Chen, M.; Luo, Y.; Ju, Z.; Nesser, N.; Johnson-Camacho, K.; Boniface, C.; Lawrence, Y.; Pande, N.; et al. Large-scale characterization of drug responses of clinically relevant proteins in cancer cell lines. Cancer Cell 2020, 38, 829–843. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhai, X.; Li, X.; Zhong, C.; Guo, C.; Yang, F.; Yuan, Y.; Zheng, S. Identification of mst1 as a potential early detection biomarker for colorectal cancer through a proteomic approach. Sci. Rep. 2017, 7, 14265. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Deng, X.; Zhong, J.; Yuan, L.; Tao, X.; Zhang, S.; Zeng, Y.; He, G.; Tan, P.; Tao, Y. Prognostic value of biomarkers epcam and αb-crystallin associated with lymphatic metastasis in breast cancer by itraq analysis. BMC Cancer 2019, 19, 831. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Shen, J.; Mannoor, K.; Guarnera, M.; Jiang, F. Identification of eno1 as a potential sputum biomarker for early-stage lung cancer by shotgun proteomics. Clin. Lung Cancer 2014, 15, 372–378. [Google Scholar] [CrossRef]
- Dai, L.; Qu, Y.; Li, J.; Wang, X.; Wang, K.; Wang, P.; Jiang, B.; Zhang, J. Serological proteome analysis approach-based identification of eno1 as a tumor-associated antigen and its autoantibody could enhance the sensitivity of cea and cyfra 21-1 in the detection of non-small cell lung cancer. Oncotarget 2017, 8, 36664–36673. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Xu, W.; Xu, J.; Shi, Q.; Li, J.; Weng, Y.; Jiang, Z.; Feng, L.; Wang, X.; Zhou, J.; et al. Enolase 1 stimulates glycolysis to promote chemoresistance in gastric cancer. Oncotarget 2017, 8, 47691–47708. [Google Scholar] [CrossRef]
- Capello, M.; Ferri-Borgogno, S.; Riganti, C.; Chattaragada, M.; Principe, M.; Roux, C.; Zhou, W.; Petricoin, E.; Cappello, P.; Novelli, F. Targeting the warburg effect in cancer cells through eno1 knockdown rescues oxidative phosphorylation and induces growth arrest. Oncotarget 2016, 7, 5598–5612. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.; Rizza, P.; Donà, A.; Nigro, A.; Ricci, E.; Fiorillo, M.; Perrotta, I.; Lanzino, M.; Giordano, C.; Bonofiglio, D.; et al. Foxo3a as a positive prognostic marker and a therapeutic target in tamoxifen-resistant breast cancer. Cancers 2019, 11, 1858. [Google Scholar] [CrossRef]
- Lin, H.; He, Q.; Shi, L.; Sleeman, M.; Baker, M.; Nice, E. Proteomics and the microbiome: Pitfalls and potential. Expert Rev. Proteom. 2019, 16, 501–511. [Google Scholar] [CrossRef]
- Qiu, Q.; Lin, Y.; Ma, Y.; Li, X.; Liang, J.; Chen, Z.; Liu, K.; Huang, Y.; Luo, H.; Huang, R.; et al. Exploring the emerging role of the gut microbiota and tumor microenvironment in cancer immunotherapy. Front. Immunol. 2020, 11, 612202. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
- Rubinstein, M.; Baik, J.; Lagana, S.; Han, R.; Raab, W.; Sahoo, D.; Dalerba, P.; Wang, T.; Han, Y. Fusobacterium nucleatum promotes colorectal cancer by inducing wnt/β-catenin modulator annexin a1. EMBO Rep. 2019, 20, e47638. [Google Scholar] [CrossRef] [PubMed]
- Parhi, L.; Alon-Maimon, T.; Sol, A.; Nejman, D.; Shhadeh, A.; Fainsod-Levi, T.; Yajuk, O.; Isaacson, B.; Abed, J.; Maalouf, N.; et al. Breast cancer colonization by fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 2020, 11, 3259. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, M. Bacterial invaders drive crc progression. Sci. Signal. 2020, 13, eabc4218. [Google Scholar] [CrossRef]
- Guo, S.; Chen, J.; Chen, F.; Zeng, Q.; Liu, W.; Zhang, G. Exosomes derived from fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying mir-1246/92b-3p/27a-3p and cxcl16. Gut 2020. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Lagoudas, G.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.; Mazzilli, S.; et al. Commensal microbiota promote lung cancer development via γδ t cells. Cell 2019, 176, 998–1013. [Google Scholar] [CrossRef] [PubMed]
- Díaz, P.; Valenzuela Valderrama, M.; Bravo, J.; Quest, A. Helicobacter pylori and gastric cancer: Adaptive cellular mechanisms involved in disease progression. Front. Microbiol. 2018, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Fujiya, M.; Tanaka, H.; Ueno, N.; Moriichi, K.; Sasajima, J.; Ikuta, K.; Akutsu, H.; Tanabe, H.; Kohgo, Y. Probiotic-derived ferrichrome inhibits colon cancer progression via jnk-mediated apoptosis. Nat. Commun. 2016, 7, 12365. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, K.; Vitetta, L. Effects of intestinal microbial-elaborated butyrate on oncogenic signaling pathways. Nutrients 2019, 11, 1026. [Google Scholar] [CrossRef]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.; Rolain, J.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef]
- Burckhardt, I.; Zimmermann, S. Susceptibility testing of bacteria using maldi-tof mass spectrometry. Front. Microbiol. 2018, 9, 1744. [Google Scholar] [CrossRef] [PubMed]
- Felgner, S.; Kocijancic, D.; Frahm, M.; Weiss, S. Bacteria in cancer therapy: Renaissance of an old concept. Int. J. Microbiol. 2016, 2016, 8451728. [Google Scholar] [CrossRef] [PubMed]
- Droller, M. Intracavitary bacillus calmette-guérin for superficial bladder tumors. J. Urol. 2017, 197, S146–S147. [Google Scholar] [CrossRef]
- Herr, H.; Morales, A. History of bacillus calmette-guerin and bladder cancer: An immunotherapy success story. J. Urol. 2008, 179, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Kamat, A.M.; Hahn, N.M.; Efstathiou, J.A.; Lerner, S.P.; Malmström, P.-U.; Choi, W.; Guo, C.C.; Lotan, Y.; Kassouf, W. Bladder cancer. Lancet 2016, 388, 2796–2810. [Google Scholar] [CrossRef]
- Biot, C.; Rentsch, C.; Gsponer, J.; Birkhäuser, F.; Jusforgues-Saklani, H.; Lemaître, F.; Auriau, C.; Bachmann, A.; Bousso, P.; Demangel, C.; et al. Preexisting bcg-specific t cells improve intravesical immunotherapy for bladder cancer. Sci. Transl. Med. 2012, 4, 137ra172. [Google Scholar] [CrossRef] [PubMed]
- Ohta, N.; Fukase, S.; Watanabe, T.; Ito, T.; Aoyagi, M. Effects and mechanism of ok-432 therapy in various neck cystic lesions. Acta Oto-Laryngol. 2010, 130, 1287–1292. [Google Scholar] [CrossRef] [PubMed]
- Ohta, N.; Fukase, S.; Suzuki, Y.; Ishida, A.; Aoyagi, M. Treatments of various otolaryngological cystic diseases by ok-4321: Its indications and limitations. Laryngoscope 2010, 120, 2193–2196. [Google Scholar] [CrossRef]
- Kono, M.; Satomi, T.; Abukawa, H.; Hasegawa, O.; Watanabe, M.; Chikazu, D. Evaluation of ok-432 injection therapy as possible primary treatment of intraoral ranula. J. Oral Maxillofac. Surg. 2017, 75, 336–342. [Google Scholar] [CrossRef]
- Olivieri, C.; Nanni, L.; De Gaetano, A.M.; Manganaro, L.; Pintus, C. Complete resolution of retroperitoneal lymphangioma with a single trial of ok-432 in an infant. Pediatrics Neonatol. 2016, 57, 240–243. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Paton, A.W.; Morona, R.; Paton, J.C. Bioengineered microbes in disease therapy. Trends Mol. Med. 2012, 18, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, X.; Zeng, X.; Li, L.; Chen, Q.; Li, J. Tumor-targeting bacterial therapy: A potential treatment for oral cancer (review). Oncol. Lett. 2014, 8, 2359–2366. [Google Scholar] [CrossRef] [PubMed]
- Staedtke, V.; Roberts, N.J.; Bai, R.-Y.; Zhou, S. Clostridium novyi-nt in cancer therapy. Genes Dis. 2016, 3, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Bereta, M.; Hayhurst, A.; Gajda, M.; Chorobik, P.; Targosz, M.; Marcinkiewicz, J.; Kaufman, H.L. Improving tumor targeting and therapeutic potential of salmonella vnp20009 by displaying cell surface cea-specific antibodies. Vaccine 2007, 25, 4183–4192. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Winnenthal, F.H.; Hohmann, N.; Schmidt, T.; Podola, L.; Friedrich, T.; Lubenau, H.; Springer, M.; Wieckowski, S.; Breiner, K.M.; Mikus, G.; et al. A phase 1 trial extension to assess immunologic efficacy and safety of prime-boost vaccination with vxm01, an oral t cell vaccine against vegfr2, in patients with advanced pancreatic cancer. OncoImmunology 2018, 7, e1303584. [Google Scholar] [CrossRef]
- Felfoul, O.; Mohammadi, M.; Taherkhani, S.; de Lanauze, D.; Zhong Xu, Y.; Loghin, D.; Essa, S.; Jancik, S.; Houle, D.; Lafleur, M.; et al. Magneto-aerotactic bacteria deliver drug-containing nanoliposomes to tumour hypoxic regions. Nat. Nanotechnol. 2016, 11, 941–947. [Google Scholar] [CrossRef]
- Martel, S. Targeting active cancer cells with smart bullets. Ther. Deliv. 2017, 8, 301–312. [Google Scholar] [CrossRef]
- Lee, J.; Shin, J.; Kim, E.; Kang, H.; Yim, I.; Kim, J.; Joo, H.; Woo, H. Immunomodulatory and antitumor effects in vivo by the cytoplasmic fraction of lactobacillus casei and bifidobacterium longum. J. Vet. Sci. 2004, 5, 41–48. [Google Scholar] [CrossRef]
- Ewaschuk, J.; Diaz, H.; Meddings, L.; Diederichs, B.; Dmytrash, A.; Backer, J.; Looijer-van Langen, M.; Madsen, K. Secreted bioactive factors from bifidobacterium infantis enhance epithelial cell barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, G1025–G1034. [Google Scholar] [CrossRef]
- Bergmann, K.; Liu, S.; Tian, R.; Kushnir, A.; Turner, J.; Li, H.; Chou, P.; Weber, C.; De Plaen, I. Bifidobacteria stabilize claudins at tight junctions and prevent intestinal barrier dysfunction in mouse necrotizing enterocolitis. Am. J. Pathol. 2013, 182, 1595–1606. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.; Aquino-Michaels, K.; Earley, Z.; Benyamin, F.; Lei, Y.; Jabri, B.; Alegre, M.; et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-pd-l1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Uccello, M.; Malaguarnera, G.; Basile, F.; D’agata, V.; Malaguarnera, M.; Bertino, G.; Vacante, M.; Drago, F.; Biondi, A. Potential role of probiotics on colorectal cancer prevention. BMC Surg. 2012, 12, S35. [Google Scholar] [CrossRef] [PubMed]
- Flickinger, J.; Rodeck, U.; Snook, A. Listeria monocytogenes as a vector for cancer immunotherapy: Current understanding and progress. Vaccines 2018, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Tangney, M.; Gahan, C. Listeria monocytogenes as a vector for anti-cancer therapies. Curr. Gene Ther. 2010, 10, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Bolhassani, A.; Naderi, N.; Soleymani, S. Prospects and progress of listeria-based cancer vaccines. Expert Opin. Biol. Ther. 2017, 17, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Yang, H.; Tang, J.; Liu, Z.; Chen, Y.; Lu, B.; He, H.; Tang, S.; Sun, Y.; Liu, F.; et al. Intestinal probiotics e. Coli nissle 1917 as a targeted vehicle for delivery of p53 and tum-5 to solid tumors for cancer therapy. J. Biol. Eng. 2019, 13, 58. [Google Scholar] [CrossRef]
- Parker, W.B.; King, S.A.; Allan, P.W.; Bennett, L.L.; Secrist, J.A.; Montgomery, J.A.; Gilbert, K.S.; Waud, W.R.; Wells, A.H.; Gillespie, G.Y.; et al. In vivo gene therapy of cancer with e. Coli purine nucleoside phosphorylase. Hum. Gene Ther. 1997, 8, 1637–1644. [Google Scholar] [CrossRef]
- Dapito, D.; Mencin, A.; Gwak, G.; Pradere, J.; Jang, M.; Mederacke, I.; Caviglia, J.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and tlr4. Cancer Cell 2012, 21, 504–516. [Google Scholar] [CrossRef]
- Grivennikov, S.; Wang, K.; Mucida, D.; Stewart, C.; Schnabl, B.; Jauch, D.; Taniguchi, K.; Yu, G.; Osterreicher, C.; Hung, K.; et al. Adenoma-linked barrier defects and microbial products drive il-23/il-17-mediated tumour growth. Nature 2012, 491, 254–258. [Google Scholar] [CrossRef]
- Zitvogel, L.; Ayyoub, M.; Routy, B.; Kroemer, G. Microbiome and anticancer immunosurveillance. Cell 2016, 165, 276–287. [Google Scholar] [CrossRef]
- Graham, D. Helicobacter pylori update: Gastric cancer, reliable therapy, and possible benefits. Gastroenterology 2015, 148, 719–731. [Google Scholar] [CrossRef]
- Peek, R.M.; Blaser, M.J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2002, 2, 28–37. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Loo, T.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Rhee, K.-J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.-R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of t helper type 17 t cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- Kostic, A.; Chun, E.; Robertson, L.; Glickman, J.; Gallini, C.; Michaud, M.; Clancy, T.; Chung, D.; Lochhead, P.; Hold, G.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating e-cadherin/β-catenin signaling via its fada adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the fap2 protein of fusobacterium nucleatum to human inhibitory receptor tigit protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.; Fan, T.; Campbell, B.; Abujamel, T.; Dogan, B.; Rogers, A.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Alshamsan, A.; Khan, S.; Imran, A.; Aljuffali, I.A.; Alsaleh, K. Prediction of chlamydia pneumoniae protein localization in host mitochondria and cytoplasm and possible involvements in lung cancer etiology: A computational approach. Saudi Pharm. J. 2017, 25, 1151–1157. [Google Scholar] [CrossRef]
- Chumduri, C.; Gurumurthy, R.; Zadora, P.; Mi, Y.; Meyer, T. Chlamydia infection promotes host DNA damage and proliferation but impairs the DNA damage response. Cell Host Microbe 2013, 13, 746–758. [Google Scholar] [CrossRef]
- Verberkmoes, N.; Russell, A.; Shah, M.; Godzik, A.; Rosenquist, M.; Halfvarson, J.; Lefsrud, M.; Apajalahti, J.; Tysk, C.; Hettich, R.; et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009, 3, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Bosch, L.; de Wit, M.; Pham, T.; Coupé, V.; Hiemstra, A.; Piersma, S.; Oudgenoeg, G.; Scheffer, G.; Mongera, S.; Sive Droste, J.; et al. Novel stool-based protein biomarkers for improved colorectal cancer screening: A case-control study. Ann. Intern. Med. 2017, 167, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Yang, Y.; Shen, C.; Wang, Y.; Deng, A.; Qin, Q.; Qiao, L. Metaproteomics characterizes human gut microbiome function in colorectal cancer. NPJ Biofilms Microbiomes 2020, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Sung, J.; Yong, D.; Chun, J.; Kim, S.; Song, J.; Chung, K.; Kim, E.; Jung, J.; Kang, Y.; et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 2016, 102, 89–95. [Google Scholar] [CrossRef]
- Wei, W.; Sun, W.; Yu, S.; Yang, Y.; Ai, L. Butyrate production from high-fiber diet protects against lymphoma tumor. Leuk. Lymphoma 2016, 57, 2401–2408. [Google Scholar] [CrossRef] [PubMed]
- Aranda, F.; Bloy, N.; Pesquet, J.; Petit, B.; Chaba, K.; Sauvat, A.; Kepp, O.; Khadra, N.; Enot, D.; Pfirschke, C.; et al. Immune-dependent antineoplastic effects of cisplatin plus pyridoxine in non-small-cell lung cancer. Oncogene 2015, 34, 3053–3062. [Google Scholar] [CrossRef] [PubMed]
- Ijiri, M.; Fujiya, M.; Konishi, H.; Tanaka, H.; Ueno, N.; Kashima, S.; Moriichi, K.; Sasajima, J.; Ikuta, K.; Okumura, T. Ferrichrome identified from lactobacillus casei atcc334 induces apoptosis through its iron-binding site in gastric cancer cells. Tumour Biol. 2017, 39, 1010428317711311. [Google Scholar] [CrossRef]
- Han, D.; Kim, J.; Park, S.; Yang, M.; Kim, H. Growth inhibition of hepatocellular carcinoma huh7 cells by lactobacillus casei extract. Yonsei Med. J. 2013, 54, 1186–1193. [Google Scholar] [CrossRef]
- Kita, A.; Fujiya, M.; Konishi, H.; Tanaka, H.; Kashima, S.; Iwama, T.; Ijiri, M.; Murakami, Y.; Takauji, S.; Goto, T.; et al. Probiotic-derived ferrichrome inhibits the growth of refractory pancreatic cancer cells. Int. J. Oncol. 2020, 57, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, J.; Jin, D.; Wang, B.; Cao, H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int. J. Cancer 2019, 145, 2021–2031. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Fu, L.; Wang, J. Protocol for fecal microbiota transplantation in inflammatory bowel disease: A systematic review and meta-analysis. BioMed Res. Int. 2018, 2018, 8941340. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Ma, X.; Geng, S.; Jiang, X.; Li, Y.; Hu, L.; Li, J.; Wang, Y.; Han, X. Fecal microbiota transplantation beneficially regulates intestinal mucosal autophagy and alleviates gut barrier injury. mSystems 2018, 3, e00137-18. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Guo, K.; Zeng, L.; Zeng, B.; Huo, R.; Luo, Y.; Wang, H.; Dong, M.; Zheng, P.; Zhou, C.; et al. Metabolite identification in fecal microbiota transplantation mouse livers and combined proteomics with chronic unpredictive mild stress mouse livers. Transl. Psychiatry 2018, 8, 34. [Google Scholar] [CrossRef]
- Baruch, E.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021, 371, 602–609. [Google Scholar] [CrossRef]
- Davar, D.; Dzutsev, A.; McCulloch, J.; Rodrigues, R.; Chauvin, J.; Morrison, R.; Deblasio, R.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-pd-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.; Anderson, I.; Cheng, J.; Darling, A.; Malfatti, S.; Swan, B.; Gies, E.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.; Armougom, F.; Million, M.; Hugon, P.; Pagnier, I.; Robert, C.; Bittar, F.; Fournous, G.; Gimenez, G.; Maraninchi, M.; et al. Microbial culturomics: Paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012, 18, 1185–1193. [Google Scholar] [CrossRef]
- Picardo, S.; Coburn, B.; Hansen, A. The microbiome and cancer for clinicians. Crit. Rev. Oncol. Hematol. 2019, 141, 1–12. [Google Scholar] [CrossRef]
- Cifani, P.; Kentsis, A. High sensitivity quantitative proteomics using automated multidimensional nano-flow chromatography and accumulated ion monitoring on quadrupole-orbitrap-linear ion trap mass spectrometer. Mol. Cell. Proteom. 2017, 16, 2006–2016. [Google Scholar] [CrossRef]
- Poulos, R.; Hains, P.; Shah, R.; Lucas, N.; Xavier, D.; Manda, S.; Anees, A.; Koh, J.; Mahboob, S.; Wittman, M.; et al. Strategies to enable large-scale proteomics for reproducible research. Nat. Commun. 2020, 11, 3793. [Google Scholar] [CrossRef]
- Uhlen, M.; Bandrowski, A.; Carr, S.; Edwards, A.; Ellenberg, J.; Lundberg, E.; Rimm, D.; Rodriguez, H.; Hiltke, T.; Snyder, M.; et al. A proposal for validation of antibodies. Nat. Methods 2016, 13, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Björling, E.; Uhlén, M. Antibodypedia, a portal for sharing antibody and antigen validation data. Mol. Cell. Proteom. 2008, 7, 2028–2037. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; Melby, J.; Roberts, D.; Ge, Y. Top-down proteomics: Challenges, innovations, and applications in basic and clinical research. Expert Rev. Proteom. 2020, 17, 719–733. [Google Scholar] [CrossRef] [PubMed]
- Timp, W.; Timp, G. Beyond mass spectrometry, the next step in proteomics. Sci. Adv. 2020, 6, eaax8978. [Google Scholar] [CrossRef]
- Winter, D.; Wilkins, M.; Donald, W. Differential ion mobility-mass spectrometry for detailed analysis of the proteome. Trends Biotechnol. 2019, 37, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Anttalainen, O.; Puton, J.; Kontunen, A.; Karjalainen, M.; Kumpulainen, P.; Oksala, N.; Safaei, Z.; Roine, A. Possible strategy to use differential mobility spectrometry in real time applications. Int. J. Ion Mobil. Spectrom. 2020, 23, 1–8. [Google Scholar] [CrossRef]
- Baharlou, H.; Canete, N.; Cunningham, A.; Harman, A.; Patrick, E. Mass cytometry imaging for the study of human diseases-applications and data analysis strategies. Front. Immunol. 2019, 10, 2657. [Google Scholar] [CrossRef]
- Ali, H.R.; Jackson, H.W.; Zanotelli, V.R.T.; Danenberg, E.; Fischer, J.R.; Bardwell, H.; Provenzano, E.; Ali, H.R.; Al Sa’d, M.; Alon, S.; et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Cancer 2020, 1, 163–175. [Google Scholar] [CrossRef]
- Palit, S.; Heuser, C.; de Almeida, G.; Theis, F.; Zielinski, C. Meeting the challenges of high-dimensional single-cell data analysis in immunology. Front. Immunol. 2019, 10, 1515. [Google Scholar] [CrossRef] [PubMed]
- Ramiya Ramesh Babu, H.; Gheber, L. Fluorescence-based kinetic analysis of miniaturized protein microarrays. Biosens. Bioelectron. 2018, 122, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Sauer, U. Analytical protein microarrays: Advancements towards clinical applications. Sensors 2017, 17, 256. [Google Scholar] [CrossRef] [PubMed]
- Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017, 32, 185–203. [CrossRef] [PubMed]
- Li, J.; Zhao, W.; Akbani, R.; Liu, W.; Ju, Z.; Ling, S.; Vellano, C.; Roebuck, P.; Yu, Q.; Eterovic, A.; et al. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell 2017, 31, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Huang, T. Translating proteomics big data into biomedical applications with computational modeling. Biochim. Biophys. Acta Proteins Proteom. 2021, 1869, 140560. [Google Scholar] [CrossRef]
- Moldogazieva, N.; Mokhosoev, I.; Zavadskiy, S.; Terentiev, A. Proteomic profiling and artificial intelligence for hepatocellular carcinoma translational medicine. Biomedicines 2021, 9, 159. [Google Scholar] [CrossRef]
- Giulietti, M.; Cecati, M.; Sabanovic, B.; Scirè, A.; Cimadamore, A.; Santoni, M.; Montironi, R.; Piva, F. The role of artificial intelligence in the diagnosis and prognosis of renal cell tumors. Diagnostics 2021, 11, 206. [Google Scholar] [CrossRef]
- Tunali, I.; Gillies, R.; Schabath, M. Application of radiomics and artificial intelligence for lung cancer precision medicine. Cold Spring Harb. Perspect. Med. 2021, 039537. [Google Scholar] [CrossRef]
- Gessulat, S.; Schmidt, T.; Zolg, D.; Samaras, P.; Schnatbaum, K.; Zerweck, J.; Knaute, T.; Rechenberger, J.; Delanghe, B.; Huhmer, A.; et al. Prosit: Proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods 2019, 16, 509–518. [Google Scholar] [CrossRef]
- Callaway, E. ‘It will change everything’: Deepmind’s ai makes gigantic leap in solving protein structures. Nature 2020, 588, 203–204. [Google Scholar] [CrossRef] [PubMed]
- Specht, H.; Slavov, N. Transformative opportunities for single-cell proteomics. J. Proteome Res. 2018, 17, 2565–2571. [Google Scholar] [CrossRef]
- Labib, M.; Kelley, S.O. Single-cell analysis targeting the proteome. Nat. Rev. Chem. 2020, 4, 143–158. [Google Scholar] [CrossRef]
- Lim, Z.; Ma, P. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J. Hematol. Oncol. 2019, 12, 134. [Google Scholar] [CrossRef] [PubMed]
- Reuben, A.; Gittelman, R.; Gao, J.; Zhang, J.; Yusko, E.; Wu, C.; Emerson, R.; Zhang, J.; Tipton, C.; Li, J.; et al. Tcr repertoire intratumor heterogeneity in localized lung adenocarcinomas: An association with predicted neoantigen heterogeneity and postsurgical recurrence. Cancer Discov. 2017, 7, 1088–1097. [Google Scholar] [CrossRef]
- Lavin, Y.; Kobayashi, S.; Leader, A.; Amir, E.; Elefant, N.; Bigenwald, C.; Remark, R.; Sweeney, R.; Becker, C.; Levine, J.; et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 2017, 169, 750–765. [Google Scholar] [CrossRef]
- Swaminathan, J.; Boulgakov, A.; Hernandez, E.; Bardo, A.; Bachman, J.; Marotta, J.; Johnson, A.; Anslyn, E.; Marcotte, E. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat. Biotechnol. 2018, 36, 1076–1082. [Google Scholar] [CrossRef]
- van Ginkel, J.; Filius, M.; Szczepaniak, M.; Tulinski, P.; Meyer, A.; Joo, C. Single-molecule peptide fingerprinting. Proc. Natl. Acad. Sci. USA 2018, 115, 3338–3343. [Google Scholar] [CrossRef] [PubMed]
- Ohayon, S.; Girsault, A.; Nasser, M.; Shen-Orr, S.; Meller, A. Simulation of single-protein nanopore sensing shows feasibility for whole-proteome identification. PLoS Comput. Biol. 2019, 15, e1007067. [Google Scholar] [CrossRef]
- Mitra, R.; Müller, P.; Qiu, P.; Ji, Y. Bayesian hierarchical models for protein networks in single-cell mass cytometry. Cancer Inform. 2014, 13, 79–89. [Google Scholar] [CrossRef]
- Amir, E.-A.; Davis, K.; Tadmor, M.; Simonds, E.; Levine, J.; Bendall, S.; Shenfeld, D.; Krishnaswamy, S.; Nolan, G.; Pe’er, D. Visne enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013, 31, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Lähnemann, D.; Köster, J.; Szczurek, E.; McCarthy, D.; Hicks, S.; Robinson, M.; Vallejos, C.; Campbell, K.; Beerenwinkel, N.; Mahfouz, A.; et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020, 21, 31. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Bergan, R. Clinical trial design: Past, present, and future in the context of big data and precision medicine. Cancer 2020, 126, 4838–4846. [Google Scholar] [CrossRef] [PubMed]
- Garralda, E.; Dienstmann, R.; Piris-Giménez, A.; Braña, I.; Rodon, J.; Tabernero, J. New clinical trial designs in the era of precision medicine. Mol. Oncol. 2019, 13, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Madhusoodanan, J. Health-care inequality could deepen with precision oncology. Nature 2020, 585, S13–S15. [Google Scholar] [CrossRef]
| Proteomics Toolbox | Technical Process | Strengths | Limitations | Reference | ||
|---|---|---|---|---|---|---|
| Targeted Approaches | MRM, SRM | (1) Primary mass spectrometry scans to screen out parent ions that are consistent with the specificity of the target molecule. (2) Collision and fragmentation of parent ions to remove interfering ions. (3) Mass spectrometry signals collected from selected specific ions | (1) High sensitivity (2) Good accuracy (3) Good reproducibility (4) High-throughput | (1) Only preselected proteins can be detected (2) No screening analysis | [33,34] | |
| PRM | (1) Select targeted peptides. (2) Define acquisition method. (3) Preliminary experiment to adjust the parameters and target peptide fragment. (4) Perform PRM analysis. (5) Analyze data | (1) Simpler and cheaper. (2) Wider linear detection range. (3) Higher selectivity, better sensitivity and better reproducibility | (1) Only preselected proteins can be detected (2) No screening analysis (3) When the number of peptides to be analyzed is large it is necessary to fine-tune the MS collection parameters | [35,36] | ||
| Untargeted Approaches | DDA | iTRAQ/TMT | (1) Enzymatic or chemical fragmentation. (2) Differential labeling using iTRAQ/TMT reagent. (3) Mixed labeled protein samples analyzed by tandem mass spectrometry. (4) Data analysis | (1) Good repeatability. (2) Mature method. (3) Wide range of sample sources | (1) Easily contaminated by other proteins in the sample (2) Maximum 12 channels at a time | [37] |
| Label-free | (1) Protein extraction. (2) Protein digestion. (3) LC–MS/MS analysis. (4) Data analysis | (1) Low cost (2) Not limited by the number of samples. (3) Wide range of application | (1) Highly dependent on machine stability. (2) Quantitative results can be unreliable | [38] | ||
| DIA | SWATH | (1) Sample digestion (2) Consecutive, adjacent precursor ion windows (SWATHs) scanned using a Triple TOF MS (3) Data analyzed using bioinformatics and a relevant reference spectral library | (1) Good reproducibility. (2) Less affected by high-abundance proteins | Quality of spectral library for data analysis | [32,39,40,41] | |
| Cancer Biomarkers | Cancer Type | Use | Proteomic Technology | Reference |
|---|---|---|---|---|
| APOAII-2 | Pancreatic cancer | Detection of early-stage pancreatic cancer and risk factors for pancreatic malignancy | 2D-PAGE, QTOF MS system and ELISA | [121,122] |
| Ezrin | Colorectal cancer | Predicting lymph node metastasis in colorectal cancer | iTRAQ in a comparative proteomics approach | [124,126,127] |
| QSOX1 | Lung cancer | A potential therapeutic target | LC/MS-MS analysis | [128] |
| Serine/threonine kinase 4 | Colorectal cancer | An early detection biomarker | MALDI-TOF-MS | [134] |
| αB-crystallin | Breast cancer | A potential prognostic biomarker | Quantitative iTRAQ proteomics | [135] |
| ENO1 | Lung cancer | A Potential Sputum Biomarker for Early-Stage Lung Cancer and a potential therapeutic target | Shotgun proteomics; liquid chromatography–tandem mass spectrometry technology | [136,137,138,139] |
| CT45 | High-grade serous ovarian cancer | A Chemosensitivity Mediator and Immunotherapy Target | Liquid Chromatography–MS analysis; Phosphatase activity assay and phosphoproteomics; Immunofluorescence | [132] |
| FoxO3a | Breast cancer | A Positive Prognostic Marker and a Therapeutic Target in Tamoxifen-Resistant | Label-Free Semiquantitative Proteomic Analysis and ingenuity pathway analysis (IPA) | [140] |
| HSP 90β | Lung adenocarcinoma | A potential prognostic biomarker | Nano-LC–MS/MS analysis; ELISA; label-free quantification | [80] |
| SAA2, APCS, APOA4, F2 and AMBP | Colorectal cancer | Potential early diagnosis biomarkers | SWATH-MS and ELISA | [26] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, M.; Zhang, Z.; Zhou, L.; Han, C.; Huang, C.; Nice, E.C. Proteomics, Personalized Medicine and Cancer. Cancers 2021, 13, 2512. https://doi.org/10.3390/cancers13112512
Su M, Zhang Z, Zhou L, Han C, Huang C, Nice EC. Proteomics, Personalized Medicine and Cancer. Cancers. 2021; 13(11):2512. https://doi.org/10.3390/cancers13112512
Chicago/Turabian StyleSu, Miao, Zhe Zhang, Li Zhou, Chao Han, Canhua Huang, and Edouard C. Nice. 2021. "Proteomics, Personalized Medicine and Cancer" Cancers 13, no. 11: 2512. https://doi.org/10.3390/cancers13112512
APA StyleSu, M., Zhang, Z., Zhou, L., Han, C., Huang, C., & Nice, E. C. (2021). Proteomics, Personalized Medicine and Cancer. Cancers, 13(11), 2512. https://doi.org/10.3390/cancers13112512








