High-Risk Clinicopathological and Genetic Features and Outcomes in Patients Receiving Neoadjuvant Radiochemotherapy for Locally Advanced Rectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. SNP Array Studies
2.3. Survival Analysis
2.4. Prediction of Response to Preoperative Radiochemotherapy (RCT)
2.5. Interphase Fluorescence In Situ Hybridization (FISH) Studies
2.6. Other Statistical Methods
3. Results
3.1. Clinical and Biological Characteristics of Locally Advanced Rectal Cancer (LARC) before and after Preoperative Radiochemotherapy (RCT)
3.2. Distribution of Chromosomal Alterations in LARC before Preoperative RCT
3.3. Chromosomal Alterations and Response to Preoperative RCT
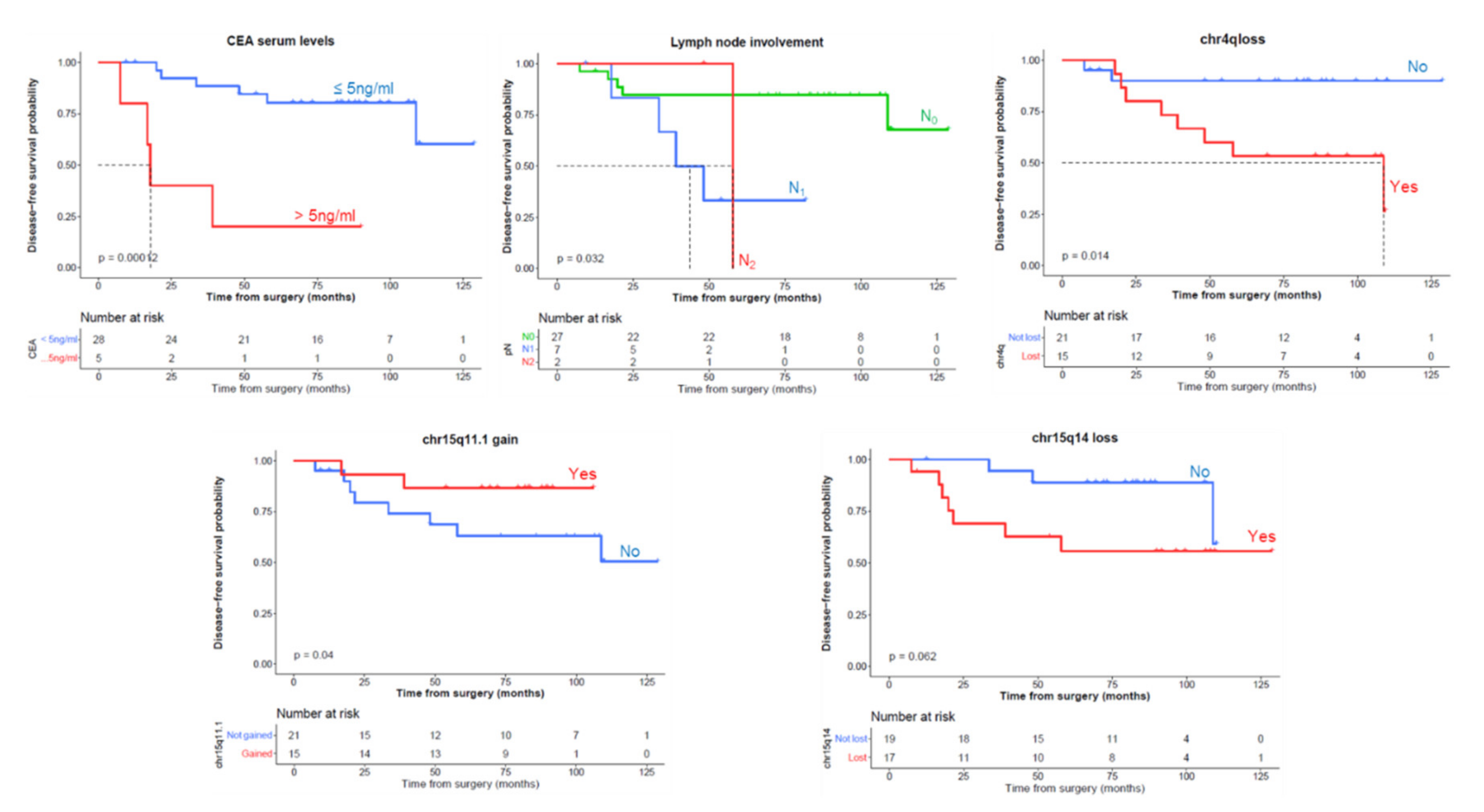
3.4. Analysis of Prognostic Impact and Predictiveness of Clinical-Biologic Features and Chromosomal Alterations on Disease-Free Survival (DFS) an Overall Survival (OS)
3.5. Correlation between the Chromosomal Changes Detected by the SNP Array and FISH Techniques
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sauer, R.; Becker, H.; Hohenberger, W.; Rödel, C.; Wittekind, C.; Fietkau, R.; Martus, P.; Tschmelitsch, J.; Hager, E.; Hess, C.F.; et al. Preoperative versus Postoperative Chemoradiotherapy for Rectal Cancer. N. Engl. J. Med. 2004, 351, 1731–1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, F.M.; Reynolds, J.V.; Miller, N.; Stephens, R.B.; Kennedy, M.J. Pathological and molecular predictors of the response of rectal cancer to neoadjuvant radiochemotherapy. Eur. J. Surg. Oncol. 2006, 32, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Therasse, P.; Arbuck, S.G.; Eisenhauer, E.A.; Wanders, J.; Kaplan, R.S.; Rubinstein, L.; Verweij, J.; Van Glabbeke, M.; Van Oosterom, A.T.; Christian, M.C.; et al. New guidelines to evaluate the response to treatment in solid tumors. J. Natl. Cancer Inst. 2000, 92, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Rengo, M.; Picchia, S.; Marzi, S.; Bellini, D.; Caruso, D.; Caterino, M.; Ciolina, M.; De Santis, D.; Musio, D.; Tombolini, V.; et al. Magnetic resonance tumor regression grade (MR-TRG) to assess pathological complete response following neoadjuvant radiochemotherapy in locally advanced rectal cancer. Oncotarget 2017, 8, 114746–114755. [Google Scholar] [CrossRef]
- Reginelli, A.; Clemente, A.; Sangiovanni, A.; Nardone, V.; Selvaggi, F.; Sciaudone, G.; Ciardiello, F.; Martinelli, E.; Grassi, R.; Cappabianca, S. Endorectal Ultrasound and Magnetic Resonance Imaging for Rectal Cancer Staging: A Modern Multimodality Approach. J. Clin. Med. 2021, 10, 641. [Google Scholar] [CrossRef] [PubMed]
- Murahashi, S.; Akiyoshi, T.; Sano, T.; Fukunaga, Y.; Noda, T.; Ueno, M.; Zembutsu, H. Serial circulating tumour DNA analysis for locally advanced rectal cancer treated with preoperative therapy: Prediction of pathological response and postoperative recurrence. Br. J. Cancer 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, C.; Lin, G.; Xiao, Y.; Jia, W.; Xiao, G.; Liu, Q.; Wu, B.; Wu, A.; Qiu, H.; et al. Serial Circulating Tumor DNA in Predicting and Monitoring the Effect of Neoadjuvant Chemoradiotherapy in Patients with Rectal Cancer: A Prospective Multicenter Study. Clin. Cancer Res. 2021. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Wang, Y.; Li, L.; Christie, M.; Simons, K.; Elsaleh, H.; Kosmider, S.; Wong, R.; Yip, D.; et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 2019. [Google Scholar] [CrossRef] [PubMed]
- Habr-Gama, A.; Perez, R.O.; Nadalin, W.; Sabbaga, J.; Ribeiro, U.; Silva, E.; Sousa, A.H.; Campos, F.G.; Kiss, D.R.; Gama-Rodrigues, J.; et al. Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: Long-term results. Ann. Surg. 2004, 240, 711–718. [Google Scholar] [CrossRef]
- Saw, R.P.M.; Morgan, M.; Koorey, D.; Painter, D.; Findlay, M.; Stevens, G.; Clarke, S.; Chapuis, P.; Solomon, M.J. p53, deleted in colorectal cancer gene, and thymidylate synthase as predictors of histopathologic response and survival in low, locally advanced rectal cancer treated with preoperative adjuvant therapy. Dis. Colon Rectum 2003. [Google Scholar] [CrossRef]
- Frydrych, L.M.; Ulintz, P.; Bankhead, A.; Sifuentes, C.; Greenson, J.; Maguire, L.; Irwin, R.; Fearon, E.R.; Hardiman, K.M. Rectal cancer sub-clones respond differentially to neoadjuvant therapy. Neoplasia 2019. [Google Scholar] [CrossRef] [PubMed]
- González-González, M.; Garcia, J.; Alcazar, J.A.; Gutiérrez, M.L.; Gónzalez, L.M.; Bengoechea, O.; Abad, M.M.; Santos-Briz, A.; Blanco, O.; Martín, M.; et al. Association between the cytogenetic profile of tumor cells and response to preoperative radiochemotherapy in locally advanced rectal cancer. Medicine 2014. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, Z.; Li, W.; Qu, K.; Deng, X.; Varma, M.G.; Fichera, A.; Pigazzi, A.; Garcia-Aguilar, J. Chromosomal copy number alterations are associated with tumor response to chemoradiation in locally advanced rectal cancer. Genes Chromosom. Cancer 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molinari, C.; Ballardini, M.; Teodorani, N.; Giannini, M.; Zoli, W.; Emiliani, E.; Lucci, E.; Passardi, A.; Rosetti, P.; Saragoni, L.; et al. Genomic alterations in rectal tumors and response to neoadjuvant chemoradiotherapy: An exploratory study. Radiat. Oncol. 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dworak, O.; Keilholz, L.; Hoffmann, A. Pathological features of rectal cancer after preoperative radiochemotherapy. Int. J. Colorectal Dis. 1997. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression: Appendices, 3rd ed.; Sage Publications: Newbury Park, CA, USA, 2019; pp. 1–17. ISBN 978-1-5443-3647-3. [Google Scholar]
- Kassambara, A.; Kosinski, M.; Biecek, P.; Fabian, S. Drawing Survival Curves Using “ggplot2” [R Package Survminer Version 0.4. 2]. Compr. R Arch. Netw. 2018. Available online: https://CRAN.R-project.org/package=survminer (accessed on 23 June 2021).
- Terry, M.; Therneau, P.M.G. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000. [Google Scholar]
- Mogensen, U.B.; Ishwaran, H.; Gerds, T.A. Evaluating Random Forests for Survival Analysis Using Prediction Error Curves. J. Stat. Softw. 2012, 50. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the boruta package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Grömping, U. Relative importance for linear regression in R: The package relaimpo. J. Stat. Softw. 2006, 17, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Law Biecek, P. DALEX: Explainers for Complex Predictive Models in R. J. Mach. Learn. Res. 2018, 19, 3245–3249. [Google Scholar]
- Janitza, S.; Celik, E.; Boulesteix, A.L. A computationally fast variable importance test for random forests for high-dimensional data. Adv. Data Anal. Classif. 2018. [Google Scholar] [CrossRef]
- Dimitriadou, E.; Hornik, K.; Leisch, F.; Meyer, D.; Weingessel, A. Misc functions of the Department of Statistics (e1071). TU Wien. R Package 2008, 1, 5–24. [Google Scholar]
- Burguillo, F.; Corchete, L.; Martin, J.; Barrera, I.; Bardsley, W. A Partial Least Squares Algorithm for Microarray Data Analysis Using the VIP Statistic for Gene Selection and Binary Classification. Curr. Bioinform. 2014. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, M. Classification and Regression Training [R Package Caret Version 6.0-86]. Compr. R Arch. Netw. 2020. [Google Scholar]
- Agustin, P.-M.; Agustin, P.-T.; Marta, V.-L.; Verdu-Jover, A.J. Create the Best Train for Classification Models [R package OptimClassifier version 0.1.5]; 2020. Available online: https://mran.microsoft.com/snapshot/2020-02-28/web/packages/OptimClassifier/index.html (accessed on 23 June 2021).
- Sayagués, J.M.; Fontanillo, C.; Abad, M.; del Mar Abad, M.; González-González, M.; Sarasquete, M.E.; del Carmen Chillon, M.; Garcia, E.; Bengoechea, O.; Fonseca, E.; et al. Mapping of genetic abnormalities of primary tumours from metastatic CRC by high-resolution SNP arrays. PLoS ONE 2010, 5, e13752. [Google Scholar] [CrossRef] [Green Version]
- Sayagués, J.M.; Abad, M.D.M.; Melchor, H.B.; Gutiérrez, M.L.; González-González, M.; Jensen, E.; Bengoechea, O.; Fonseca, E.; Orfao, A.; Muñoz-Bellvis, L. Intratumoural cytogenetic heterogeneity of sporadic colorectal carcinomas suggests several pathways to liver metastasis. J. Pathol. 2010, 221, 308–319. [Google Scholar] [CrossRef]
- Benson, A.B.; Bekaii-Saab, T.; Chan, E.; Chen, Y.J.; Choti, M.A.; Cooper, H.S.; Engstrom, P.F.; Enzinger, P.C.; Fakih, M.G.; Fuchs, C.S.; et al. Rectal cancer. JNCCN J. Natl. Compr. Cancer Netw. 2012, 10, 1528–1564. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulos, G.; Wise, W.E.; Padmanabhan, A.; Kerner, B.A.; Taylor, C.W.; Aguilar, P.S.; Khanduja, K.S. T-level downstaging and complete pathologic response after preoperative chemoradiation for advanced rectal cancer result in decreased recurrence and improved disease-free survival. Dis. Colon Rectum 2002, 45, 895–903. [Google Scholar] [CrossRef]
- Liu, M.; Liu, Y.; Di, J.; Su, Z.; Yang, H.; Jiang, B.; Wang, Z.; Zhuang, M.; Bai, F.; Su, X. Multi-region and single-cell sequencing reveal variable genomic heterogeneity in rectal cancer. BMC Cancer 2017, 17. [Google Scholar] [CrossRef]
- Bettoni, F.; Masotti, C.; Corrêa, B.R.; Donnard, E.; Dos Santos, F.F.; São Julião, G.P.; Vailati, B.B.; Habr-Gama, A.; Galante, P.A.F.; Perez, R.O.; et al. The effects of neoadjuvant chemoradiation in locally advanced rectal cancer—The impact in intratumoral heterogeneity. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Yuan, L.; Cai, Y.; Chen, X.; Chi, Y.; Wei, P.; Zhou, X.; Shi, D. Identification of chromosomal copy number variations and novel candidate loci in hereditary nonpolyposis colorectal cancer with mismatch repair proficiency. Genomics 2013, 102, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Ding, J.; Jiang, L.; Liu, Z.; Zhou, X.; Shi, D. DNA copy number profiling in microsatellite-stable and microsatellite-unstable hereditary non-polyposis colorectal cancers by targeted CNV array. Funct. Integr. Genomics 2017, 17, 85–96. [Google Scholar] [CrossRef]
- Almutairi, M.; Rouabhia, M.; Sahab Almutairi, M.; Al-Zahrani, M.; Al-Numair, N.S.; Mohammad Alhadeq, A.; Reddy Parine, N.; Semlali, A. Correlation between genetic variation in thymine DNA glycosylase and smoking behavior. Gene 2021, 766, 145092. [Google Scholar] [CrossRef] [PubMed]
- Goossens-Beumer, I.J.; Oosting, J.; Corver, W.E.; Janssen, M.J.F.W.; Janssen, B.; Workum, W.; Zeestraten, E.C.M.; Velde, C.J.H.; Morreau, H.; Kuppen, P.J.K.; et al. Copy number alterations and allelic ratio in relation to recurrence of rectal cancer. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.F.; Li, X.Q.; Zhang, J.; Yang, L.F.; Zhu, J.I.; Li, G.C.; Liang, L.P.; Shen, L.J.; Zhang, H.; Li, J.; et al. Aneuploidy of chromosome 8 and mutation of circulating tumor cells predict pathologic complete response in the treatment of locally advanced rectal cancer. Oncol. Lett. 2018, 16, 1863–1868. [Google Scholar] [CrossRef]
- Fang, T.; Lv, H.; Wu, F.; Wang, C.; Li, T.; Lv, G.; Tang, L.; Guo, L.; Tang, S.; Cao, D.; et al. Musashi 2 contributes to the stemness and chemoresistance of liver cancer stem cells via LIN28A activation. Cancer Lett. 2017, 384, 50–59. [Google Scholar] [CrossRef]
- Wang, N.; Wang, W.; Mao, W.; Kuerbantayi, N.; Jia, N.; Chen, Y.; Zhou, F.; Yin, L.; Wang, Y. RBBP4 Enhances Platinum Chemo Resistance in Lung Adenocarcinoma. Biomed Res. Int. 2021, 2021. [Google Scholar] [CrossRef]
- Jiang, F.; Desper, R.; Papadimitriou, C.H.; Schäffer, A.A.; Kallioniemi, O.P.; Richter, J.; Schraml, P.; Sauter, G.; Mihatsch, M.J.; Moch, H. Construction of evolutionary tree models for renal cell carcinoma from comparative genomic hybridization data. Cancer Res. 2000, 60, 6503–6509. [Google Scholar] [PubMed]
- Luebke, A.M.; Baudis, M.; Matthaei, H.; Vashist, Y.K.; Verde, P.E.; Hosch, S.B.; Erbersdobler, A.; Klein, C.A.; Izbicki, J.R.; Knoefel, W.T.; et al. Losses at chromosome 4q are associated with poor survival in operable ductal pancreatic adenocarcinoma. Pancreatology 2012. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cai, H.; Xie, Y.; Jiang, H. Targeting long non-coding RNA HERC2P3 inhibits cell growth and migration in human gastric cancer cells. Int. J. Clin. Exp. Pathol. 2017, 10, 7632–7639. [Google Scholar]
- Bonanno, L.; Costa, C.; Majem, M.; Sanchez, J.J.; Rodriguez, I.; Gimenez-Capitan, A.; Molina-Vila, M.A.; Vergnenegre, A.; Massuti, B.; Favaretto, A.; et al. Combinatory effect of BRCA1 and HERC2 expression on outcome in advanced non-small-cell lung cancer. BMC Cancer 2016. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, G.; Zheng, K.; Lou, Z.; Gao, X.H.; Meng, R.G.; Furnée, E.J.B.; Zhang, W. Prognostic factors in patients with complete response of the tumour (ypT0) after neoadjuvant chemoradiotherapy and radical resection of rectal cancer. ANZ J. Surg. 2021. [Google Scholar] [CrossRef]
- Cai, Z.; Huang, L.; Chen, Y.; Xie, X.; Zou, Y.; Lan, P.; Wu, X. CEA decline predicts tumor regression and prognosis in locally advanced rectal cancer patients with elevated baseline CEA. J. Cancer 2020. [Google Scholar] [CrossRef] [PubMed]
- Cetin, E.; Cengiz, B.; Gunduz, E.; Gunduz, M.; Nagatsuka, H.; Bekir Beder, L.; Fukushima, K.; Pehlivan, D.; Ozaslan, M.; Yamanaka, N.; et al. Deletion mapping of chromosome 4q22-35 and identification of four frequently deleted regions in head and neck cancers. Neoplasma 2008, 55, 299–304. [Google Scholar]
- Shivapurkar, N.; Sood, S.; Wistuba, I.I.; Virmani, A.K.; Maitra, A.; Milchgrub, S.; Minna, J.D.; Gazdar, A.F. Multiple regions of chromosome 4 demonstrating allelic losses in breast carcinomas. Cancer Res. 1999, 59, 3576–3580. [Google Scholar] [PubMed]
- Brosens, R.P.M.; Belt, E.J.T.H.; Haan, J.C.; Buffart, T.E.; Carvalho, B.; Grabsch, H.; Quirke, P.; Cuesta, M.A.; Engel, A.F.; Ylstra, B.; et al. Deletion of chromosome 4q predicts outcome in Stage II colon cancer patients. Anal. Cell. Pathol. 2011, 33, 95–104. [Google Scholar] [CrossRef] [Green Version]
- Bardi, G.; Fenger, C.; Johansson, B.; Mitelman, F.; Heim, S. Tumor karyotype predicts clinical outcome in colorectal cancer patients. J. Clin. Oncol. 2004, 22, 2623–2634. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kumamoto, K.; Okayama, H.; Matsumoto, T.; Nakano, H.; Saito, K.; Matsumoto, Y.; Endo, E.; Kanke, Y.; Watanabe, Y.; et al. Downregulation of PAICS due to loss of chromosome 4q is associated with poor survival in stage III colorectal cancer. PLoS ONE 2021. [Google Scholar] [CrossRef] [PubMed]
- Hermsen, M.; Postma, C.; Baak, J.; Weiss, M.; Rapallo, A.; Sciutto, A.; Roemen, G.; Arends, J.-W.; Williams, R.; Giaretti, W.; et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology 2002, 123, 1109–1119. [Google Scholar] [CrossRef] [Green Version]
- Sheffer, M.; Bacolod, M.D.; Zuk, O.; Giardina, S.F.; Pincas, H.; Barany, F.; Paty, P.B.; Gerald, W.L.; Notterman, D.A.; Domany, E. Association of survival and disease progression with chromosomal instability: A genomic exploration of colorectal cancer. Proc. Natl. Acad. Sci. USA 2009. [Google Scholar] [CrossRef] [Green Version]
- El Otmani, I.; El Agy, F.; El Baradai, S.; Bouguenouch, L.; Lahmidani, N.; El Abkari, M.; Benajah, D.A.; Toughrai, I.; El Bouhaddouti, H.; Mouaqit, O.; et al. Analysis of molecular pretreated tumor profiles as predictive biomarkers of therapeutic response and survival outcomes after neoadjuvant therapy for rectal cancer in moroccan population. Dis. Markers 2020. [Google Scholar] [CrossRef]
- Caramés, C.; Cristobal, I.; Moreno, V.; Marín, J.P.; González-Alonso, P.; Torrejón, B.; Minguez, P.; Leon, A.; Martín, J.I.; Hernández, R.; et al. Microrna-31 emerges as a predictive biomarker of pathological response and outcome in locally advanced rectal cancer. Int. J. Mol. Sci. 2016, 17, 878. [Google Scholar] [CrossRef]
- Yang, D.; Schneider, S.; Azuma, M.; Iqbal, S.; El-Khoueiry, A.; Groshen, S.; Agafitei, D.; Danenberg, K.D.; Danenberg, P.V.; Ladner, R.D.; et al. Gene expression levels of epidermal growth factor receptor, survivin, and vascular endothelial growth factor as molecularmarkers of lymph node involvement in patients with locally advanced rectal cancer. Clin. Colorectal Cancer 2006. [Google Scholar] [CrossRef]
- Yu, J.; Lee, S.H.; Jeung, T.S.; Chang, H.K. Expression of vascular endothelial growth factor as a predictor of complete response for preoperative chemoradiotherapy in rectal cancer. Medicine 2019, 98. [Google Scholar] [CrossRef]
- Huh, J.W.; Kim, H.C.; Kim, S.H.; Park, Y.A.; Cho, Y.B.; Yun, S.H.; Lee, W.Y.; Park, H.C.; Choi, D.H.; Park, J.O.; et al. Mismatch repair gene expression as a predictor of tumor responses in patients with rectal cancer treated with preoperative chemoradiation. Medicine 2016. [Google Scholar] [CrossRef]
- Ryan, É.J.; Creavin, B.; Sheahan, K. Delivery of Personalized Care for Locally Advanced Rectal Cancer: Incorporating Pathological, Molecular Genetic, and Immunological Biomarkers Into the Multimodal Paradigm. Front. Oncol. 2020, 10, 1369. [Google Scholar] [CrossRef] [PubMed]
- Bottarelli, L.; De’angelis, G.L.; Azzoni, C.; Di Mario, F.; De’ Angelis, N.; Leandro, G.; Fornaroli, F.; Gaiani, F.; Negri, F. Potential predictive biomarkers in locally advanced rectal cancer treated with preoperative chemo-radiotherapy. Acta Biomed. 2018, 21, 1768. [Google Scholar]
- Riesco-Martinez, M.C.; Fernandez-Martos, C.; Gravalos-Castro, C.; Espinosa-Olarte, P.; La Salvia, A.; Robles-Diaz, L.; Modrego-Sanchez, A.; Garcia-Carbonero, R. Impact of total neoadjuvant therapy vs. Standard chemoradiotherapy in locally advanced rectal cancer: A systematic review and meta-analysis of randomized trials. Cancers 2020, 12, 3655. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jiang, T.; Xiao, L.; Yang, S.; Liu, Q.; Gao, Y.; Chen, G.; Xiao, W. Total neoadjuvant therapy ( TNT ) versus standard neoadjuvant chemoradiotherapy for locally advanced rectal cancer: A systematic review and meta-analysis. Oncologist 2021. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Zhou, Y.; Cui, W.; Su, X.; Guo, Z.; Hidasa, I.; Li, Q.; Wang, Z.; Song, Y. The Addition of EGFR Inhibitors in Neoadjuvant Therapy for KRAS-Wild Type Locally Advanced Rectal Cancer Patients: A Systematic Review and Meta-Analysis. Front. Pharmacol. 2020. [Google Scholar] [CrossRef]
- Mutalib, N.S.; Yusof, N.F.; Abdul, S.N.; Jamal, R. Pharmacogenomics DNA Biomarkers in Colorectal Cancer: Current Update. Front. Pharmacol. 2017, 8, 736. [Google Scholar] [CrossRef] [Green Version]




| Clinical Features | Pre-Treatment | Post-Treatment | p |
|---|---|---|---|
| Age (years) * | 69 (39–88) | 69 (39–88) | NS |
| Gender Female Male | 11 (28%) 28 (72%) | NA NA | NA |
| Tumor Size (cm) * | 4 (1–5) | 1.92 (0–4) | 0.02 |
| Localization in the rectum Lower Medium Upper | 4 (10%) 20 (51%) 15 (39%) | NA NA NA | NA |
| TNM T0 T1 T2 T3 T4 | 0 (0%) 0 (0%) 1 (2%) 26 (67%) 12 (31%) | 5 (13%) 3 (8%) 14 (36%) 17 (43%) 0 (0%) | <0.0001 |
| N0 N1 N2 | 8 (20%) 30 (77%) 1 (3%) | 27 (69%) 10 (26%) 2 (5%) | <0.0001 |
| M0 M1 | 39 (100%) 0 (0%) | 38 (97%) 1 (3%) | NS |
| Tumor stage Stage 0 Stage I Stage II Stage III Stage IV | 0 (0%) 1 (3%) 6 (15%) 32 (82%) 0 (0%) | 4 (10%) 15 (39%) 8 (20%) 11 (28%) 1 (3%) | <0.0001 |
| Dworak regression grade G0 G1 G2 G3 G4 | NA NA NA NA NA | 3 (8%) 13 (33%) 13 (33%) 5 (13%) 5 (13%) | NA |
| Type of surgery APR AR | NA NA | 13 (33%) 26 (67%) | NA |
| Type of tumor resection R0 R1 R2 | NA NA NA | 36 (92%) 1 (3%) 2 (5%) | NA |
| CEA serum levels ≤5 ng/mL ≥5 ng/mL | 23 (59%) 16 (41%) | 34 (87%) 5 (13%) | 0.005 |
| KRAS mutation Wild-type Mutated G12D G12V G13D | 26 (67%) 1 (3%) 3 (8%) 4 (10%) 5 (12%) | NA NA NA NA NA | NA |
| Local recurrence | NA | 2 (5%) | NA |
| Minimal Common Altered Regions (bp) | Region Length (bp) | N. of SNPs | Chr. Band | Event | Altered Cases (%) | Gene List |
|---|---|---|---|---|---|---|
| Chr18: 48351659-48920677 | 569018 | 2340 | 18q21.2 | Loss | 69 | RN7SL695P, SRSF10P1, RNU1-46P, MRO, ME2, MEX3C, ELAC1, SMAD4 |
| Chr20: 1560988-1585059 | 24071 | 18 | 20p13 | Gain | 56 | SIRPB1 |
| Chr1: 7829422-10869532 | 3040110 | 14577 | 1p36.23 | Loss | 56 | RNU1-7P, RN7SL729P, RNU6-991P, RPL7P11, RPL7P7, ENO1-IT1, ENO1-AS1, RNU6-304P, HMGN2P17, RN7SL451P, MIR34A, RNA5SP40, C1orf200, RN7SKP269, MIR5697, PGAM1P11, RNU6-828P, MIR1273D, RNU6-37P, RN7SL731P, RN7SL721P, CORT, RN7SL614P, VAMP3, UTS2, PARK7, ERRFI1, ENO1, CA6, SLC2A7, SLC2A5, SPSB1, SLC25A33, TMEM201, PIK3CD, LZIC, NMNAT1, RBP7, PGD, APITD1, APITD1-CORT, DFFA, PER3, TNFRSF9, RERE, GPR157, H6PD, CLSTN1, CTNNBIP1, UBE4B, KIF1B, PEX14, CASZ1, CAMTA1, SLC45A1 |
| Chr1: 26284282-31197400 | 4913118 | 20676 | 1p35.3 | Loss | 54 | RNU6-110P, SLC30A2, FAM110D, ZNF593, CD52, RN7SL490P, HMGN2, DPPA2P2, MIR1976, RN7SL679P, RN7SL501P, RN7SL165P, SFN, GPATCH3, NR0B2, OSTCP2, TRNP1, FAM46B, CHCHD3P3, NPM1P39, SNRPEP7, RNU6-48P, FCN3, CD164L2, IFI6, RNU6-949P, CHMP1AP1, RNU6-424P, RPEP3, RNU6-1245P, SCARNA1, THEMIS2, XKR8, RN7SL559P, SPCS2P4, RNU6-176P, RNU7-29P, ATPIF1, RNU6ATAC27P, SNORA73B, PRDX3P2, SNHG12, SNORD99, RAB42, RNU11, TMEM200B, PAFAH2, EXTL1, TRIM63, PDIK1L, CNKSR1, CATSPER4, CEP85, UBXN11, AIM1L, ZNF683, DHDDS, ARID1A, PIGV, ZDHHC18, GPN2, C1orf172, SLC9A1, WDTC1, SYTL1, MAP3K6, GPR3, FGR, FAM76A, STX12, PPP1R8, RPA2, SMPDL3B, PTAFR, DNAJC8, SESN2, MED18, TRNAU1AP, GMEB1, YTHDF2, OPRD1, MECR, SH3BGRL3, LIN28A, RPS6KA1, TMEM222, WASF2, AHDC1, PHACTR4, RCC1, SNHG3, TAF12, SRSF4, PTPRU, MATN1, MATN1-AS1, NUDC, EYA3, EPB41 |
| Chr1: 23401844-25226751 | 1824907 | 7976 | 1p36.11 | Loss | 54 | RNU6-514P, RNU6-135P, HTR1D, C1orf213, ID3, RN7SL532P, PITHD1, LYPLA2, GALE, RN7SL24P, MIR378F, PNRC2, RN7SL857P, RNU6-1208P, KDM1A, HNRNPR, ZNF436, ASAP3, MDS2, RPL11, TCEB3, HMGCL, FUCA1, SRSF10, MYOM3, IL22RA1, GRHL3, STPG1, RCAN3, SRRM1, RUNX3, LUZP1, TCEA3, E2F2, CNR2, IFNLR1, NIPAL3, NCMAP, CLIC4 |
| Chr8: 39235592-39384956 | 149364 | 964 | 8p11.22 | Gain | 51 | ADAM5, ADAM3A |
| Chr1: 20830489-20979684 | 149195 | 963 | 1p36.12 | Loss | 51 | MUL1, RPS4XP4, FAM43B, CDA, DDOST, PINK1, PINK1-AS |
| Chr1: 31457917-31735879 | 277962 | 1756 | 1p35.2 | Loss | 51 | SEPW1P, NKAIN1, SNRNP40, PUM1 |
| Chr15: 34670991-34830240 | 159249 | 1137 | 15q14 | Loss | 49 | MIR1233-1, HNRNPLP2, MIR1233-2, GOLGA8A, GOLGA8B |
| Chr15: 50557160-51352248 | 795088 | 3936 | 15q21.2 | Loss | 49 | MIR4712, AHCYP7, RNA5SP395, RN7SL354P, DCAF13P3, HDC, GABPB1-AS1, USP50, SPPL2A, GABPB1, USP8, TRPM7, AP4E1, TNFAIP8L3 |
| Chr1: 32278463-33614161 | 1335698 | 4306 | 1p35.1 | Loss | 48 | MIR5585, IQCC, DCDC2B, EIF3I, FAM167B, FAM229A, GAPDHP20, LRRC37A12P, RN7SL122P, FNDC5, TMEM54, SPOCD1, TMEM39B, TXLNA, CCDC28B, TMEM234, MTMR9LP, LCK, MARCKSL1, TSSK3, BSDC1, ZBTB8B, ZBTB8OS, RBBP4, KIAA1522, YARS, HPCA, AK2, TRIM62, PTP4A2, KPNA6, HDAC1, ZBTB8A, SYNC, S100PBP, RNF19B, KHDRBS1, ADC |
| Chr8: 2784419-6422612 | 3638193 | 44962 | 8p23.1 | Loss | 48 | RNA5SP251, RN7SL872P, PAICSP4, RN7SL318P, RPL23AP54, RN7SKP159, ANGPT2, CSMD1, MCPH1 |
| Chr8: 32577483-35655135 | 3077652 | 13569 | 8p12 | Loss | 48 | RNU6-663P, MTND1P6, MTND2P32, RANP9, RNU6-528P, SNORD13, RN7SL621P, RN7SL457P, VENTXP5, LSM12P1, TTI2, MAK16, DUSP26, FUT10, RNF122, NRG1, UNC5D |
| Chr1: 17005967-17253362 | 247395 | 1356 | 1p36.13 | Loss | 46 | EIF1AXP1, FAM231C, RNU1-4, CROCCP4, MIR3675, RNU1-2, MST1L, ESPNP, CROCC |
| Chr15: 35085898-35540410 | 454512 | 2309 | 15q14 | Loss | 46 | ACTC1, NANOGP8, PRELID1P4, ZNF770, AQR, ANP32AP1, DPH6 |
| Chr4: 113427910-113740790 | 312880 | 1219 | 4q25 | Loss | 46 | NEUROG2, MIR302B, MIR367, MIR302D, MIR302A, MIR302C, WRBP1, RPL7AP30, LARP7, OSTCP4, C4orf21, ANK2 |
| Chr4: 165303804-166130292 | 826488 | 5907 | 4q32.3 | Loss | 46 | RNU6-284P, RNU6-668P, TRIM60P14, FAM218BP, NACA3P, FAM218A, TRIM61, TRIM60, TMEM192, KLHL2, MARCH1 |
| Chr22: 29192671-29455689 | 263018 | 1166 | 22q12.1 | Loss | 46 | C22orf31, XBP1, ZNRF3-IT1, ZNRF3-AS1, ZNRF3 |
| Chr17: 44267864-44276547 | 8683 | 56 | 17q21.31 | Loss | 44 | KANSL1-AS1, KANSL1 |
| Chr14:1-20456201 | 20456200 | 4929 | 14q11.2 | Loss | 44 | RNU6-458P, OR11H12, ARHGAP42P5, NF1P4, MED15P1, RNU6-1239P, GRAMD4P3, DUXAP10, OR11H13P, GRAMD4P4, RNU6-1268P, MED15P6, ARHGAP42P4, OR11H2, OR4Q3, OR4H12P, OR4M1, OR4N1P, OR4K3, OR4K2, OR4K4P, OR4K5, OR4K1, OR4K16P, OR4K15, POTEG, BMS1P17, BMS1P18, POTEM, OR4N2, OR11K2P, OR4K6P |
| Chr4: 128751602-129198401 | 446799 | 1425 | 4q28.2 | Loss | 44 | RNU6-583P, FOSL1P1, PLK4, C4orf29, PGRMC2, HSPA4L, MFSD8, LARP1B |
| Chr1: 152552808-152586527 | 33719 | 100 | 1q21.3 | Loss | 41 | LCE3D, LCE3C, LCE3B |
| Chr1: 22455143-22963470 | 508327 | 2714 | 1p36.12 | Loss | 41 | MIR4418, ZBTB40-IT1, C1QA, WNT4, EPHA8, ZBTB40 |
| Chr15: 20586675-20717373 | 130698 | 443 | 15q11.1 | Gain | 41 | HERC2P3 |
| Chr8: 7290942-7771549 | 480607 | 514 | 8p23.1 | Gain | 41 | DEFB104B, DEFB105B, PRR23D1, FAM90A6P, FAM90A7P, FAM90A22P, OR7E157P, OR7E154P, FAM90A14P, FAM90A16P, FAM90A8P, FAM90A17P, FAM90A19P, FAM90A9P, FAM90A10P, PRR23D2, DEFB107A, DEFB105A, DEFB104A, DEFB103A, DEFB4A, SPAG11B, DEFB107B, FAM90A21, FAM90A23P, FAM90A18P, DEFB106A, SPAG11, HSPD1P2, DEFB106B |
| Non-Responders (G0 and G1) (n = 17) | Partial Responders (G2) (n = 9) | Responders (G3 and G4) (n = 13) | q-Value | Total Cases (n = 39) | |
|---|---|---|---|---|---|
| 1p36.12 | |||||
| Deleted | 7 (39%) | 4 (44%) | 10 (75%) | <0.001 | 21 (54%) |
| 3q22 | |||||
| Deleted | 1 (7%) | 0 (0%) | 0 (0%) | <0.001 | 1 (3%) |
| 7q34 | |||||
| Deleted | 1 (7%) | 1 (11%) | 0 (0%) | <0.001 | 2 (5%) |
| 7q35 | |||||
| Deleted | 4 (22%) | 2 (22%) | 0 (0%) | 0.03 | 6 (15%) |
| 12p11.23 | |||||
| Deleted | 0 (0%) | 0 (0%) | 1 (8%) | 0.04 | 1 (2.5%) |
| 12p13.31 | |||||
| Deleted | 6 (33%) | 0 (0%) | 2 (17%) | 0.03 | 8 (21%) |
| 17q21.31 | |||||
| Deleted | 6 (33%) | 4 (44%) | 8 (58%) | <0.001 | 18 (46%) |
| Amplified | 7 (39%) | 1 (11%) | 8 (58%) | <0.001 | 16 (41%) |
| 20p12 | |||||
| Deleted | 2 (11%) | 4 (44%) | 5 (42%) | 0.001 | 11 (28%) |
| 22q12.1 | |||||
| Deleted | 5 (28%) | 5 (56%) | 9 (67%) | 0.04 | 19 (49%) |
| Variables | Importance Ranking by Method | Median Ranking | ||||
|---|---|---|---|---|---|---|
| Boruta | Xgboost | Relative Importance | DALEX | VITA | ||
| N2 | 2 | 7 | 1 | 3 | 1 | 2 |
| N1 | 4 | 5 | 4 | 4 | 3 | 4 |
| chr4q loss | 3 | 1 | 3 | 1 | 2 | 2 |
| chr15q11.1 gain | 5 | 4 | 2 | 2 | 5 | 4 |
| chr17q21.31 gain | 1 | 2 | 5 | 6 | 7 | 5 |
| chr15q14 loss | 7 | 3 | 7 | 5 | 6 | 6 |
| CEA | 6 | 6 | 6 | 7 | 4 | 6 |
| Algorithm | Parameters | Filtering Method | Nº of Variables | Hit Rate (%) | |||
|---|---|---|---|---|---|---|---|
| G0/G1 | G2 | G3/G4 | Global | ||||
| PLS | Number of factors: 3 | No | 7 | 40 | 100 | 67 | 60 |
| PLS | Number of factors: 2 | Yes | 4 | 80 | 0 | 0 | 40 |
| wSVM | Kernel: Sigmoid; gamma: 8; cost: 100 | No | 7 | 80 | 50 | 33 | 60 |
| wSVM | Kernel: polynomial; gamma: 0.25; cost: 100 | Yes | 4 | 0 | 0 | 67 | 20 |
| SVM | Kernel: Sigmoid; gamma: 0.25; cost: 0.001 | No | 7 | 100 | 0 | 0 | 50 |
| SVM | Kernel: polynomial; gamma: 0.25; cost: 0.001 | Yes | 4 | 100 | 0 | 0 | 50 |
| KNN | k neighbors: 23 | No | 7 | 100 | 0 | 0 | 50 |
| KNN | k neighbors: 23 | Yes | 4 | 100 | 0 | 0 | 50 |
| Random Forest | Number of trees: 2 | No | 7 | 40 | 100 | 67 | 60 |
| Random Forest | Number of trees: 2 | Yes | 4 | 60 | 0 | 33 | 40 |
| Validation Sample ID | Real Time and Event | Prediction at | ||||||
|---|---|---|---|---|---|---|---|---|
| DFS Censor | Time to DFS (Months) | 12 Months | 36 Months | 60 Months | ||||
| Probability of Absence of the Event | Success in Prediction? | Probability of Absence of the Event | Success in Prediction? | Probability of Absence of the Event | Success in Prediction? | |||
| 1 | 1 | 34 | 1 | YES | 0.8 | NO | 0.5 | YES |
| 2 | 1 | 18 | 1 | YES | 0.0 | YES | 0.0 | YES |
| 3 | 0 | 129 | 1 | YES | 0.9 | YES | 0.7 | YES |
| 4 | 1 | 109 | 1 | YES | 0.9 | YES | 0.9 | YES |
| 5 | 1 | 8 | 1 | NO | 0.0 | YES | 0.0 | YES |
| 6 | 0 | 54 | 1 | YES | 1.0 | YES | 1.0 | NC |
| 7 | 0 | 89 | 1 | YES | 1.0 | YES | 1.0 | YES |
| 8 | 0 | 86 | 1 | YES | 0.9 | YES | 0.8 | YES |
| 9 | 0 | 84 | 1 | YES | 1.0 | YES | 1.0 | YES |
| 10 | 0 | 110 | 1 | YES | 1.0 | YES | 0.9 | YES |
| Sucess rate | 90% | 90% | 100% | |||||
| Sensitivity | 0% | 67% | 100% | |||||
| Specificity | 100% | 100% | 100% | |||||
| Positive predictor value | NC | 100% | 100% | |||||
| Negative predictor value | 90% | 88% | 100% | |||||
| Validation Simple ID | Real Time and Event | Prediction at | ||||||
|---|---|---|---|---|---|---|---|---|
| OS Censor | Time to OS (Months) | 12 Months | 36 Months | 60 Months | ||||
| Probability of Absence of Event | Success in Prediction | Probability of Absence of the Event | Success in Prediction | Probability of Absence of Event | Success in Prediction | |||
| 1 | 1 | 52 | 0.9 | YES | 0.6 | YES | 0.3 | YES |
| 2 | 1 | 36 | 0.0 | NO | 0.0 | YES | 0.0 | YES |
| 3 | 0 | 129 | 1.0 | YES | 0.9 | YES | 0.8 | YES |
| 4 | 0 | 121 | 1.0 | YES | 0.9 | YES | 0.8 | YES |
| 6 | 1 | 17 | 0.0 | NO | 0.0 | YES | 0.0 | YES |
| 6 | 0 | 54 | 1.0 | YES | 1.0 | YES | 1.0 | NC |
| 7 | 1 | 89 | 1.0 | YES | 1.0 | YES | 1.0 | YES |
| 8 | 0 | 86 | 1.0 | YES | 0.9 | YES | 0.8 | YES |
| 9 | 0 | 84 | 1.0 | YES | 1.0 | YES | 1.0 | YES |
| 10 | 0 | 110 | 1.0 | YES | 0.9 | YES | 0.8 | YES |
| Success rate | NC | 100% | 100% | |||||
| Sensitivity | NC | 100% | 100% | |||||
| Specificity | NC | 100% | 100% | |||||
| Positive predictor value | NC | 100% | 100% | |||||
| Negative predictor value | NC | 100% | 100% | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
del Carmen, S.; Corchete, L.A.; González Velasco, C.; Sanz, J.; Alcazar, J.A.; García, J.; Rodríguez, A.I.; Vidal Tocino, R.; Rodriguez, A.; Pérez-Romasanta, L.A.; et al. High-Risk Clinicopathological and Genetic Features and Outcomes in Patients Receiving Neoadjuvant Radiochemotherapy for Locally Advanced Rectal Cancer. Cancers 2021, 13, 3166. https://doi.org/10.3390/cancers13133166
del Carmen S, Corchete LA, González Velasco C, Sanz J, Alcazar JA, García J, Rodríguez AI, Vidal Tocino R, Rodriguez A, Pérez-Romasanta LA, et al. High-Risk Clinicopathological and Genetic Features and Outcomes in Patients Receiving Neoadjuvant Radiochemotherapy for Locally Advanced Rectal Cancer. Cancers. 2021; 13(13):3166. https://doi.org/10.3390/cancers13133166
Chicago/Turabian Styledel Carmen, Sofía, Luís Antonio Corchete, Cristina González Velasco, Julia Sanz, José Antonio Alcazar, Jacinto García, Ana Isabel Rodríguez, Rosario Vidal Tocino, Alba Rodriguez, Luis Alberto Pérez-Romasanta, and et al. 2021. "High-Risk Clinicopathological and Genetic Features and Outcomes in Patients Receiving Neoadjuvant Radiochemotherapy for Locally Advanced Rectal Cancer" Cancers 13, no. 13: 3166. https://doi.org/10.3390/cancers13133166
APA Styledel Carmen, S., Corchete, L. A., González Velasco, C., Sanz, J., Alcazar, J. A., García, J., Rodríguez, A. I., Vidal Tocino, R., Rodriguez, A., Pérez-Romasanta, L. A., Sayagués, J. M., & Abad, M. (2021). High-Risk Clinicopathological and Genetic Features and Outcomes in Patients Receiving Neoadjuvant Radiochemotherapy for Locally Advanced Rectal Cancer. Cancers, 13(13), 3166. https://doi.org/10.3390/cancers13133166







