Metabolic Reprogramming by Malat1 Depletion in Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
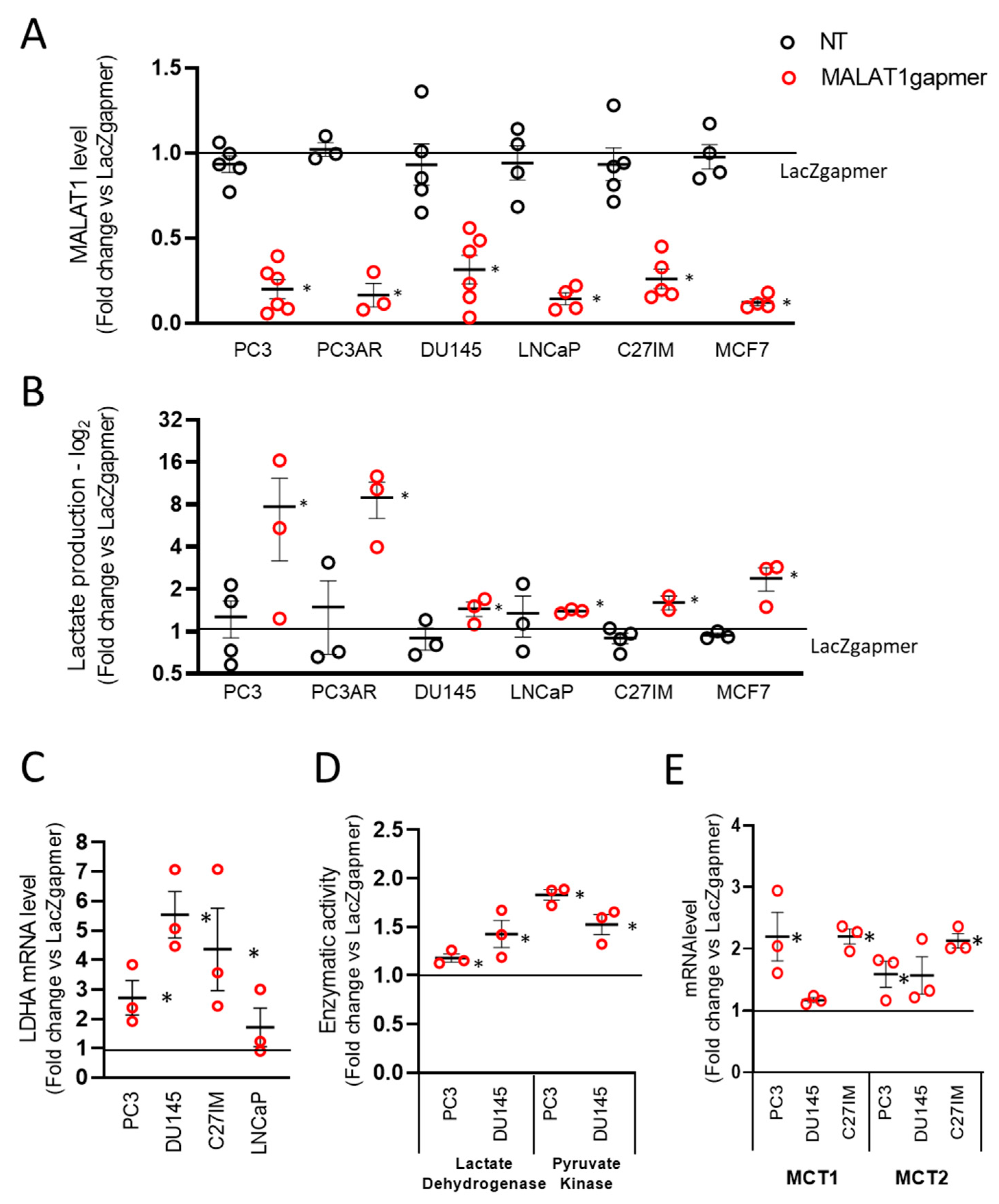
2.1. Metabolic Perturbation upon MALAT1 Depletion in Prostate Cancer Cell Lines
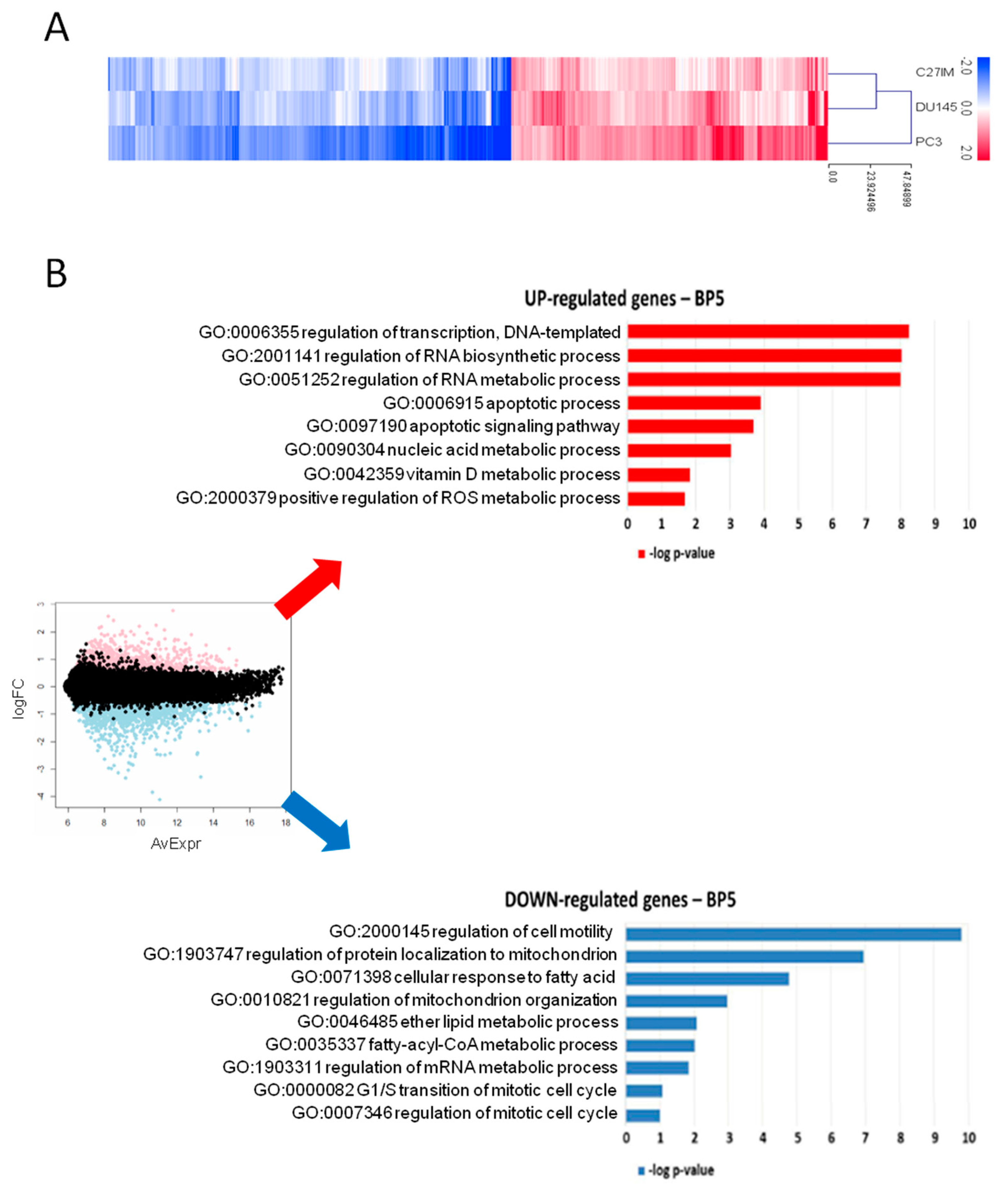
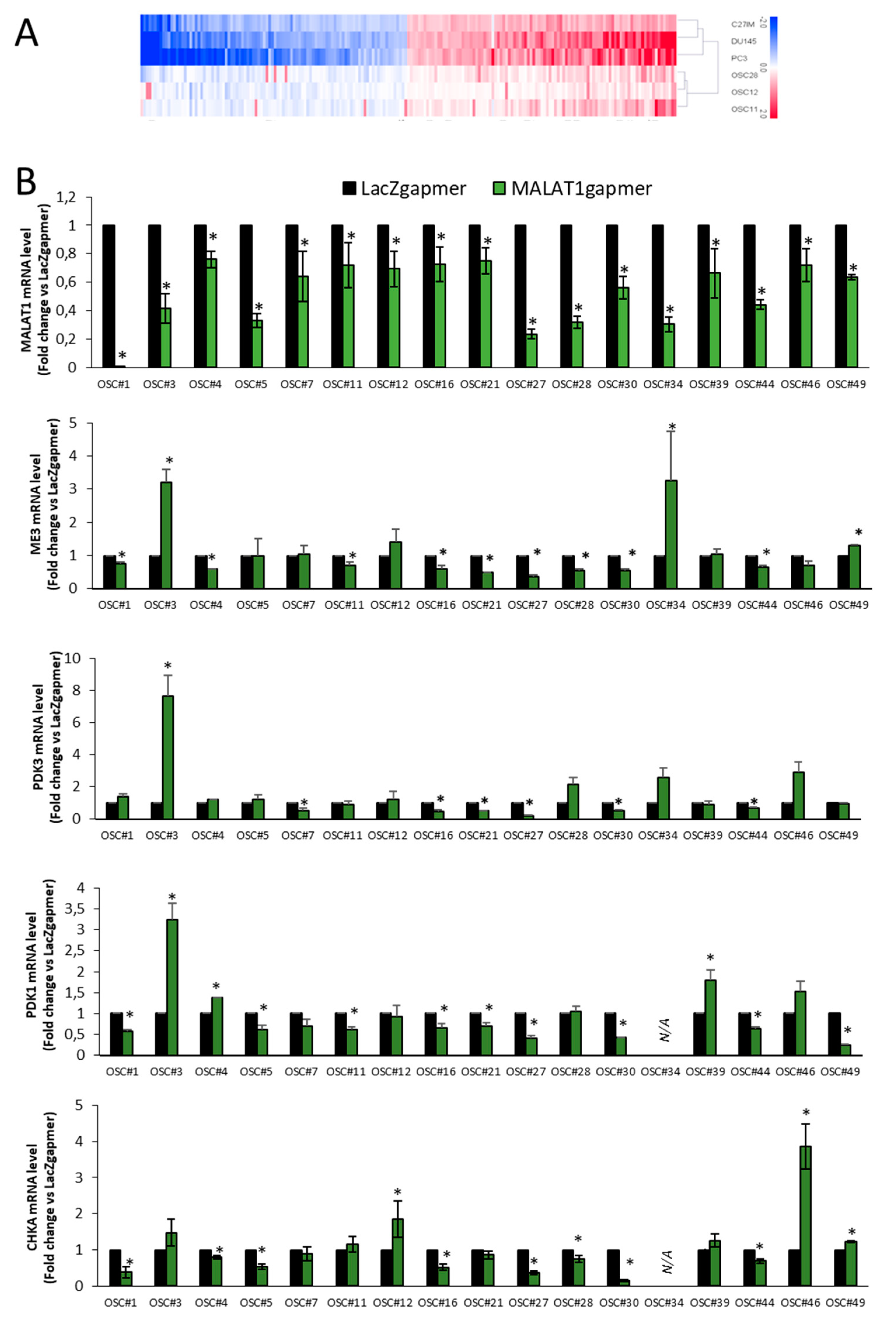
2.1.2. Gene Profiling after MALAT1 Depletion
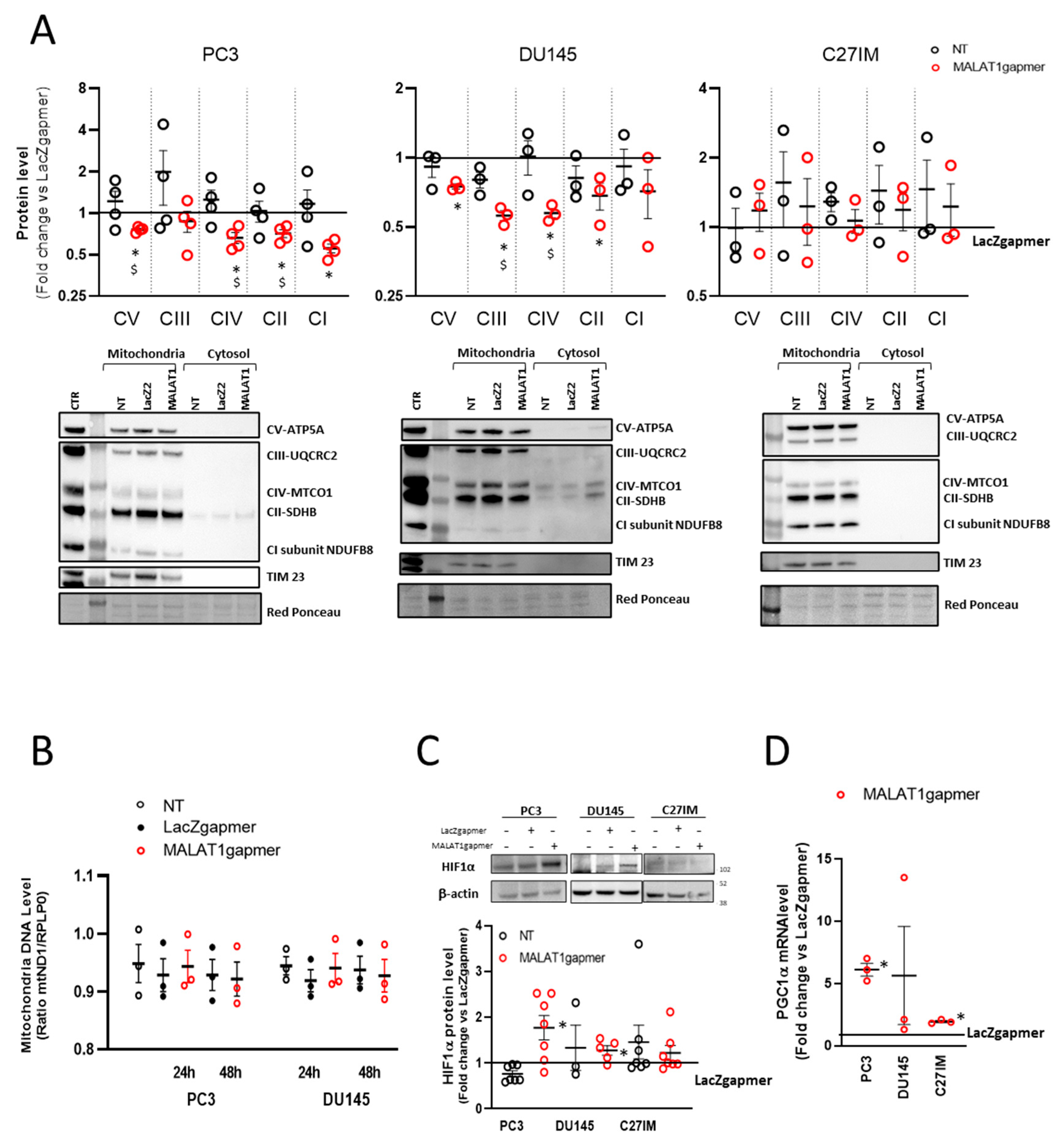
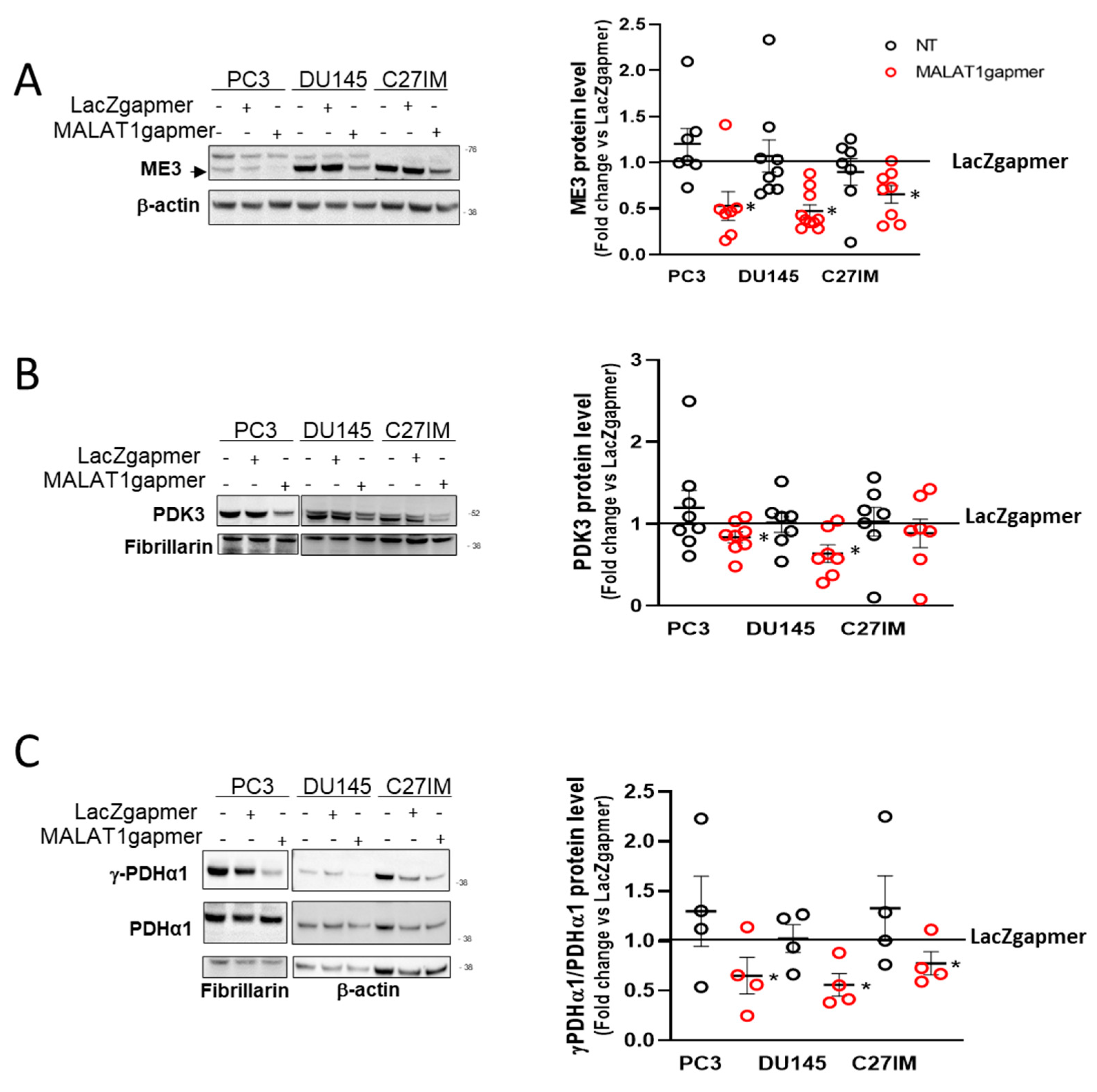
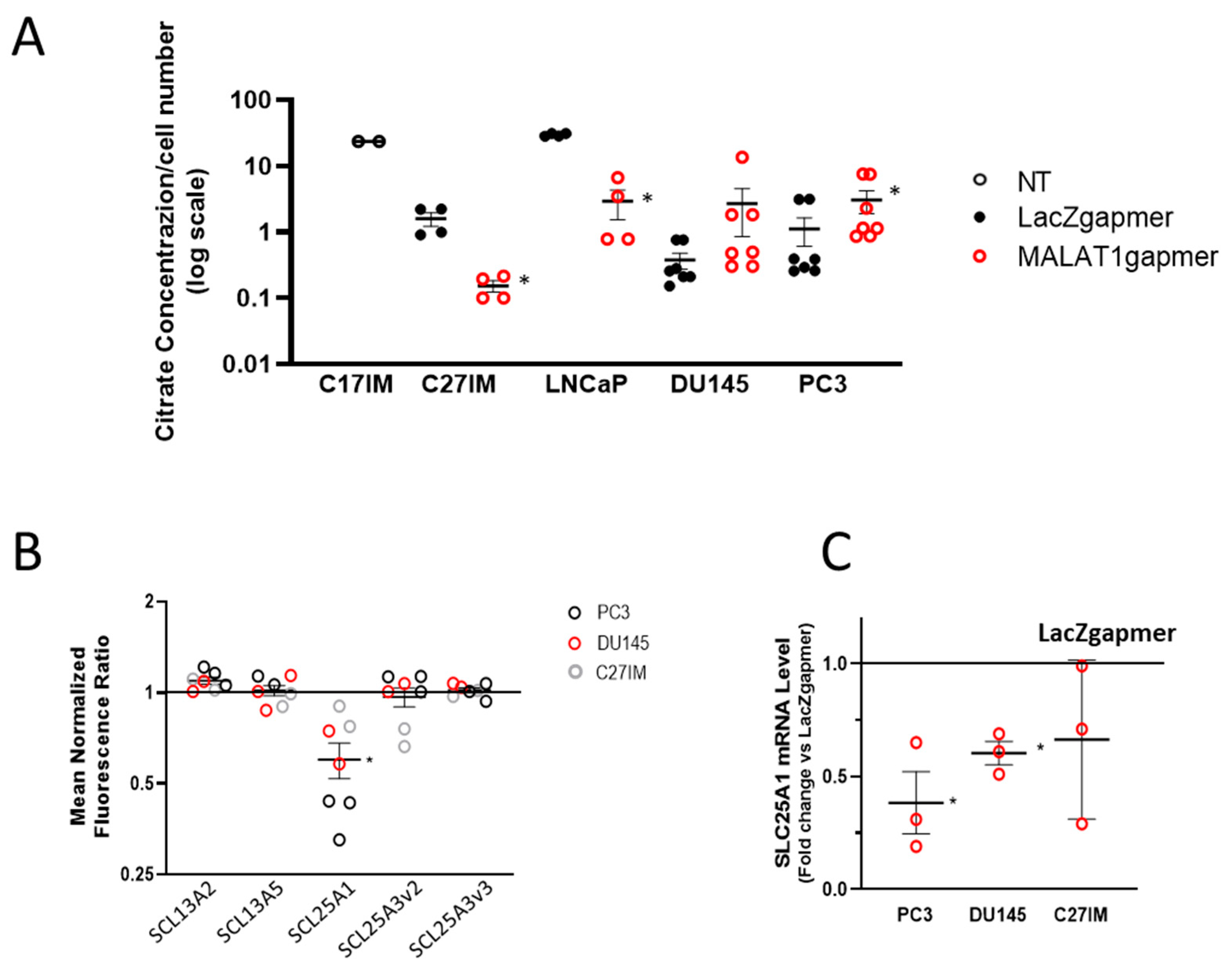
2.1.3. Expression of the Electron Transport-Chain Protein Complexes and Citrate Production in MALAT1-Depleted Metastatic Prostate Cancer Cell Lines
2.1.4. MALAT1 Knockdown Compromises the TCA Cycle
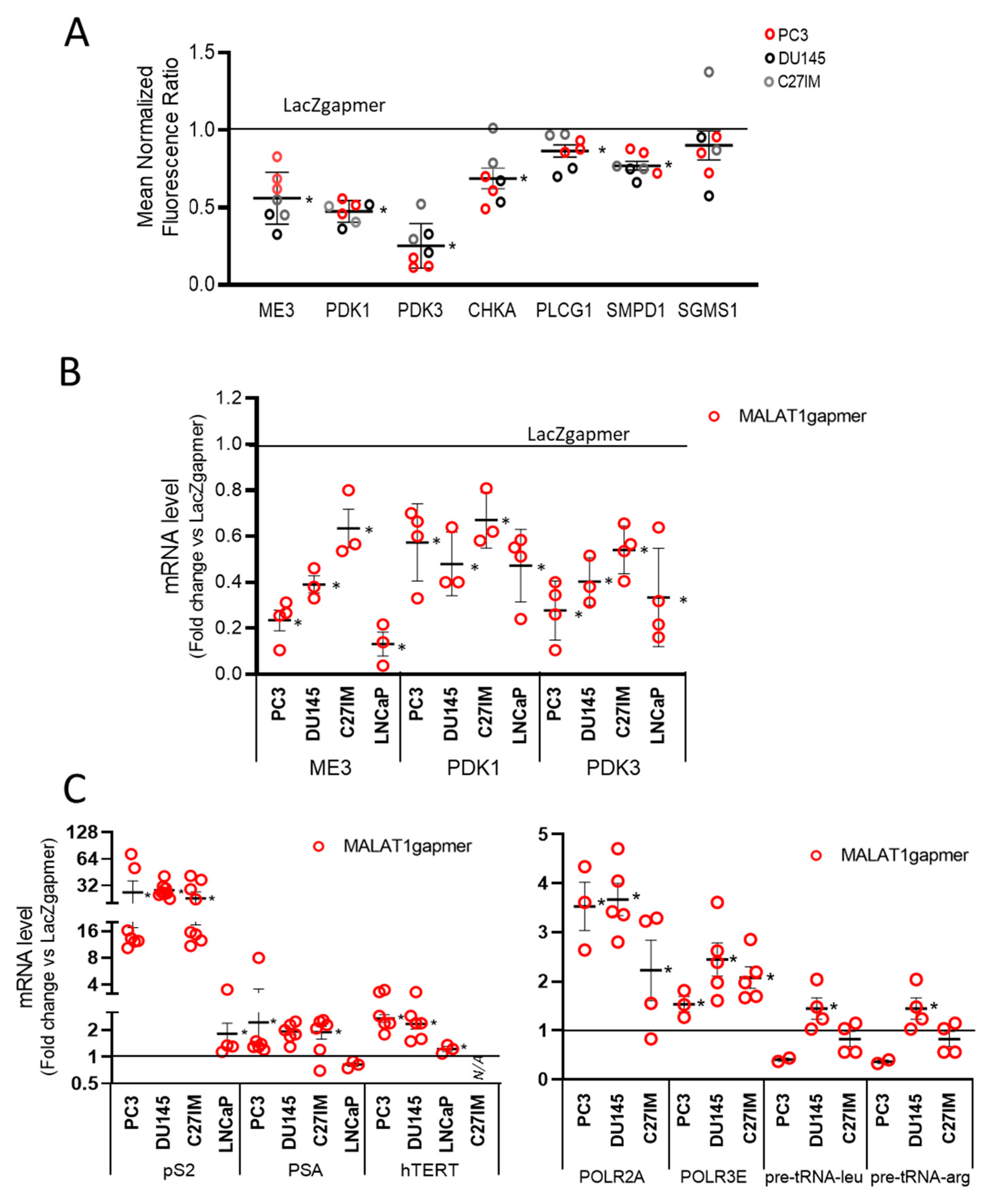
2.1.5. Silencing ME3 Increases Lactate Production
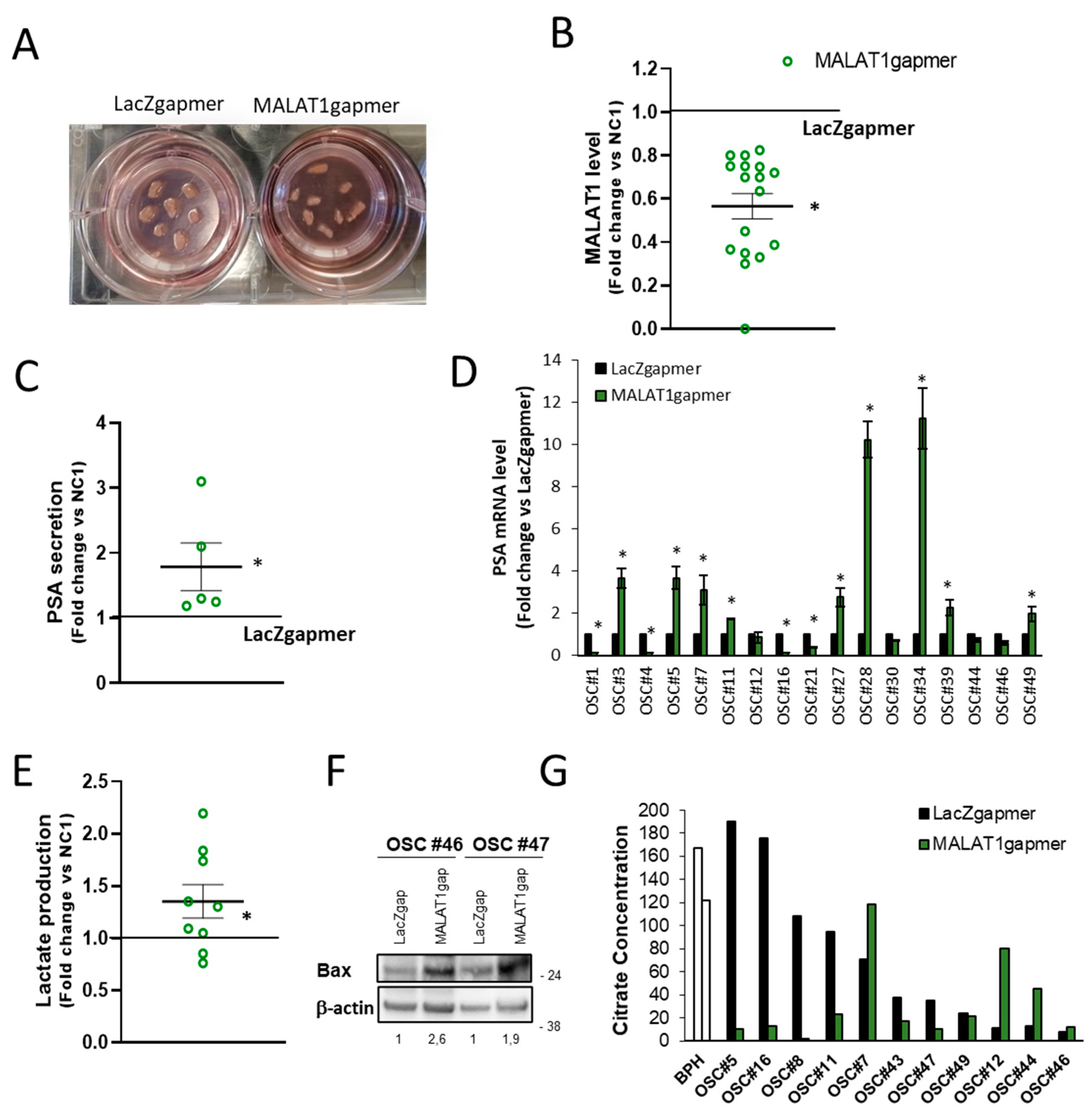
2.1.6. Consequences of MALAT1 Depletion in Human PCa-Derived Organotypic Slice Cultures (OSCs)
2.1.7. Validation of Arrays Data Set in OSCs
2.1.8. Partial Least Squares Discriminant Analysis (PLS_DA)
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef]
- Giunchi, F.; Fiorentino, M.; Loda, M. The Metabolic Landscape of Prostate Cancer. Eur. Urol. Oncol. 2019, 2, 28–36. [Google Scholar] [CrossRef]
- Cutruzzolà, F.; Giardina, G.; Marani, M.; Macone, A.; Paiardini, A.; Rinaldo, S.; Paone, A. Glucose Metabolism in the Progression of Prostate Cancer. Front. Physiol. 2017, 8, 97. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B. Concepts of citrate production and secretion by prostate. 1. Metabolic relationships. Prostate 1991, 18, 25–46. [Google Scholar] [CrossRef]
- Bader, D.A.; McGuire, S.E. Tumour metabolism and its unique properties in prostate adenocarcinoma. Nat. Rev. Urol. 2020, 17, 214–231. [Google Scholar] [CrossRef]
- Bose, S.; Le, A. Glucose Metabolism in Cancer. Adv. Exp. Med. Biol. 2018, 1063, 3–12. [Google Scholar] [CrossRef]
- Jadvar, H. Prostate Cancer. Methods Mol. Biol. 2011, 727, 265–290. [Google Scholar]
- De Paepe, B.; Lefever, S.; Mestdagh, P. How long noncoding RNAs enforce their will on mitochondrial activity: Regulation of mitochondrial respiration, reactive oxygen species production, apoptosis, and metabolic reprogramming in cancer. Curr. Genet. 2018, 64, 163–172. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Pinweha, P.; Rattanapornsompong, K.; Charoensawan, V.; Jitrapakdee, S. MicroRNAs and oncogenic transcriptional regulatory networks controlling metabolic reprogramming in cancers. Comput. Struct. Biotechnol. J. 2016, 14, 223–233. [Google Scholar] [CrossRef]
- Schulze, A.; Harris, A.L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nat. Cell Biol. 2012, 491, 364–373. [Google Scholar] [CrossRef]
- Li, X.; Zhao, Q.; Qi, J.; Wang, W.; Zhang, D.; Li, Z.; Qin, C. lnc RNA Ftx Promotes Aerobic Glycolysis and Tumor Progression through the PPARgamma Pathway in Hepatocellular Carcinoma. Int. J. Oncol. 2018, 53, 551–566. [Google Scholar]
- Liu, X.; Gan, B. lnc RNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle 2016, 15, 3471–3481. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, Q.; Liu, X.; Sun, Q.; Zhao, X.; Deng, R.; Wang, Y.; Huang, J.; Xu, M.; Yan, J.; et al. lnc RNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014, 281, 3766–3775. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA Gas5 Is a Growth Arrest- and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Ji, G.; Le, X.; Wang, C.; Xu, L.; Feng, M.; Zhang, Y.; Yang, H.; Xuan, Y.; Yang, Y.; et al. Long Noncoding RNA LINC00092 Acts in Cancer-Associated Fibroblasts to Drive Glycolysis and Progression of Ovarian Cancer. Cancer Res. 2017, 77, 1369–1382. [Google Scholar] [CrossRef]
- Luo, D. MALAT1-mediated tumorigenesis. Front. Biosci. 2017, 22, 66–80. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, X.; Hu, X.; Zhou, W.; Zhang, P.; Zhang, J.; Yang, S.; Liu, Y. The effects of lnc RNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol. Med. 2018, 24, 1–15. [Google Scholar] [CrossRef]
- Malakar, P.; Stein, I.; Saragovi, A.; Winkler, R.; Stern-Ginossar, N.; Berger, M.; Pikarsky, E.; Karni, R. Long Noncoding RNA MALAT1 Regulates Cancer Glucose Metabolism by Enhancing mTOR-Mediated Translation of TCF7L2. Cancer Res. 2019, 79, 2480–2493. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xia, Z.; Chen, C.; Hu, W.; Yuan, Y. Lnc RNA MALAT1 promotes epithelial-to-mesenchymal transition of esophageal cancer through Ezh2-Notch1 signaling pathway. Anti-Cancer Drugs 2018, 29, 767–773. [Google Scholar] [CrossRef]
- Bai, L.; Wang, A.; Zhang, Y.; Xu, X.; Zhang, X. Knockdown of MALAT1 enhances chemosensitivity of ovarian cancer cells to cisplatin through inhibiting the Notch1 signaling pathway. Exp. Cell Res. 2018, 366, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Chen, W.; Yuan, Z.; Liu, X.; Jiang, H. Lnc RNA-MALAT1 Contributes to the Cisplatin-Resistance of Lung Cancer by Upregulating MRP1 and MDR1 via STAT3 Activation. Biomed. Pharmacother. 2018, 101, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef]
- Chang, J.; Xu, W.; Du, X.; Hou, J. MALAT1 silencing suppresses prostate cancer progression by upregulating miR-1 and downregulating KRAS. Onco Targets Ther. 2018, 11, 3461–3473. [Google Scholar] [CrossRef]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zörnig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long Noncoding RNA MALAT1 Regulates Endothelial Cell Function and Vessel Growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef]
- Wang, X.; Li, M.; Wang, Z.; Han, S.; Tang, X.; Ge, Y.; Zhou, L.; Zhou, C.; Yuan, Q.; Yang, M. Silencing of Long Noncoding RNA MALAT1 by miR-101 and miR-217 Inhibits Proliferation, Migration, and Invasion of Esophageal Squamous Cell Carcinoma Cells. J. Biol. Chem. 2015, 290, 3925–3935. [Google Scholar] [CrossRef]
- Asim, M.; Massie, C.E.; Orafidiya, F.; Pértega-Gomes, N.; Warren, A.Y.; Esmaeili, M.; Selth, L.A.; Zecchini, H.I.; Luko, K.; Qureshi, A.; et al. Choline Kinase Alpha as an Androgen Receptor Chaperone and Prostate Cancer Therapeutic Target. J. Natl. Cancer Inst. 2016, 108, djv371. [Google Scholar] [CrossRef]
- Austin, S.; St-Pierre, J. PGC1 and mitochondrial metabolism—Emerging concepts and relevance in ageing and neurodegenerative disorders. J. Cell Sci. 2012, 125, 4963–4971. [Google Scholar] [CrossRef]
- Nanni, S.; Priolo, C.; Grasselli, A.; D’Eletto, M.; Merola, R.; Moretti, F.; Gallucci, M.; De Carli, P.; Sentinelli, S.; Cianciulli, A.M.; et al. Epithelial-Restricted Gene Profile of Primary Cultures from Human Prostate Tumors: A Molecular Approach to Predict Clinical Behavior of Prostate Cancer. Mol. Cancer Res. 2006, 4, 79–92. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Majd, H.; King, M.S.; Smith, A.C.; Kunji, E.R.S. Pathogenic mutations of the human mitochondrial citrate carrier SLC25A1 lead to impaired citrate export required for lipid, dolichol, ubiquinone and sterol synthesis. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pop, A.; Williams, M.; Struys, E.A.; Monné, M.; Jansen, E.E.W.; De Grassi, A.; Kanhai, W.A.; Scarcia, P.; Ojeda, M.R.F.; Porcelli, V.; et al. An overview of combined D-2- and L-2-hydroxyglutaric aciduria: Functional analysis of CIC variants. J. Inherit. Metab. Dis. 2018, 41, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, M.P.; Prasad, P.D.; Gopal, E.; Fraser, S.P.; Bolt, L.; Rizaner, N.; Palmer, C.P.; Foster, C.S.; Palmieri, F.; Ganapathy, V.; et al. Molecular origin of plasma membrane citrate transporter in human prostate epithelial cells. EMBO Rep. 2010, 11, 431–437. [Google Scholar] [CrossRef]
- González-Manchón, C.; Ferrer, M.; Ayuso-Parrilla, M.S.; Parrilla, R. Cloning, sequencing and functional expression of a cDNA encoding a NADP-dependent malic enzyme from human liver. Gene 1995, 159, 255–260. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, J.; Tan, X.P.; Zhao, Q. Effects of ME3 on the proliferation, invasion and metastasis of pancreatic cancer cells through epithelial-mesenchymal transition. Neoplasma 2019, 66, 896–907. [Google Scholar] [CrossRef]
- Aiello, A.; Bacci, L.; Re, A.; Ripoli, C.; Pierconti, F.; Pinto, F.; Masetti, R.; Grassi, C.; Gaetano, C.; Bassi, P.F.; et al. MALAT1 and HOTAIR Long Non-Coding RNAs Play Opposite Role in Estrogen-Mediated Transcriptional Regulation in Prostate Cancer Cells. Sci. Rep. 2016, 6, 38414. [Google Scholar] [CrossRef]
- Bacci, L.; Aiello, A.; Ripoli, C.; Loria, R.; Pugliese, D.; Pierconti, F.; Rotili, D.; Strigari, L.; Pinto, F.; Bassi, P.F.; et al. H19-Dependent Transcriptional Regulation of β3 and β4 Integrins Upon Estrogen and Hypoxia Favors Metastatic Potential in Prostate Cancer. Int. J. Mol. Sci. 2019, 20, 4012. [Google Scholar] [CrossRef]
- Amé, J.-C.; Spenlehauer, C.; De Murcia, G. The PARP superfamily. BioEssays 2004, 26, 882–893. [Google Scholar] [CrossRef]
- Ruopp, M.D.; Perkins, N.J.; Whitcomb, B.W.; Schisterman, E.F. Youden Index and Optimal Cut-Point Estimated from Observations Affected by a Lower Limit of Detection. Biom. J. 2008, 50, 419–430. [Google Scholar] [CrossRef]
- Roberts, W.B.; Han, M. Clinical significance and treatment of biochemical recurrence after definitive therapy for localized prostate cancer. Surg. Oncol. 2009, 18, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Partin, A.W.; Pound, C.R.; Epstein, J.I.; Walsh, P.C. LONG-TERM BIOCHEMICAL DISEASE-FREE AND CANCER-SPECIFIC SURVIVAL FOLLOWING ANATOMIC RADICAL RETROPUBIC PROSTATECTOMY. Urol. Clin. North Am. 2001, 28, 555–565. [Google Scholar] [CrossRef]
- Pang, E.-J.; Yang, R.; Fu, X.-B.; Liu, Y.-F. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumor Biol. 2015, 36, 2403–2407. [Google Scholar] [CrossRef]
- Ren, S.; Liu, Y.; Xu, W.; Sun, Y.; Lu, J.; Wang, F.; Wei, M.; Shen, J.; Hou, J.; Gao, X.; et al. Long Noncoding RNA MALAT-1 is a New Potential Therapeutic Target for Castration Resistant Prostate Cancer. J. Urol. 2013, 190, 2278–2287. [Google Scholar] [CrossRef] [PubMed]
- Zadra, G.; Loda, M. Metabolic Vulnerabilities of Prostate Cancer: Diagnostic and Therapeutic Opportunities. Cold Spring Harb. Perspect. Med. 2018, 8, a030569. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.P.; Nóbrega-Pereira, S.; Domingues-Silva, B.; Rebelo, K.; Alves-Vale, C.; Marinho, S.P.; Carvalho, T.; Dias, S.; De Jesus, B.B. An antisense transcript mediates MALAT1 response in human breast cancer. BMC Cancer 2019, 19, 1–11. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef]
- Costello, L.; Franklin, R. The Intermediary Metabolism of the Prostate: A Key to Understanding the Pathogenesis and Progression of Prostate Malignancy. Oncology 2000, 59, 269–282. [Google Scholar] [CrossRef]
- Sugden, M.C.; Holness, M.J. Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am. J. Physiol. Metab. 2003, 284, E855–E862. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Locasale, J.W.; Swanson, K.D.; Sharfi, H.; Heffron, G.J.; Amador-Noguez, D.; Christofk, H.R.; Wagner, G.; Rabinowitz, J.D.; Asara, J.M.; et al. Evidence for an Alternative Glycolytic Pathway in Rapidly Proliferating Cells. Science 2010, 329, 1492–1499. [Google Scholar] [CrossRef]
- Kelly, R.S.; Heiden, M.G.V.; Giovannucci, E.L.; Mucci, L.A. Metabolomic Biomarkers of Prostate Cancer: Prediction, Diagnosis, Progression, Prognosis, and Recurrence. Cancer Epidemiol. Biomarkers Prev. 2016, 25, 887–906. [Google Scholar] [CrossRef]
- Matheson, B.K.; Adams, J.L.; Zou, J.; Patel, R.; Franklin, R.B. Effect of metabolic inhibitors on ATP and citrate content in PC3 prostate cancer cells. Prostate 2007, 67, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Amorini, A.M.; Lazzarino, G.; Di Pietro, V.; Signoretti, S.; Lazzarino, G.; Belli, A.; Tavazzi, B. Metabolic, enzymatic and gene involvement in cerebral glucose dysmetabolism after traumatic brain injury. Biochim. Biophys. Acta 2016, 1862, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Hlouschek, J.; Hansel, C.; Jendrossek, V.; Matschke, J. The Mitochondrial Citrate Carrier (SLC25A1) Sustains Redox Homeostasis and Mitochondrial Metabolism Supporting Radioresistance of Cancer Cells With Tolerance to Cycling Severe Hypoxia. Front. Oncol. 2018, 8, 170. [Google Scholar] [CrossRef]
- Fernandez, H.R.; Gadre, S.M.; Tan, M.; Graham, G.; Mosaoa, R.; Ongkeko, M.S.; Kim, K.A.; Riggins, R.B.; Parasido, E.; Petrini, I.; et al. The mitochondrial citrate carrier, SLC25A1, drives stemness and therapy resistance in non-small cell lung cancer. Cell Death Differ. 2018, 25, 1239–1258. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; Ungaro, P.; Cimmino, A.; Lucarelli, G.; Busetto, G.M.; Cantiello, F.; Damiano, R.; Terracciano, D. Epigenetic Signature: A New Player as Predictor of Clinically Significant Prostate Cancer (PCa) in Patients on Active Surveillance (AS). Int. J. Mol. Sci. 2017, 18, 1146. [Google Scholar] [CrossRef]
- Lincoln, C.K.; Gabridge, M.G. Chapter 4 Cell Culture Contamination: Sources, Consequences, Prevention, and Elimination. Methods Cell Biol. 1998, 57, 49–65. [Google Scholar] [CrossRef]
- Matsui, M.; Corey, D.R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef]
- Khattar, E.; Kumar, P.; Liu, C.Y.; Akıncılar, S.C.; Raju, A.; Lakshmanan, M.; Maury, J.J.P.; Qiang, Y.; Li, S.; Tan, E.Y.; et al. Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J. Clin. Investig. 2016, 126, 4045–4060. [Google Scholar] [CrossRef]
- Artiss, J.D.; Karcher, R.E.; Cavanagh, K.T.; Collins, S.L.; Peterson, V.J.; Varma, S.; Zak, B. A Liquid-Stable Reagent for Lactic Acid Levels. Application to the Hitachi 911 and Beckman CX7. Am. J. Clin. Pathol. 2000, 114, 139. [Google Scholar] [CrossRef] [PubMed]
- Amorini, A.M.; Nociti, V.; Petzold, A.; Gasperini, C.; Quartuccio, E.; Lazzarino, G.; Di Pietro, V.; Belli, A.; Signoretti, S.; Vagnozzi, R.; et al. Serum lactate as a novel potential biomarker in multiple sclerosis. Biochim. Biophys. Acta 2014, 1842, 1137–1143. [Google Scholar] [CrossRef] [PubMed]
- Fresta, C.G.; Chakraborty, A.; Wijesinghe, M.B.; Amorini, A.M.; Lazzarino, G.; Lazzarino, G.; Tavazzi, B.; Lunte, S.M.; Caraci, F.; Ghazvini, S.; et al. Non-toxic engineered carbon nanodiamond concentrations induce oxidative/nitrosative stress, imbalance of energy metabolism, and mitochondrial dysfunction in microglial and alveolar basal epithelial cells. Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bregman, A. Laboratory Investigations in Cell and Molecular Biology, 4th ed.; Wiley: New York, NY, USA, 2002. [Google Scholar]
- Ostano, P.; Mello-Grand, M.; Sesia, D.; Gregnanin, I.; Peraldo-Neia, C.; Guana, F.; Jachetti, E.; Farsetti, A.; Chiorino, G. Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 1078. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Re, A.; Colussi, C.; Nanni, S.; Aiello, A.; Bacci, L.; Grassi, C.; Pontecorvi, A.; Farsetti, A. Nucleoporin 153 regulates estrogen-dependent nuclear translocation of endothelial nitric oxide synthase and estrogen receptor beta in prostate cancer. Oncotarget 2018, 9, 27985–27997. [Google Scholar] [CrossRef]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef]


| Patient (OSC#) | Age | PSA (ng/mL) | Gleason Score | Pathologic Stage | Recurrence | Time of Recurrence (mo) |
|---|---|---|---|---|---|---|
| 1 * | 66 | 11.98 | 7 (4 + 3) | T2c Nx Mx | - | - |
| 2 | 58 | 5 | 7 (3 + 4) | T2c Nx Mx | - | - |
| 3 * | 73 | 15.75 | 7 (3 + 4) | T3a N0 Mx | yes | 7 |
| 4 * | 56 | 2.15 | 7 (4 + 3) | T3a N0 Mx | yes | 6 |
| 5 * | 68 | 8.32 | 7 (4 + 3) | T2c Nx Mx | yes | 8 |
| 6 | 73 | 12.8 | 7 (3 + 4) | T2c Nx Mx | - | - |
| 7 * | 69 | 4.72 | 7 (4 + 3) | T3b N0 Mx | yes | 6 |
| 8 | 71 | 14 | 7 (3 + 4) | T2c Nx Mx | - | - |
| 9 | 69 | 18.5 | 7 (3 + 4) | T3a N0 Mx | yes | 17 |
| 10 | 75 | 10.25 | 7 (3 + 4) | T2c Nx Mx | - | - |
| 11 * | 65 | 11 | 7 (4 + 3) | T3b N0 Mx | yes | 31 |
| 12 * | 64 | 1.79 | 7 (3 + 4) | T2c Nx Mx | - | - |
| 13 | 55 | 6.12 | 6 (3 + 3) | T2 Nx Mx | - | - |
| 14 | 69 | 7.36 | 6 (3 + 3) | T2c Nx Mx | - | - |
| 15 | 60 | 5.9 | 7 (4 + 3) | T2c N0 Mx | - | - |
| 16 * | 67 | 7.9 | 7 (3 + 4) | T2a Nx Mx | - | - |
| 17 | 74 | 10.88 | 7 (4 + 3) | T2c N0 Mx | yes | 7 |
| 18 | 74 | 6.2 | 7 (4 + 3) | T2c N0 Mx | - | - |
| 19 | 63 | 4.7 | 6 (3 + 3) | T2c Nx Mx | - | - |
| 20 | 75 | 4.88 | 7 (3 + 4) | T3a N0 Mx | - | - |
| 21 * | 59 | 6.8 | 6 (3 + 3) | T2c Nx Mx | - | - |
| 22 | 60 | 4.9 | 6 (3 + 3) | T2c Nx Mx | - | - |
| 23 | 75 | 6.3 | 7 (4 + 3) | T2c Nx Mx | yes | 5 |
| 24 | 69 | 4.38 | 7 (3 + 4) | T2c N0 Mx | - | - |
| 25 | 66 | 5.28 | 8 (4 + 4) | T2b Nx Mx | - | - |
| 26 | 57 | 5.8 | 7 (3 + 4) | T2c Nx Mx | - | - |
| 27 * | 57 | 5.9 | 7 (3 + 4) | T2c Nx Mx | - | - |
| 28 * | 59 | 8.19 | 9 (4 + 5) | T2c N0 Mx | yes | 9 |
| 29 | 72 | 15.04 | 7 (4 + 3) | T2 N0 Mx | - | - |
| 30 * | 75 | 11.48 | 7 (3 + 4) | T3a N0 Mx | yes | 13 |
| 31 | 75 | 16.5 | 7 (3 + 4) | T2c N0 Mx | - | - |
| 32 | 69 | 7.4 | 7 (4 + 3) | T3a N0 Mx | - | - |
| 33 | 63 | 5.36 | 7 (3 + 4) | T2c Nx Mx | - | - |
| 34 * | 70 | 18.04 | 7 (3 + 4) | T2c N0 Mx | - | - |
| 35 | 68 | 5.75 | 7 (4 + 3) | T3b N0 Mx | yes | 6 |
| 36 | 63 | 14 | 7 (3 + 4) | T2c N0 Mx | - | - |
| 37 | 61 | 8.5 | 7 (4 + 3) | T2c Nx Mx | - | 5 |
| 38 | 57 | 18.69 | 7 (3 + 4) | T2c N0 Mx | - | - |
| 39 * | 55 | 6 | 7 (4 + 3) | T3b N0 Mx | yes | 6 |
| 40 | 74 | 8.4 | 7 (3 + 4) | T2c Nx Mx | - | - |
| 41 | 69 | 6 | 7 (3 + 4) | T2c Nx Mx | yes | 7 |
| 42 | 78 | 15 | 7 (4 + 3) | T3a Nx Mx | - | - |
| 43 | 67 | 5.5 | 7 (3 + 4) | T2c N0 Mx | - | - |
| 44 * | 67 | 13.5 | 7 (4 + 3) | T2c N0 Mx | - | - |
| 45 | 69 | 11.3 | 9 (4 + 5) | T3b N0 Mx | yes | 7 |
| 46 * | 70 | 8.68 | 7 (3 + 4) | T2c N0 Mx | - | - |
| 47 | 66 | 8.5 | 7 (4 + 3) | T3b N0 Mx | yes | 5 |
| 48 | 61 | 18 | 7 (4 + 3) | T3a N0 Mx | - | - |
| 49 * | 65 | 6.6 | 7 (4 + 3) | T2c N0 Mx | - | - |
| 50 | 69 | 11 | 7 (3 + 4) | T2c Nx Mx | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nanni, S.; Aiello, A.; Salis, C.; Re, A.; Cencioni, C.; Bacci, L.; Pierconti, F.; Pinto, F.; Ripoli, C.; Ostano, P.; et al. Metabolic Reprogramming by Malat1 Depletion in Prostate Cancer. Cancers 2021, 13, 15. https://doi.org/10.3390/cancers13010015
Nanni S, Aiello A, Salis C, Re A, Cencioni C, Bacci L, Pierconti F, Pinto F, Ripoli C, Ostano P, et al. Metabolic Reprogramming by Malat1 Depletion in Prostate Cancer. Cancers. 2021; 13(1):15. https://doi.org/10.3390/cancers13010015
Chicago/Turabian StyleNanni, Simona, Aurora Aiello, Chiara Salis, Agnese Re, Chiara Cencioni, Lorenza Bacci, Francesco Pierconti, Francesco Pinto, Cristian Ripoli, Paola Ostano, and et al. 2021. "Metabolic Reprogramming by Malat1 Depletion in Prostate Cancer" Cancers 13, no. 1: 15. https://doi.org/10.3390/cancers13010015
APA StyleNanni, S., Aiello, A., Salis, C., Re, A., Cencioni, C., Bacci, L., Pierconti, F., Pinto, F., Ripoli, C., Ostano, P., Baroni, S., Lazzarino, G., Tavazzi, B., Pugliese, D., Bassi, P., Grassi, C., Panunzi, S., Chiorino, G., Pontecorvi, A., ... Farsetti, A. (2021). Metabolic Reprogramming by Malat1 Depletion in Prostate Cancer. Cancers, 13(1), 15. https://doi.org/10.3390/cancers13010015











