Comprehensive Map of the Regulated Cell Death Signaling Network: A Powerful Analytical Tool for Studying Diseases
Abstract
1. Introduction
2. Results
2.1. General Characteristics of Regulated Cell Death Map
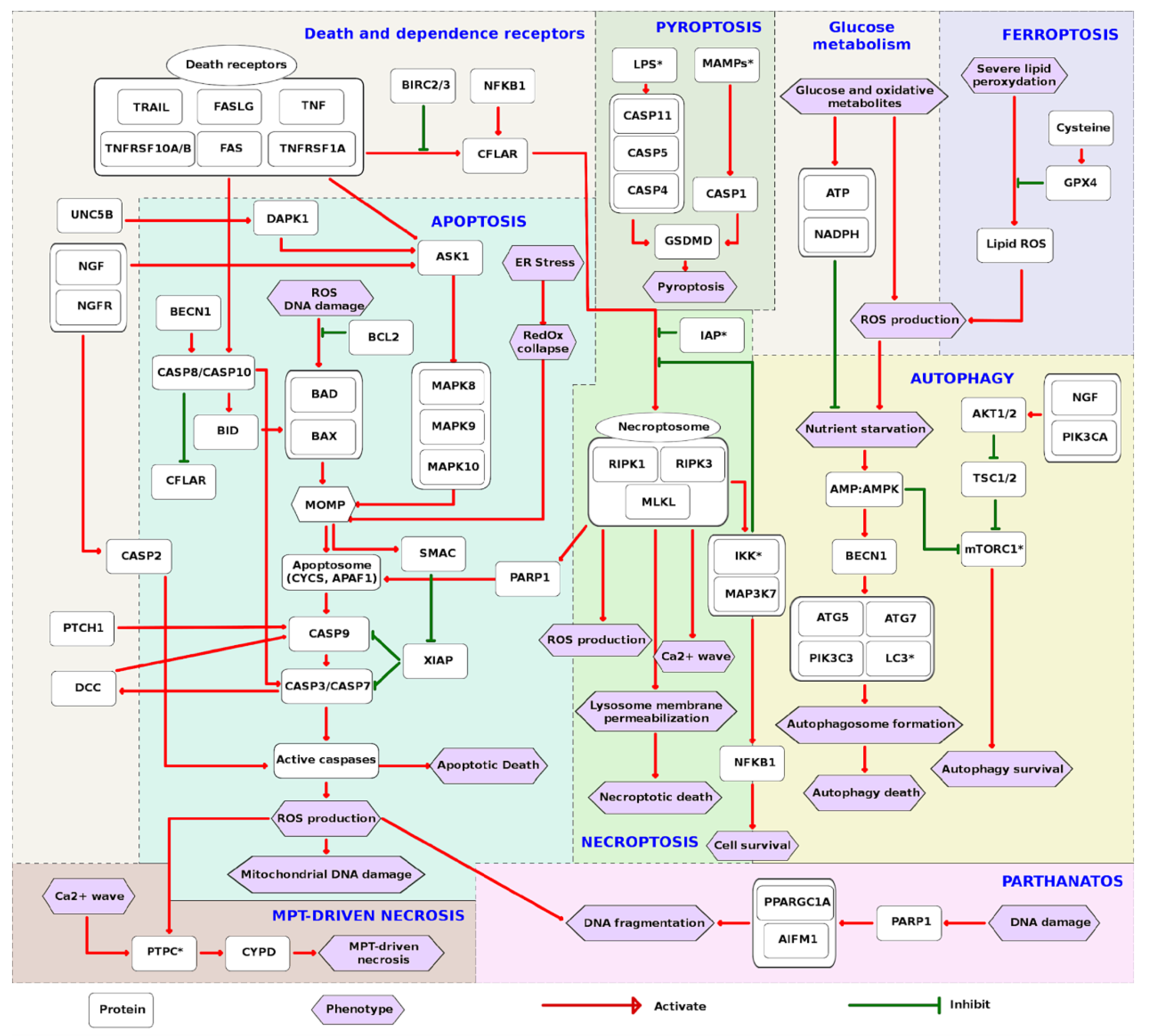
2.2. Structure of the Regulated Cell Death Map
2.3. Contents of the RCD Map
2.4. The Interplay between Modes of Cell Death and Switch Points
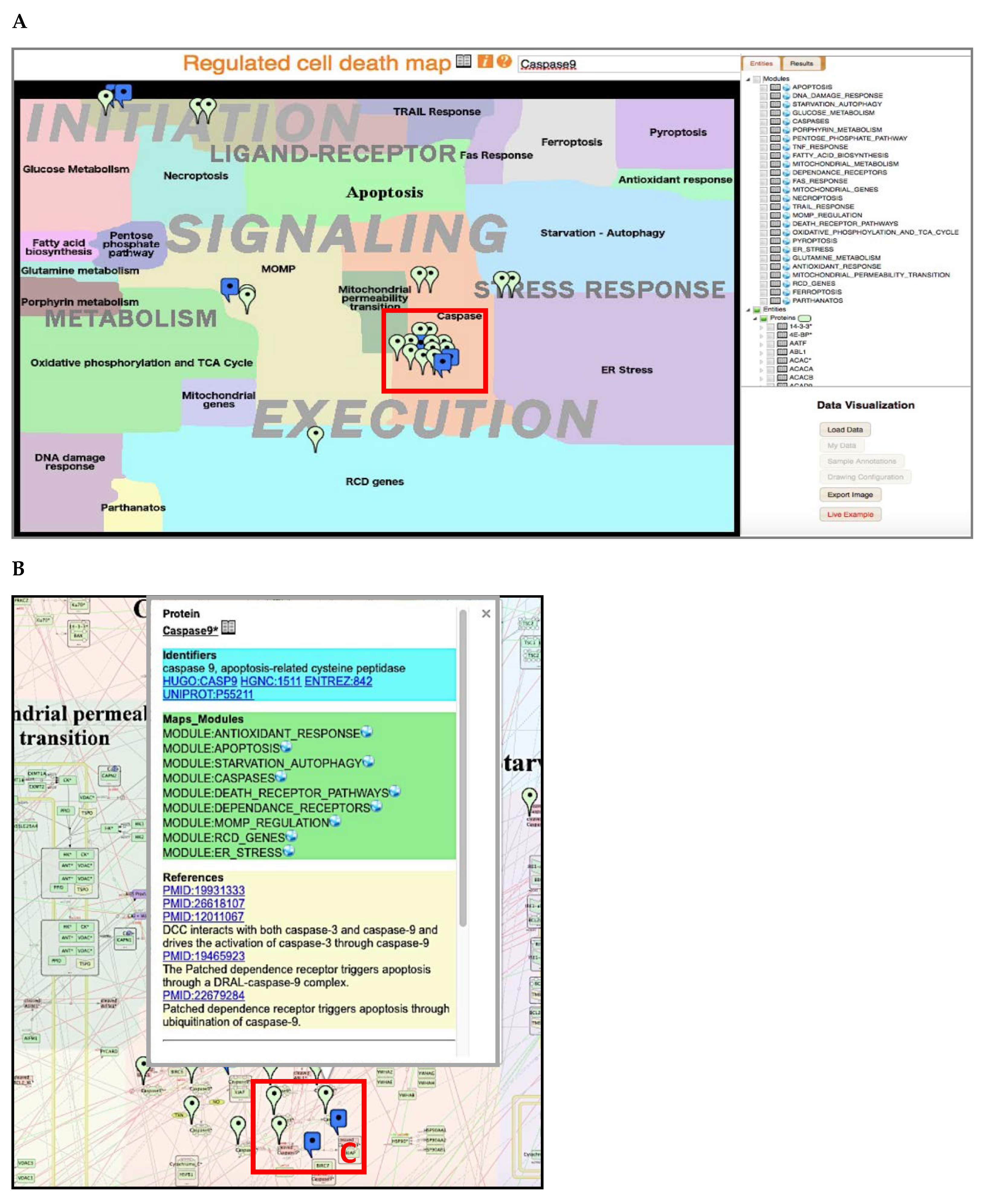
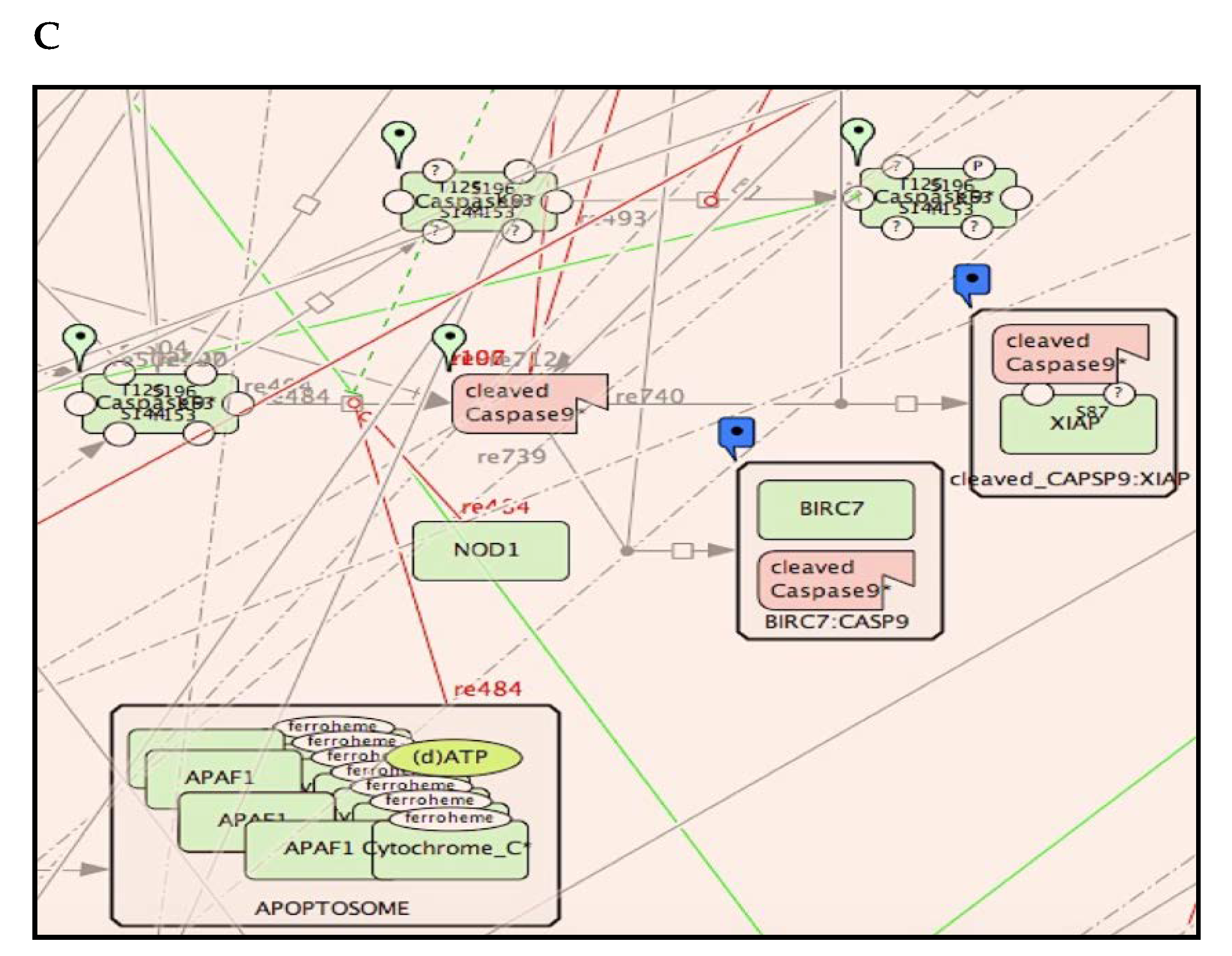
2.5. Access and Navigation of RCD Map
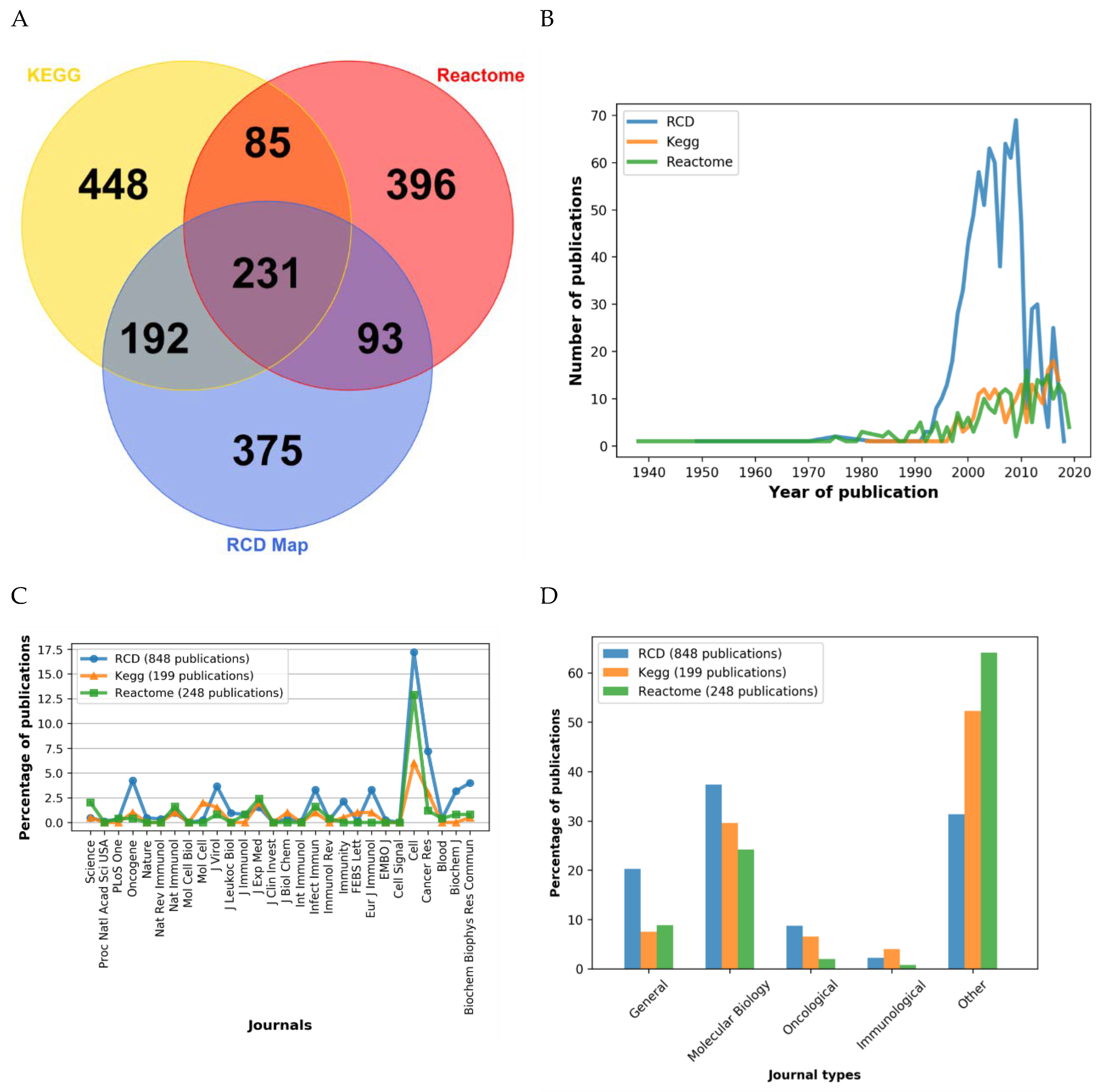
2.6. Comparison of RCD Map with Similar Pathway Resources
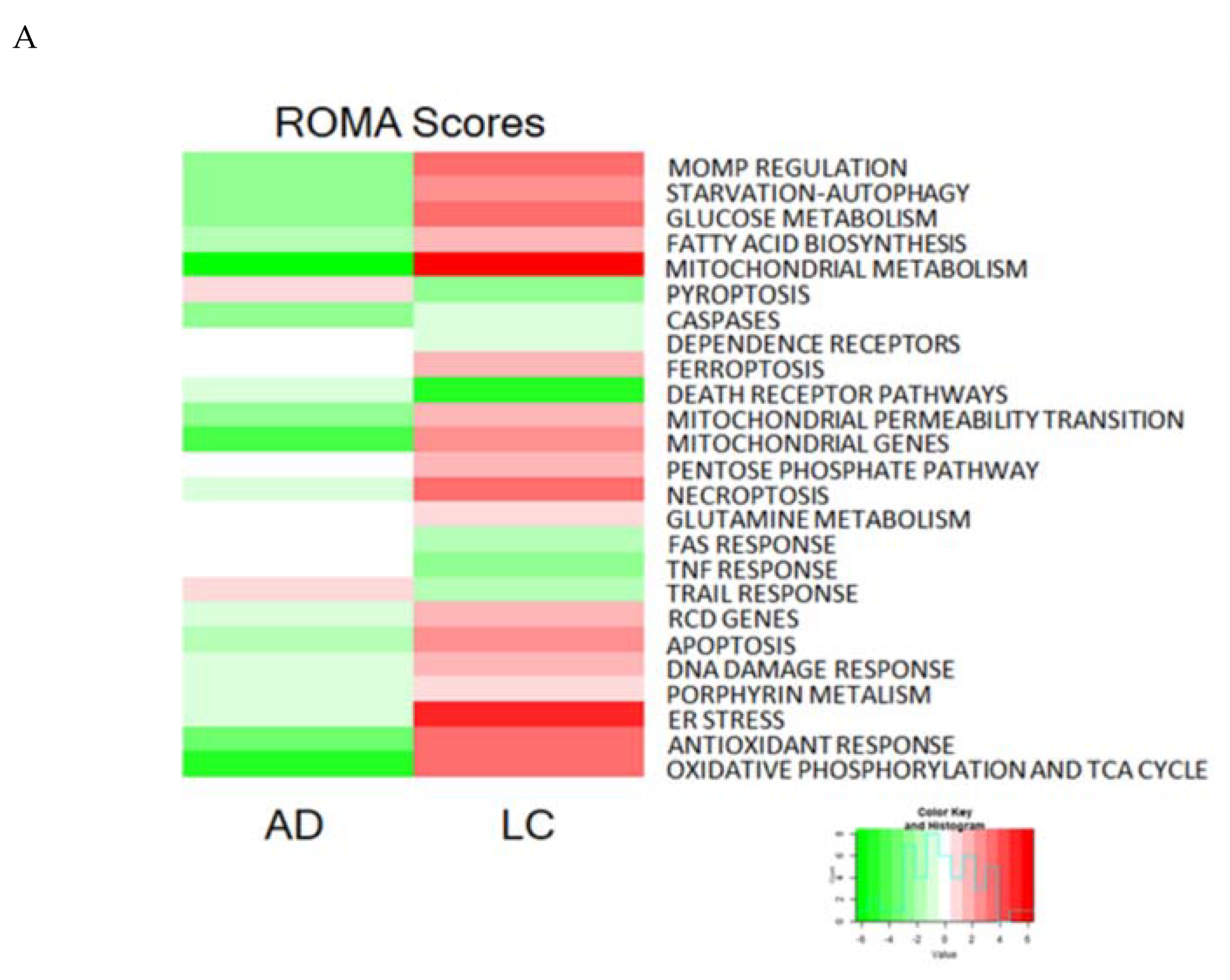
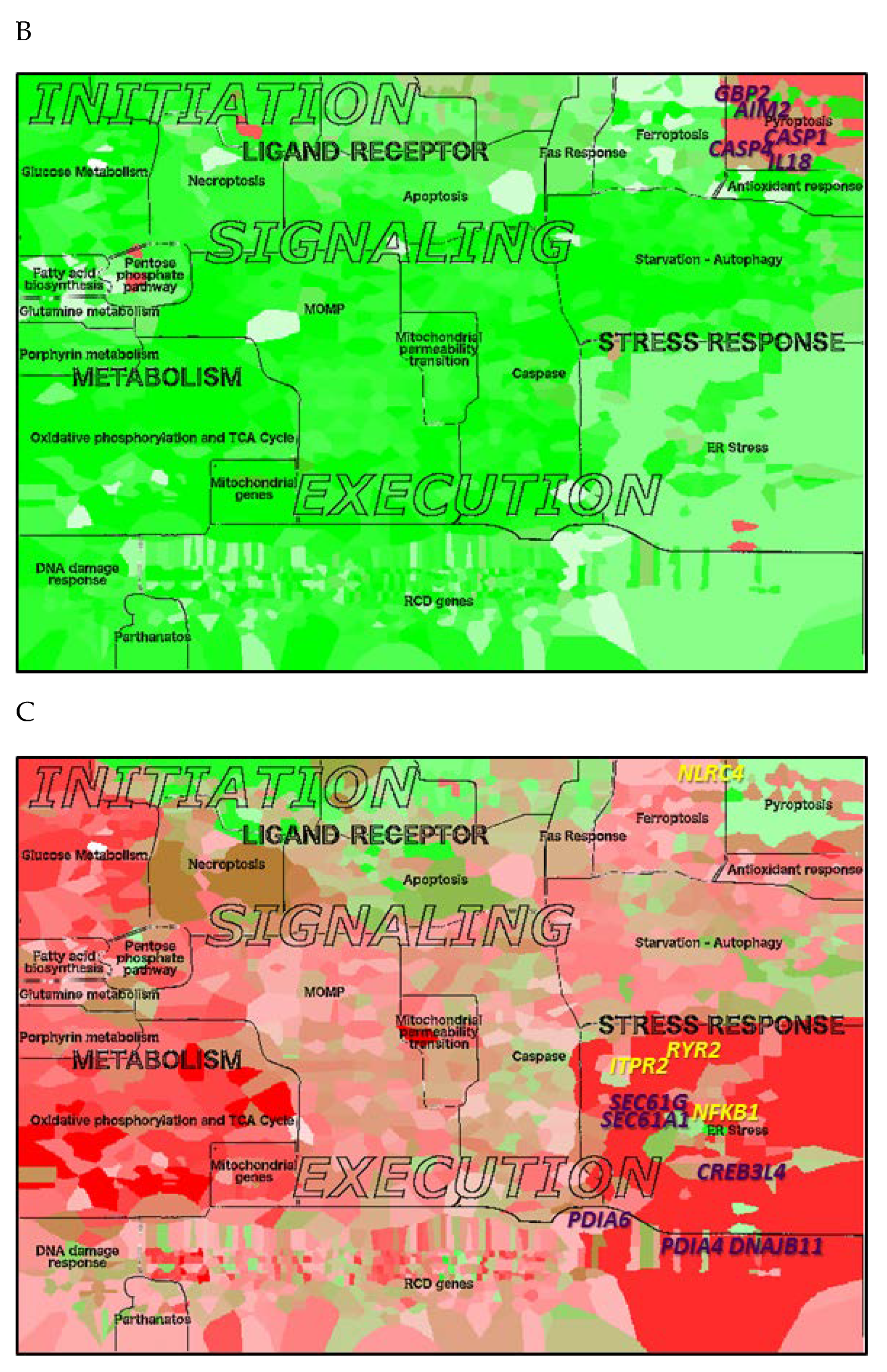
2.7. Application of the RCD Map to Neurodegenerative and Malignant Diseases
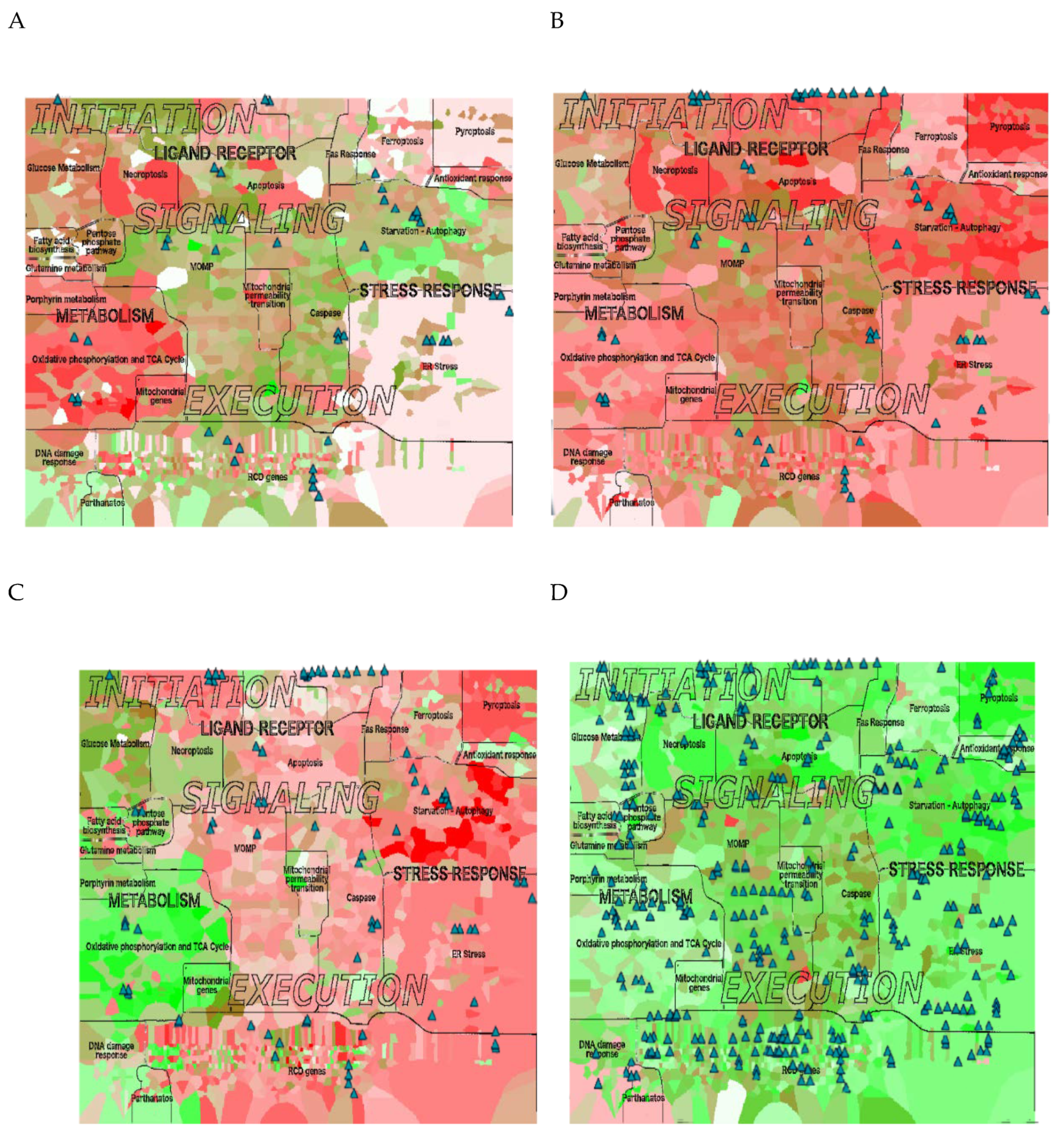
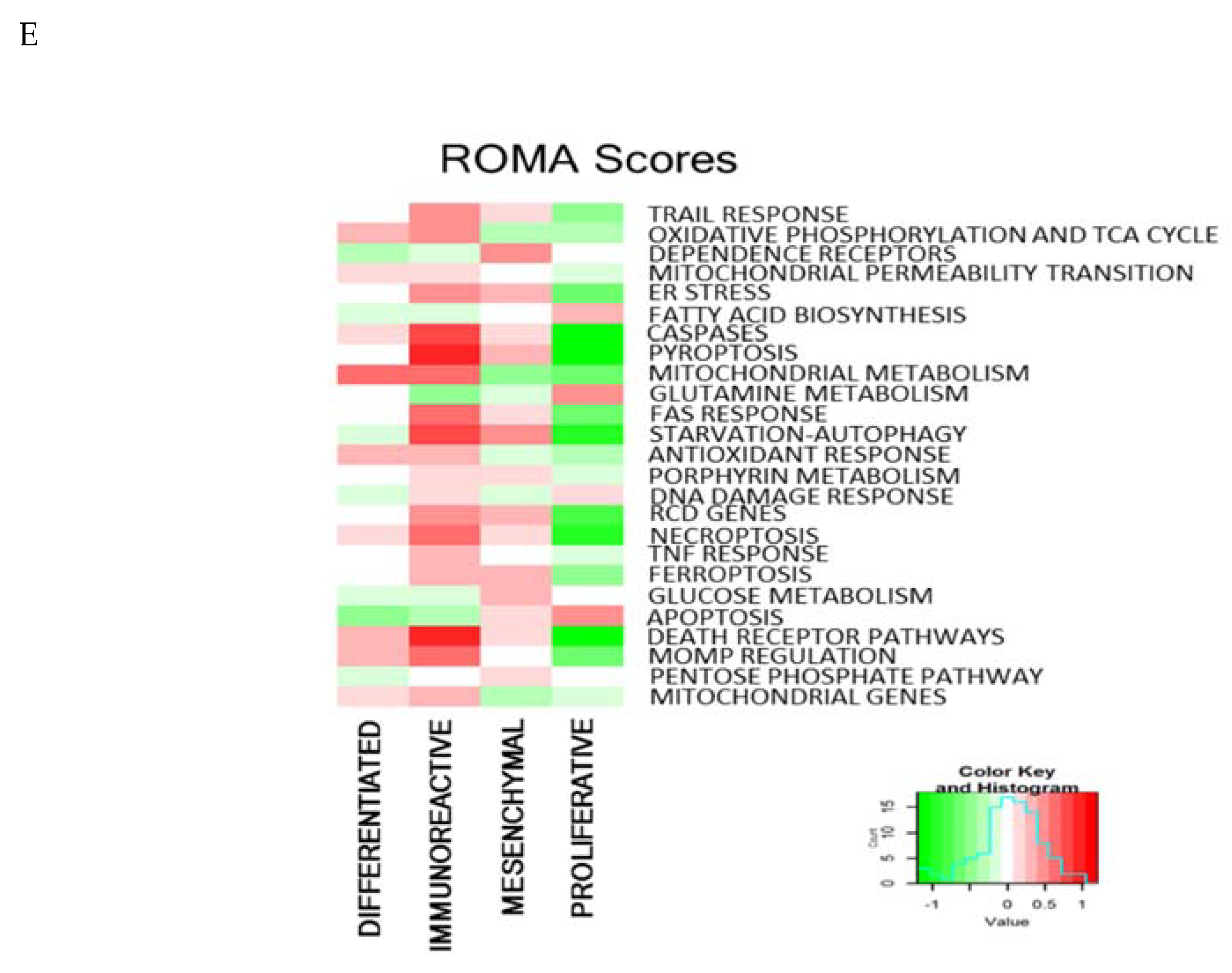
2.8. RCD Signatures in Different Ovarian Cancer Subtypes
3. Discussion
4. Materials and Methods
4.1. Map Availability
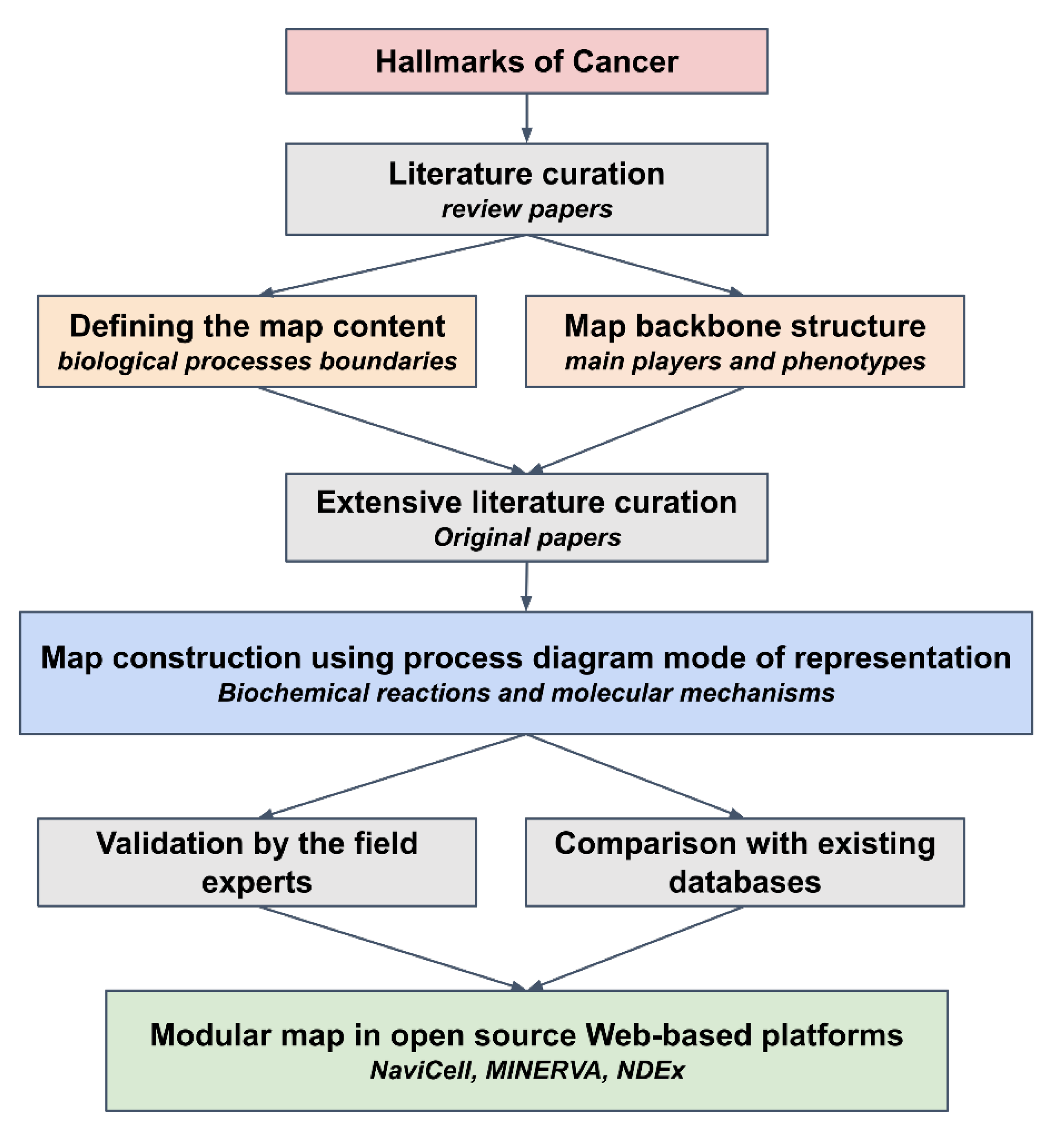
4.2. Map Construction
4.3. Data Model
4.4. Literature Selection Rules
4.5. Map Annotation Format
4.6. Module Tagging
4.7. Generation of NaviCell Map with NaviCell Factory
4.8. Map Navigation
4.9. Web-Based Platforms Support
4.10. Map Structural Analysis
4.11. Data Acquisition and Groups Definition
4.12. Module Scores Calculation and Visualization
5. Conlusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| RCD | programmed cell death |
| MOMP | Mitochondrial outer membrane permeabilization |
| ER | Endoplasmic reticulum |
References
- Kang, K.; Lee, S.-R.; Piao, X.; Hur, G.M. Post-translational modification of the death receptor complex as a potential therapeutic target in cancer. Arch Pharm Res. 2019, 42, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Pentimalli, F.; Grelli, S.; Di Daniele, N.; Melino, G.; Amelio, I. Cell death pathologies: Targeting death pathways and the immune system for cancer therapy. Genes Immun. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.T.; Zhou, T.T.; Liu, B.; Bao, J.K. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012, 45, 487–498. [Google Scholar] [CrossRef]
- Stevens, J.B.; Abdallah, B.Y.; Liu, G.; Horne, S.D.; Bremer, S.W.; Ye, K.J.; Huang, J.Y.; Kurkinen, M.; Ye, C.J.; Heng, H.H. Heterogeneity of cell death. Cytogenet Genome Res. 2013, 139, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, A.; Vandenabeele, P.; Krysko, D.V. Necroptosis: The release of damage-associated molecular patterns and its physiological relevance. Immunity 2013, 38, 209–223. [Google Scholar] [CrossRef]
- Lalaoui, N.; Lindqvist, L.M.; Sandow, J.J.; Ekert, P.G. The molecular relationships between apoptosis, autophagy and necroptosis. Semin. Cell Dev. Biol. 2015. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Yatim, N.; Cullen, S.; Albert, M.L. Dying cells actively regulate adaptive immune responses. Nat. Rev. Immunol. 2017, 17, 262–275. [Google Scholar] [CrossRef]
- Ellis, R.E.; Yuan, J.Y.; Horvitz, H.R. Mechanisms and functions of cell death. Annu. Rev. Cell Biol. 1991, 7, 663–698. [Google Scholar] [CrossRef]
- Russell, R.C.; Yuan, H.-X.; Guan, K.-L. Autophagy regulation by nutrient signalling. Cell Res. 2014, 24, 42–57. [Google Scholar] [CrossRef]
- Xie, Y.; Hou, W.; Song, X.; Yu, Y.; Huang, J.; Sun, X.; Kang, R.; Tang, D. Ferroptosis: Process and function. Cell Death Differ. 2016. [Google Scholar] [CrossRef] [PubMed]
- Fatokun, A.A.; Dawson, V.L.; Dawson, T.M. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 2014, 171, 2000–2016. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. The KEGG database. Novartis Found Symp. 2002, 247, 244–252. [Google Scholar]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Croft, D.; Mundo, A.F.; Haw, R.; Milacic, M.; Weiser, J.; Wu, G.; Caudy, M.; Garapati, P.; Gillespie, M.; Kamdar, M.R.; et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014, 42, D472–D477. [Google Scholar] [CrossRef]
- Hoffmann, R. A wiki for the life sciences where authorship matters. Nat. Genet. 2008, 40, 1047–1051. [Google Scholar] [CrossRef]
- Perfetto, L.; Briganti, L.; Calderone, A.; Perpetuini, A.C.; Iannuccelli, M.; Langone, F.; Licata, L.; Marinkovic, M.; Mattioni, A.; Pavlidou, T.; et al. SIGNOR: A database of causal relationships between biological entities. Nucleic Acids Res. 2016, 44, D548. [Google Scholar] [CrossRef]
- Calzone, L.; Gelay, A.; Zinovyev, A.; Radvanyi, F.; Barillot, E. A comprehensive modular map of molecular interactions in RB/E2F pathway. Mol. Syst. Biol. 2008, 4, 173. [Google Scholar] [CrossRef]
- Caron, E.; Ghosh, S.; Matsuoka, Y.; Ashton-Beaucage, D.; Therrien, M.; Lemieux, S.; Perreault, C.; Roux, P.P.; Kitano, H. A comprehensive map of the mTOR signalling network. Mol. Syst. Biol. 2010, 6, 453. [Google Scholar] [CrossRef]
- Mizuno, S.; Iijima, R.; Ogishima, S.; Kikuchi, M.; Matsuoka, Y.; Ghosh, S.; Miyamoto, T.; Miyashita, A.; Kuwano, R.; Tanaka, H. AlzPathway: A comprehensive map of signalling pathways of Alzheimer’s disease. BMC Syst. Biol. 2012, 6, 52. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Matsumae, H.; Katoh, M.; Eisfeld, A.J.; Neumann, G.; Hase, T.; Ghosh, S.; Shoemaker, J.E.; Lopes, T.J.; Watanabe, T.; et al. A comprehensive map of the influenza A virus replication cycle. BMC Syst. Biol. 2013, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Kuperstein, I.; Bonnet, E.; Nguyen, H.-A.; Cohen, D.; Viara, E.; Grieco, L.; Fourquet, S.; Calzone, L.; Russo, C.; Kondratova, M.; et al. Atlas of Cancer Signalling Network: A systems biology resource for integrative analysis of cancer data with Google Maps. Oncogenesis 2015, 4, e160. [Google Scholar] [CrossRef] [PubMed]
- Mazein, A.; Ostaszewski, M.; Kuperstein, I.; Watterson, S.; Le Novère, N.; Lefaudeux, D.; De Meulder, B.; Pellet, J.; Balaur, I.; Saqi, M.; et al. Systems medicine disease maps: Community-driven comprehensive representation of disease mechanisms. NPJ Syst. Biol. Appl. 2018, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Dorel, M.; Viara, E.; Barillot, E.; Zinovyev, A.; Kuperstein, I. NaviCom: A web application to create interactive molecular network portraits using multi-level omics data. Database 2017, 2017. [Google Scholar] [CrossRef]
- Gawron, P.; Ostaszewski, M.; Satagopam, V.; Gebel, S.; Mazein, A.; Kuzma, M.; Zorzan, S.; McGee, F.; Otjacques, B.; Balling, R.; et al. MINERVA-a platform for visualization and curation of molecular interaction networks. NPJ Syst. Biol. Appl. 2016, 2, 16020. [Google Scholar] [CrossRef]
- Pratt, D.; Chen, J.; Pillich, R.; Rynkov, V.; Gary, A.; Demchak, B.; Ideker, T. NDEx 2.0: A Clearinghouse for Research on Cancer Pathways. Cancer Res. 2017, 77, e58–e61. [Google Scholar] [CrossRef]
- Kondratova, M.; Sompairac, N.; Barillot, E.; Zinovyev, A.; Kuperstein, I. Signalling maps in cancer research: Construction and data analysis. Database 2018, 2018. [Google Scholar] [CrossRef]
- Le Novère, N.; Hucka, M.; Mi, H.; Moodie, S.; Schreiber, F.; Sorokin, A.; Demir, E.; Wegner, K.; Aladjem, M.I.; Wimalaratne, S.M.; et al. The Systems Biology Graphical Notation. Nat. Biotechnol. 2009, 27, 735–741. [Google Scholar] [CrossRef]
- Kitano, H.; Funahashi, A.; Matsuoka, Y.; Oda, K. Using process diagrams for the graphical representation of biological networks. Nat. Biotechnol. 2005, 23, 961–966. [Google Scholar] [CrossRef]
- Ashkenazi, A.; Dixit, V.M. Death receptors: Signalling and modulation. Science 1998, 281, 1305–1308. [Google Scholar] [CrossRef]
- Dickens, L.S.; Powley, I.R.; Hughes, M.A.; MacFarlane, M. The ‘complexities’ of life and death: Death receptor signalling platforms. Exp. Cell Res. 2012, 318, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Czabotar, P.E.; Lessene, G.; Strasser, A.; Adams, J.M. Control of apoptosis by the BCL-2 protein family: Implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014, 15, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Vandenabeele, P.; Galluzzi, L.; Vanden Berghe, T.; Kroemer, G. Molecular mechanisms of necroptosis: An ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 2010, 11, 700–714. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Kepp, O.; Trojel-Hansen, C.; Kroemer, G. Mitochondrial control of cellular life, stress, and death. Circ. Res. 2012, 111, 1198–1207. [Google Scholar] [CrossRef] [PubMed]
- David, K.K.; Andrabi, S.A.; Dawson, T.M.; Dawson, V.L. Parthanatos, a messenger of death. Front. Biosci. 2009, 14, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, W.; Shao, F. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem. Sci. 2017, 42, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Mamik, M.K.; Power, C. Inflammasomes in neurological diseases: Emerging pathogenic and therapeutic concepts. Brain 2017, 140, 2273–2285. [Google Scholar] [CrossRef]
- Magtanong, L.; Ko, P.J.; Dixon, S.J. Emerging roles for lipids in non-apoptotic cell death. Cell Death Differ. 2016, 23, 1099–1109. [Google Scholar] [CrossRef]
- Latunde-Dada, G.O. Ferroptosis: Role of lipid peroxidation, iron and ferritinophagy. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1893–1900. [Google Scholar] [CrossRef]
- Geng, J.; Ito, Y.; Shi, L.; Amin, P.; Chu, J.; Ouchida, A.T.; Mookhtiar, A.K.; Zhao, H.; Xu, D.; Shan, B.; et al. Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat Commun. 2017, 8, 359. [Google Scholar] [CrossRef]
- Silke, J.; Vucic, D. IAP Family of Cell Death and Signalling Regulators, 1st ed.; Elsevier Inc.: San Diego, CA, USA, 2014; Volume 545. [Google Scholar]
- Witt, A.; Vucic, D. Diverse ubiquitin linkages regulate RIP kinases-mediated inflammatory and cell death signalling. Cell Death Differ. 2017, 24, 1160–1171. [Google Scholar] [CrossRef] [PubMed]
- Hitomi, J.; Christofferson, D.E.; Ng, A.; Yao, J.; Degterev, A.; Xavier, R.J.; Yuan, J. Identification of a Molecular Signalling Network that Regulates a Cellular Necrotic Cell Death Pathway. Cell 2008, 135, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Kepp, O.; Krautwald, S.; Kroemer, G.; Linkermann, A. Molecular mechanisms of regulated necrosis. Semin. Cell Dev. Biol. 2014, 35, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Chaabane, W.; User, S.D.; El-Gazzah, M.; Jaksik, R.; Sajjadi, E.; Rzeszowska-Wolny, J.; Los, M.J. Autophagy, apoptosis, mitoptosis and necrosis: Interdependence between those pathways and effects on cancer. Arch. Immunol. Ther. Exp. 2013, 61, 43–58. [Google Scholar] [CrossRef]
- Tsuchihashi, N.A.; Hayashi, K.; Dan, K.; Goto, F.; Nomura, Y.; Fujioka, M.; Kanzaki, S.; Komune, S.; Ogawa, K. Autophagy through 4EBP1 and AMPK regulates oxidative stress-induced premature senescence in auditory cells. Oncotarget 2015, 6, 3644–3655. [Google Scholar] [CrossRef]
- Choi, K.; Ryu, S.-W.; Song, S.; Choi, H.; Kang, S.W.; Choi, C. Caspase-dependent generation of reactive oxygen species in human astrocytoma cells contributes to resistance to TRAIL-mediated apoptosis. Cell Death Differ. 2010, 17, 833–845. [Google Scholar] [CrossRef]
- Kuperstein, I.; Cohen, D.P.; Pook, S.; Viara, E.; Calzone, L.; Barillot, E.; Zinovyev, A. NaviCell: A web-based environment for navigation, curation and maintenance of large molecular interaction maps. BMC Syst. Biol. 2013, 7, 100. [Google Scholar] [CrossRef]
- Doctor, K.S.; Reed, J.C.; Godzik, A.; Bourne, P.E. The apoptosis database. Cell Death Differ. 2003, 10, 621–633. [Google Scholar] [CrossRef]
- Arntzen, M.Ø.; Thiede, B. ApoptoProteomics, an integrated database for analysis of proteomics data obtained from apoptotic cells. Mol. Cell Proteom. 2012, 11. [Google Scholar] [CrossRef]
- Wanichthanarak, K.; Cvijovic, M.; Molt, A.; Petranovic, D. yApoptosis: Yeast apoptosis database. Database (Oxford). 2013. [Google Scholar] [CrossRef]
- Türei, D.; Földvári-Nagy, L.; Fazekas, D.; Módos, D.; Kubisch, J.; Kadlecsik, T.; Demeter, A.; Lenti, K.; Csermely, P.; Vellai, T.; et al. Autophagy Regulatory Network—a systems-level bioinformatics resource for studying the mechanism and regulation of autophagy. Autophagy 2015, 11, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Sperling, R.A.; Aisen, P.S.; Beckett, L.A.; Bennett, D.A.; Craft, S.; Fagan, A.M.; Iwatsubo, T.; Jack, C.R.; Kaye, J.; Montine, T.J.; et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011, 7, 280–292. [Google Scholar] [CrossRef]
- Raskin, J.; Cummings, J.; Hardy, J.; Schuh, K.; Dean, R.A. Neurobiology of Alzheimer’s Disease: Integrated Molecular, Physiological, Anatomical, Biomarker, and Cognitive Dimensions. Curr. Alzheimer Res. 2015, 12, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Martignetti, L.; Calzone, L.; Bonnet, E.; Barillot, E.; Zinovyev, A. ROMA: Representation and quantification of module activity from target expression data. Front Genet. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Bonnet, E.; Calzone, L.; Rovera, D.; Stoll, G.; Barillot, E.; Zinovyev, A. BiNoM 2.0, a Cytoscape plugin for accessing and analyzing pathways using standard systems biology formats. BMC Syst. Biol. 2013, 7, 18. [Google Scholar] [CrossRef]
- Hokama, M.; Oka, S.; Leon, J.; Ninomiya, T.; Honda, H.; Sasaki, K.; Iwaki, T.; Ohara, T.; Sasaki, T.; LaFerla, F.M.; et al. Altered expression of diabetes-related genes in Alzheimer’s disease brains: The Hisayama study. Cereb. Cortex 2014, 24, 2476–2488. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.S.; Reiman, E.M.; Valla, J.; Dunckley, T.; Beach, T.G.; Grover, A.; Niedzielko, T.L.; Schneider, L.E.; Mastroeni, D.; Caselli, R.; et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc. Natl. Acad. Sci. USA 2008, 105, 4441–4446. [Google Scholar] [CrossRef]
- Sánchez-Valle, J.; Tejero, H.; Ibáñez, K.; Portero, J.L.; Krallinger, M.; Al-Shahrour, F.; Tabarés-Seisdedos, R.; Baudot, A.; Valencia, A. A molecular hypothesis to explain direct and inverse co-morbidities between Alzheimer’s Disease, Glioblastoma and Lung cancer. Sci. Rep. 2017. [Google Scholar] [CrossRef]
- Musicco, M.; Adorni, F.; Di Santo, S.; Prinelli, F.; Pettenati, C.; Caltagirone, C.; Palmer, K.; Russo, A. Inverse occurrence of cancer and Alzheimer disease: A population-based incidence study. Neurology 2013, 81, 322–328. [Google Scholar] [CrossRef]
- Ou, S.M.; Lee, Y.J.; Hu, Y.W.; Liu, C.J.; Chen, T.J.; Fuh, J.L.; Wang, S.J. Does Alzheimer’s disease protect against cancers? A nationwide population-based study. Neuroepidemiology 2012, 40, 42–49. [Google Scholar] [CrossRef]
- Tabarés-Seisdedos, R.; Baudot, A. Editorial: Direct and Inverse Comorbidities between Complex Disorders. Front Physiol. 2016, 7, 117. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.N.; Carroll, R.G.; Galván-Peña, S.; Mills, E.L.; Olden, R.; Triantafilou, M.; Wolf, A.I.; Bryant, C.E.; Triantafilou, K.; Masters, S.L. Inflammasome Priming in Sterile Inflammatory Disease. Trends Mol. Med. 2017, 23, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, V.; Dye, R.; Pakavathkumar, P.; Foveau, B.; Flores, J.; Hyman, B.; Ghetti, B.; Koller, B.H.; LeBlanc, A.C. Neuronal NLRP1 inflammasome activation of Caspase-1 coordinately regulates inflammatory interleukin-1-beta production and axonal degeneration-associated Caspase-6 activation. Cell Death Differ. 2015, 22, 1676–1686. [Google Scholar] [CrossRef] [PubMed]
- Schmid-Burgk, J.L.; Chauhan, D.; Schmidt, T.; Ebert, T.S.; Reinhardt, J.; Endl, E.; Hornung, V. A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J. Biol. Chem. 2016, 291, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Ojala, J.; Alafuzoff, I.; Herukka, S.K.; van Groen, T.; Tanila, H.; Pirttilä, T. Expression of interleukin-18 is increased in the brains of Alzheimer’s disease patients. Neurobiol. Aging. 2009, 30, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, Y.; McKenzie, A.; Dorr, N.; Gama Sosa, M.A.; Elder, G.; Schmeidler, J.; Dickstein, D.L.; Bozdagi, O.; Zhang, B.; Buxbaum, J.D. The human-specific CASP4 gene product contributes to Alzheimer-related synaptic and behavioural deficits. Hum. Mol. Genet. 2016, 25, 4315–4327. [Google Scholar] [CrossRef] [PubMed]
- Pompl, P.N.; Yemul, S.; Xiang, Z.; Ho, L.; Haroutunian, V.; Purohit, D.; Mohs, R.; Pasinetti, G.M. Caspase gene expression in the brain as a function of the clinical progression of alzheimer disease. Arch Neurol. 2003, 60, 369–376. [Google Scholar] [CrossRef]
- Saresella, M.; La Rosa, F.; Piancone, F.; Zoppis, M.; Marventano, I.; Calabrese, E.; Rainone, V.; Nemni, R.; Mancuso, R.; Clerici, M. The NLRP3 and NLRP1 inflammasomes are activated in Alzheimer’s disease. Mol. Neurodegener. 2016, 11, 1–14. [Google Scholar] [CrossRef]
- Giampietri, C.; Petrungaro, S.; Conti, S.; Facchiano, A.; Filippini, A.; Ziparo, E. Cancer microenvironment and endoplasmic reticulum stress response. Mediat. Inflamm. 2015, 2015. [Google Scholar] [CrossRef]
- Avril, T.; Vauléon, E.; Chevet, E. Endoplasmic reticulum stress signalling and chemotherapy resistance in solid cancers. Oncogenesis 2017, 6, e373. [Google Scholar] [CrossRef]
- Salaroglio, I.C.; Panada, E.; Moiso, E.; Buondonno, I.; Provero, P.; Rubinstein, M.; Kopecka, J.; Riganti, C. PERK induces resistance to cell death elicited by endoplasmic reticulum stress and chemotherapy. Mol. Cancer 2017, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tufo, G.; Jones, A.W.E.; Wang, Z.; Hamelin, J.; Tajeddine, N.; Esposti, D.D.; Martel, C.; Boursier, C.; Gallerne, C.; Migdal, C.; et al. The protein disulfide isomerases PDIA4 and PDIA6 mediate resistance to cisplatin-induced cell death in lung adenocarcinoma. Cell Death Differ. 2014, 21, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, S.; Verma, I.M. NF- B, an Active Player in Human Cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Logue, S.E.; Cleary, P.; Saveljeva, S.; Samali, A. New directions in ER stress-induced cell death. Apoptosis 2013, 18, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Välk, K.; Vooder, T.; Kolde, R.; Reintam, M.-A.; Petzold, C.; Vilo, J.; Metspalu, A. Gene expression profiles of non-small cell lung cancer: Survival prediction and new biomarkers. Oncology 2010, 79, 283–292. [Google Scholar] [CrossRef]
- McCluggage, W.G. Morphological subtypes of ovarian carcinoma: A review with emphasis on new developments and pathogenesis. Pathology 2011, 43, 420–432. [Google Scholar] [CrossRef]
- Gilks, C.B.; Prat, J. Ovarian carcinoma pathology and genetics: Recent advances. Hum. Pathol. 2009, 40, 1213–1223. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. (Poznan, Poland). 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; DiSaia, P.; Gabra, H.; Glenn, P.; et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Konecny, G.E.; Wang, C.; Hamidi, H.; Winterhoff, B.; Kalli, K.R.; Dering, J.; Ginther, C.; Chen, H.-W.; Dowdy, S.; Cliby, W.; et al. Prognostic and Therapeutic Relevance of Molecular Subtypes in High-Grade Serous Ovarian Cancer. JNCI J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed]
- Etemadmoghadam, D.; deFazio, A.; Beroukhim, R.; Mermel, C.; George, J.; Getz, G.; Tothill, R.; Okamoto, A.; Raeder, M.B.; Harnett, P.; et al. Integrated Genome-Wide DNA Copy Number and Expression Analysis Identifies Distinct Mechanisms of Primary Chemoresistance in Ovarian Carcinomas. Clin. Cancer Res. 2009, 15, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Ye, J.; Fei, H.; Jiang, S.; Wu, Z.; Chen, Y.; Zhang, L.; Yang, X. YWHAZ promotes ovarian cancer metastasis by modulating glycolysis. Oncol. Rep. 2018, 41, 1101–1112. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Jiang, X.; Li, H.; Lin, Z.; Zhou, X.; Luo, X.; Liu, L.; Chen, G. Expression and prognostic value of WISP-1 in patients with endometrial endometrioid adenocarcinoma. J. Obstet. Gynaecol. Res. 2011, 37, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, J.; Ye, Z.; Li, Z.; Wu, X. N-myc downstream regulated gene 1 acts as a tumour suppressor in ovarian cancer. Oncol. Rep. 2014, 31, 2279–2285. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, P.; Zheng, C.-H.; Xia, J. Identification of ovarian cancer subtype-specific network modules and candidate drivers through an integrative genomics approach. Oncotarget 2016, 7, 4298–4309. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
| Layer/Module | Chemical Species (Entities) | Proteins | Genes | RNAs | asRNAs | Reactions |
|---|---|---|---|---|---|---|
| Initiation (reversible) | ||||||
| Stress response | ||||||
| Antioxidant response | 213 | 151 | 4 | 9 | 12 | 95 |
| DNA damage response | 128 | 59 | 3 | 3 | 5 | 80 |
| ER stress | 382 | 161 | 45 | 44 | 4 | 291 |
| Starvation-autophagy | 438 | 165 | 19 | 31 | 31 | 290 |
| Ligand receptor | ||||||
| Death receptor pathways | 640 | 287 | 23 | 27 | 32 | 469 |
| Trail response | 36 | 25 | 0 | 0 | 0 | 18 |
| Fas response | 29 | 24 | 0 | 0 | 0 | 14 |
| TNF response | 87 | 39 | 3 | 3 | 0 | 57 |
| Dependence receptors | 78 | 52 | 0 | 0 | 0 | 36 |
| metabolism | ||||||
| Fatty acid biosynthesis | 78 | 34 | 1 | 3 | 2 | 33 |
| Glucose metabolism | 190 | 112 | 0 | 0 | 0 | 104 |
| Glutamine metabolism | 42 | 20 | 0 | 0 | 0 | 20 |
| Pentose phosphate pathway | 64 | 12 | 2 | 2 | 2 | 39 |
| Porphyrin metabolism | 51 | 17 | 0 | 0 | 0 | 26 |
| Mitochondrial metabolism | 608 | 349 | 1 | 35 | 0 | 360 |
| Oxidative phosphorylation and TCA cycle | 232 | 179 | 0 | 0 | 0 | 109 |
| Mitochondrial genes | 114 | 116 | 2 | 30 | 0 | 36 |
| Signalling (rewirable) | ||||||
| Apoptosis | 584 | 246 | 34 | 40 | 57 | 401 |
| Necroptosis | 242 | 118 | 0 | 0 | 0 | 188 |
| Ferroptosis | 193 | 65 | 17 | 19 | 0 | 110 |
| Parthanatos | 80 | 17 | 1 | 0 | 0 | 64 |
| Pyroptosis | 112 | 36 | 1 | 0 | 0 | 79 |
| Execution (irreversible) | ||||||
| MOMP regulation | 630 | 319 | 19 | 48 | 17 | 396 |
| Mitochondrial permeability transition | 57 | 36 | 0 | 0 | 0 | 33 |
| Caspases | 318 | 158 | 5 | 5 | 12 | 221 |
| RCD genes | 633 | 135 | 168 | 179 | 90 | 365 |
| RCD global map | 2657 | 1008 | 215 | 260 | 93 | 2020 |
| Module | Top Contributing Genes AD | Top Contributing Genes LC |
|---|---|---|
| MOMP regulation | ATG5, VDAC3, CUL2, CYCS, HK1, MAPK6, BMF, RPS6KA1, MAPK10, VDAC2 | DIABLO,RPS6KA1, CISH, CSNK2A1, C1QBP, FOXO1, PPP1CC, BCL2L11, SLC25A5, VDAC2 |
| Starvation Autophagy | CAB39, ATG5, MAPK6, MAPK8, ACACA, PINK1, RPS6KA1, CASP8, MTOR, ATG12 | PARP1, SESN1, LDHA, BCL2L11, PI3KC3, RPS6KA1, PINK1, ATG3, ATG12 |
| Glucose Metabolism | PFKM, PRKACB, PRKAA2, LDHA, NOTCH1, H6PD, ALDOA | PGK1, LDHA, ANAPC11M ALDOA, LDHC, PKM, ANAPC2 |
| Fatty acid biosynthesis | PRKAA2, PPAT, CAB39, PRPS1, PRKACB, ACACA | ACLY, ACSS2, MLST8, PRPS1, PRPS2 |
| Mitochondrial metabolism | SLC25A14, NDUFA9, FH, DLD, NDUFAF1, HK3, TNFRSF1A, ALOX15B | UQCRH, ATP5G1, SLC25A10, CYC1, TACO1, NDUFAB1, PFKB2, RIPK3, SESN1, PLA2G4C |
| Pyroptosis | IL18, CASP4, GBP2, CASP1, AIM2 | CASP1, GBP2, IL18, CASP4, IL1B |
| Caspases | YWHAG, MAPK8, MAPK6, CAB39, LMNA, HIP1, RIPK3, MTOR | G0S2, BIRC3, PIDD, FOXO1, RNF41 |
| Dependence receptors | AATF, APPL2, PPP2CB, NGF, CARD8, NSMAF | PIK3CA, NEDD4, TRAF6, PPP2CB, NSMAF |
| Ferroptosis | NFE2L2, NQO1, FTL, ACSF2, SLC3A2, FTH1 | GCLC, GCLM, GSR, MAFG, GSS, SLC3A2 |
| Death receptor pathways | B4GALT6, GUCY1B3, CUL3, RBX1, RIPK3, HIF1A, NDUT5, CASP3 | CYLD, PARP1, BRCA1, H2AFX, PRKDC, CARD8, STAT5A, FOXO1, MAGED1, TNFAIP3 |
| Mitochondrial permeability transition | SLC25A4, HK1, VDAC3, VDAC1, PPID, CKMT1A, VDAC2 | PPID, CAPN2, VDAC2, SLC25A5 |
| Mitochondrial genes | ATP5C1, ATP5H, ATP5L, AIFM1, ATP5G1, NDUFB6, NDUFC2, ATP5A1, COX7A2, NDUFS4 | ATP5G1, NDUFAB1, COX5B, ATP5B, NDUFB9, ATP5G3, COX6A1, COX6C, TACO1, COX8A |
| Pentose phosphate pathway | ACACB, NME4 | NME2, ACACB, RRM1, ACSS2, RRM2B, NME4, TXN, TP53, NME7 |
| Necroptosis | B4GALT6, CUL3, RBX1, RIPK3, HK3, CASP10, PPP1CC | GUCY1A3, H2AFX, PARP1, CFLAR, RIPK3, GUCY1A2, DNM1L, MLKL, NCF1 |
| Glutamine metabolism | GLS, GLS2 | MLKL, RIPK3, RIPK1, PRPS1 |
| FAS response | CFLAR, CASP8, MLKL, GALNT14, CASP10 | PTPN13, TNFSF10, MLKL, CFLAR, RIPK1, SQSTM1 |
| TNF response | CYLD, MAP3K7, BIRC2, NSMAF, TAB2 | CYLD, FLAD1, RNF11, PPP6C, CIAPIN1, RIPK1, CFLAR |
| TRAIL response | CYLD, CASP8, MLKL, GALNT14, CASP10 | MLKL, TNFAIP3, CFLAR, RIPK1, TNFRSF10D |
| RCD genes | FBXO45, PKM, CDC42, ATG12, HSP90AA1, SIRT1, HIF1A, CASP3, BIRC2 | KLF8, BCL2L11, STAT5A, PGK1, HUWE1, ALAS1, ENO1, NPM1, HSP90B1, ELK1 |
| Apoptosis | RHOA, TCEB1, CYCS, SKP1, CCNB1, NKRF, BECN1, YWHAG, RPS6KA1, HIP1 | DIABLO, STAT5A, RIPK3, CISH, RPS6KA1, HUWE1, C1QBP, BCL2L11, CDK6, PPP3R1 |
| DNA damage response | PPP2R1A, CAB39, CSNK2A1, NKRF, HUWE1, RNF8, PRKDC | H2AFX, RAD50, MRE11A, ACSS2, HUWE1, BARD1, NKRF, CANK2A1, PPARGC1A, APBB1 |
| Porphyrin metabolism | UROD, COX10, UROS, CPOX, COX15 | CPOX, ALAS1, HMBS, ALAS2 |
| ER stress | DERL1, DERL2, HSPA9, ATP2A1, MAP3K5, MAP3K4, DNAJC3, VCP, EIF2S3 | NLRC4, PDIA6, RYR2, NFKB1, PDIA4, DNAJB11, SEC61G, SEC61A1, CREB3L4, ITPR1 |
| Antioxidant response | NDUFS1, AIFM1, RBX1, CUL3, NDUFC2, NDUFAF1, COX7A2, NDUFS4, NDUFB5, COX6B2 | UQCRH, MCL1, NDUFAB1, CYC1, COX5B, STAT5A, NDUFB9, COX6B1, COX4I2, COX6C |
| Oxidative phosphorylation and TCA cycle | NDUFS1, ATP5H, SLC25A14, PARK2, IDH3B, NDUFC2, FH, ATP5G1, IDH3A, SLC1A5 | ATP5B, NDUFAB1, COX5B, ATP5G3, COX8A, COX6C, COX6B1, FH, NDUFB10, COX5A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravel, J.-M.; Monraz Gomez, L.C.; Sompairac, N.; Calzone, L.; Zhivotovsky, B.; Kroemer, G.; Barillot, E.; Zinovyev, A.; Kuperstein, I. Comprehensive Map of the Regulated Cell Death Signaling Network: A Powerful Analytical Tool for Studying Diseases. Cancers 2020, 12, 990. https://doi.org/10.3390/cancers12040990
Ravel J-M, Monraz Gomez LC, Sompairac N, Calzone L, Zhivotovsky B, Kroemer G, Barillot E, Zinovyev A, Kuperstein I. Comprehensive Map of the Regulated Cell Death Signaling Network: A Powerful Analytical Tool for Studying Diseases. Cancers. 2020; 12(4):990. https://doi.org/10.3390/cancers12040990
Chicago/Turabian StyleRavel, Jean-Marie, L. Cristobal Monraz Gomez, Nicolas Sompairac, Laurence Calzone, Boris Zhivotovsky, Guido Kroemer, Emmanuel Barillot, Andrei Zinovyev, and Inna Kuperstein. 2020. "Comprehensive Map of the Regulated Cell Death Signaling Network: A Powerful Analytical Tool for Studying Diseases" Cancers 12, no. 4: 990. https://doi.org/10.3390/cancers12040990
APA StyleRavel, J.-M., Monraz Gomez, L. C., Sompairac, N., Calzone, L., Zhivotovsky, B., Kroemer, G., Barillot, E., Zinovyev, A., & Kuperstein, I. (2020). Comprehensive Map of the Regulated Cell Death Signaling Network: A Powerful Analytical Tool for Studying Diseases. Cancers, 12(4), 990. https://doi.org/10.3390/cancers12040990







