Functional Transcription Factor Target Networks Illuminate Control of Epithelial Remodelling
Abstract
1. Introduction
2. Results and Discussion
2.1. A Comprehensive Drosophila melanogaster Functional Gene Network (DroFN)
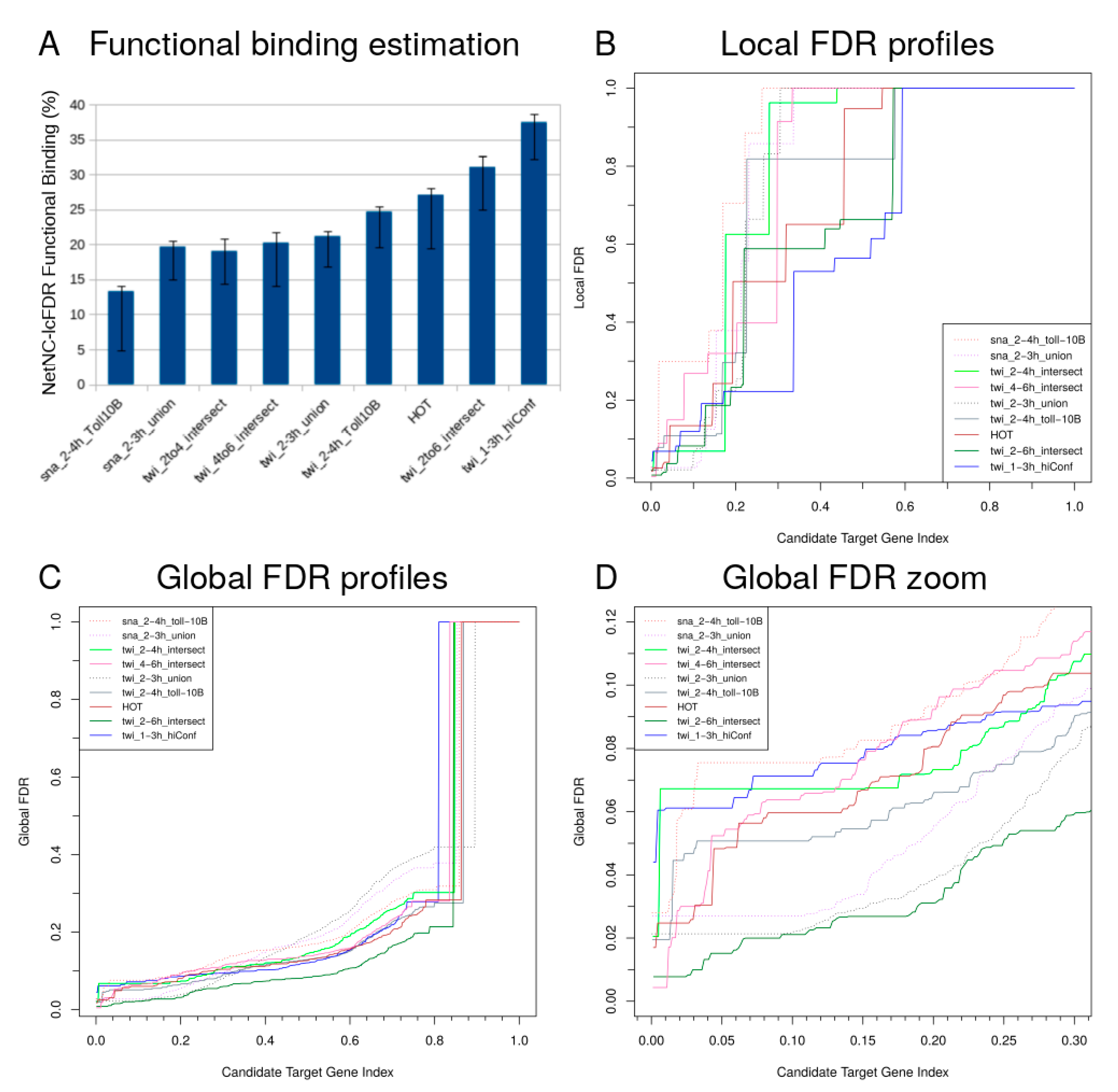
2.2. Prediction of Functional Transcription Factor Targets
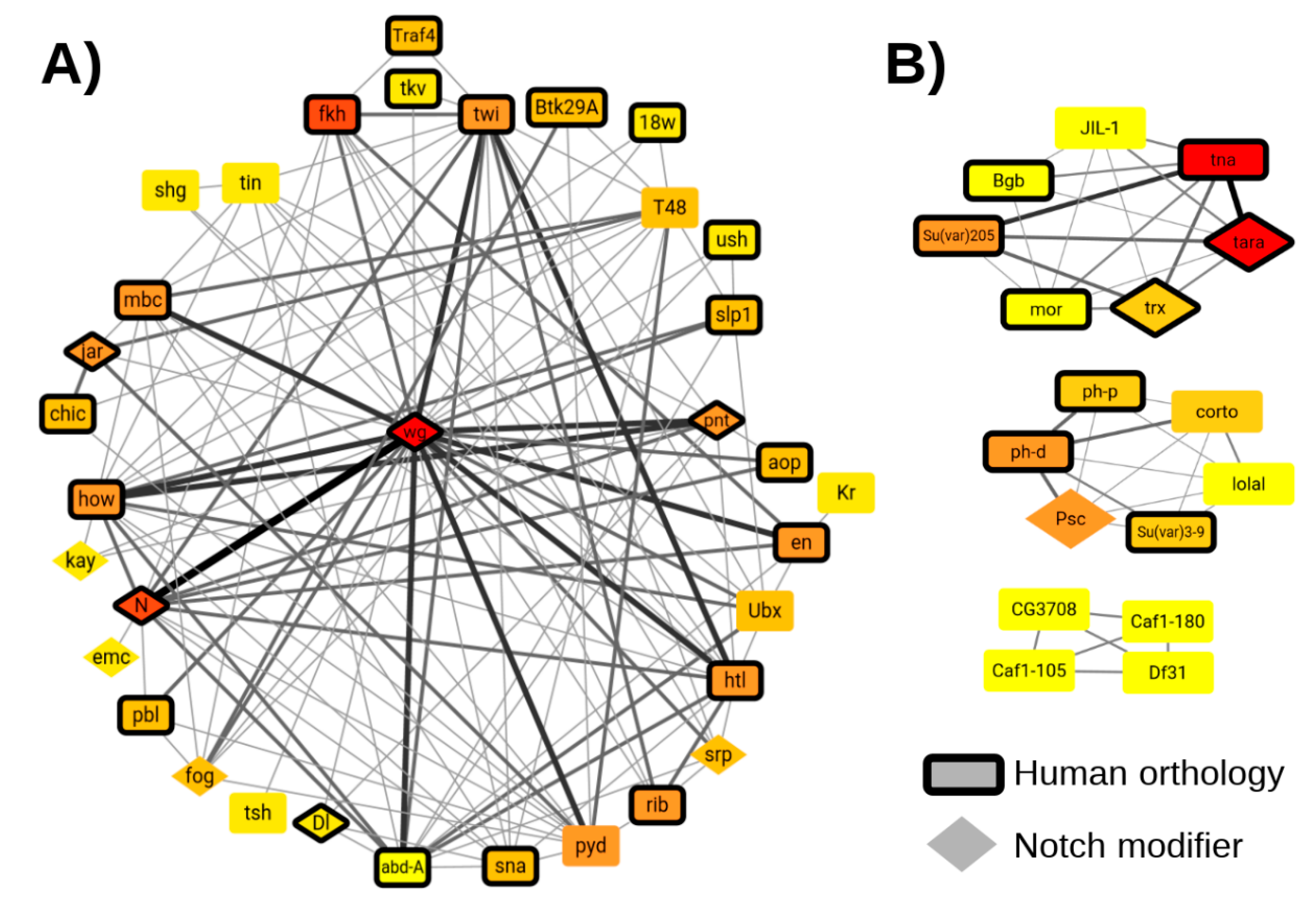
2.3. Analysis of EMT Transcription Factors and Highly Occupied Target (HOT) Regions
2.4. Genome-Scale Functional Transcription Factor Target Networks
2.5. Breast Cancer Subtypes are Recovered by Unsupervised Clustering with Orthologous Snail and Twist Functional Targets
2.6. Integrating NetNC Functional Target Networks and Breast Cancer Transcriptome Profiling
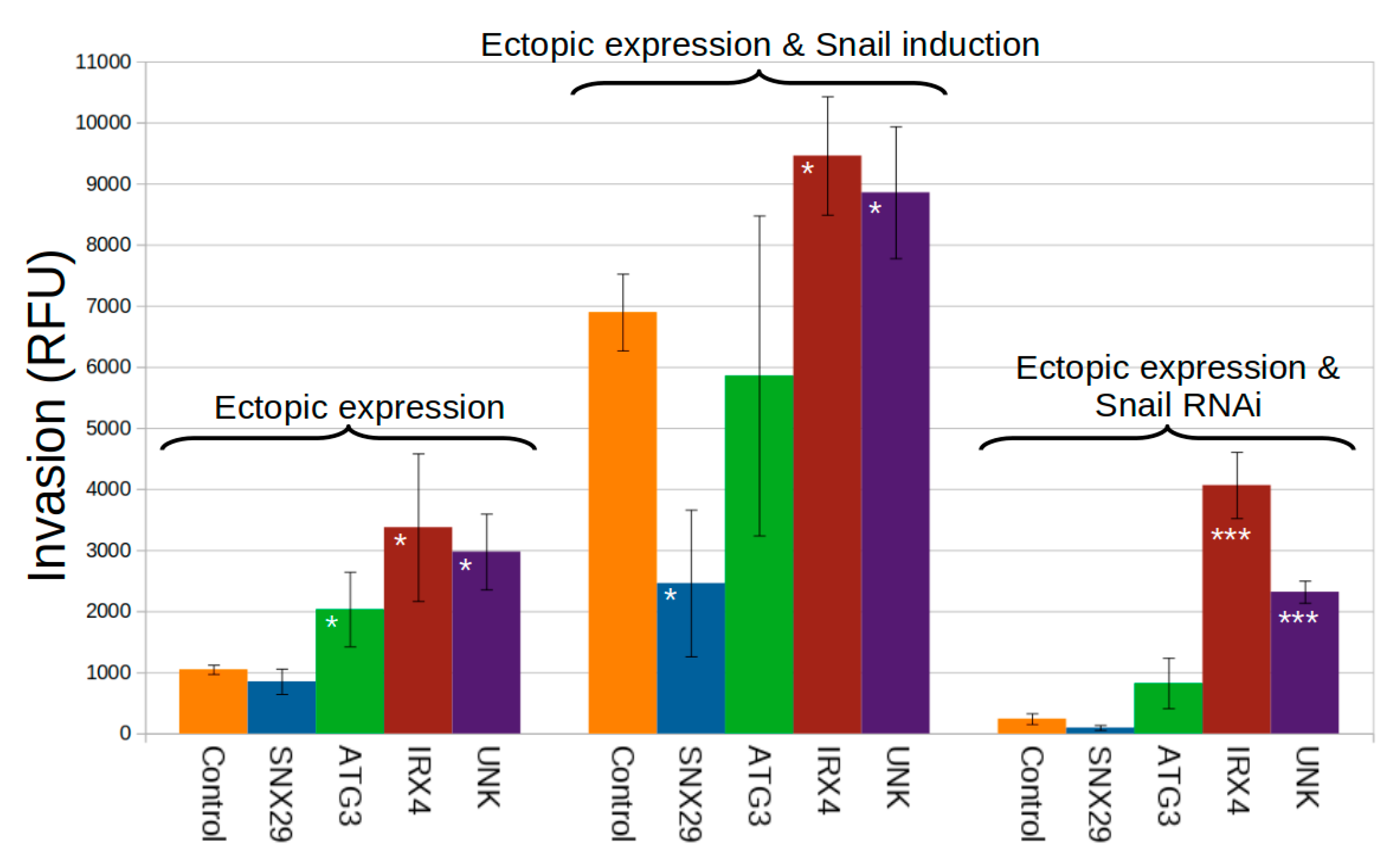
2.7. Novel Twist and Snail Functional Targets Influence Invasion in a Breast Cancer Model of EMT
3. Methods
3.1. A Comprehensive D. Melanogaster Functional Gene Network (DroFN)
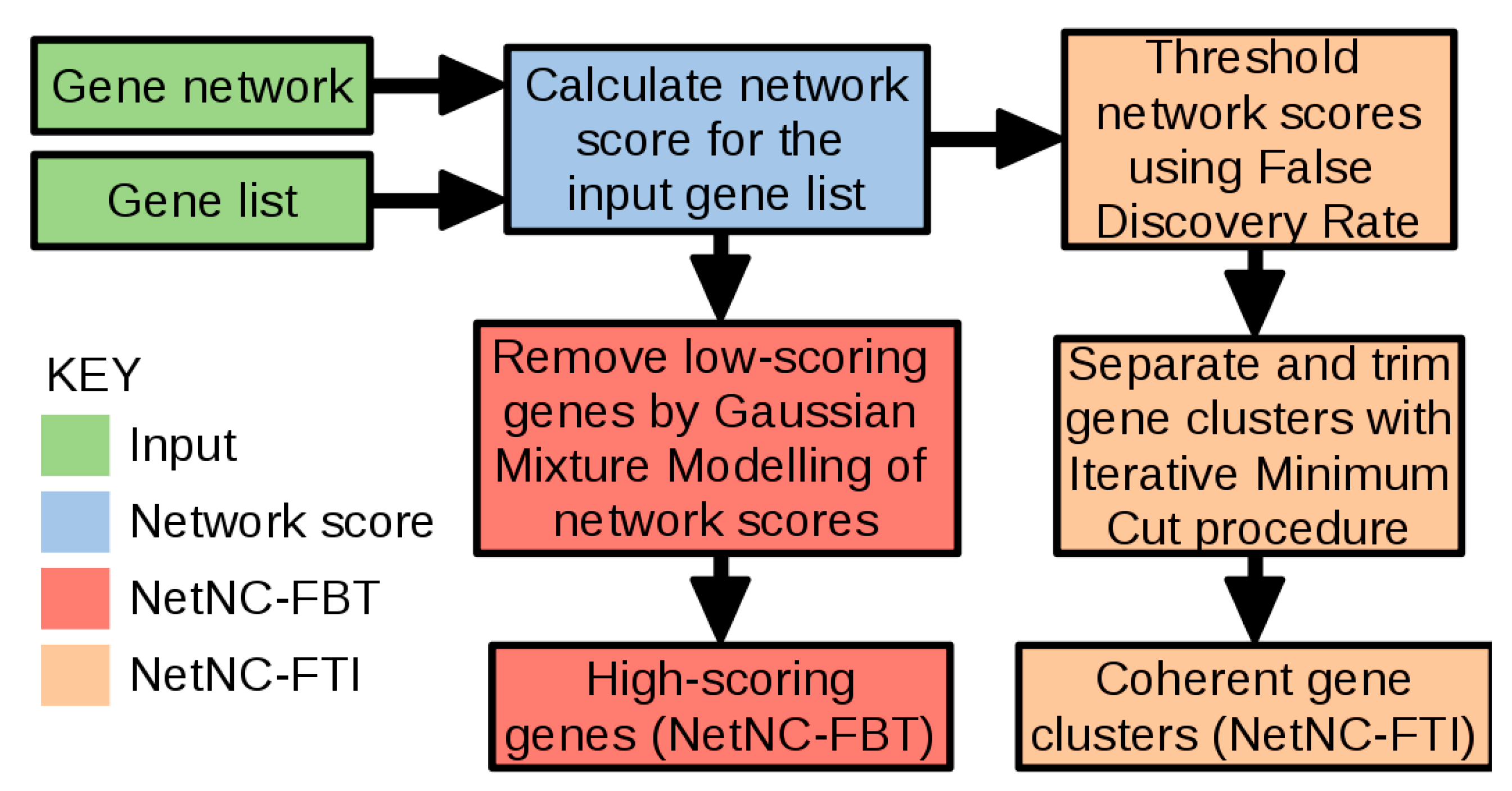
3.2. Network Neighbourhood Clustering (NetNC) Algorithm
- 1.
- A two times two contingency table is derived for each edge Sij by conditioning on the Boolean connectivity of nodes in S to Si and Sj. Nodes Si and Sj are not counted in the contingency table.
- 2.
- Exact hypergeometric p-values [46] for enrichment of the nodes in S that have edges to the nodes Si and Sj are calculated using Fisher’s Exact Test from the contingency table. Therefore, a distribution of p-values (H1) is generated for all edges Sij.
- 3.
- The NetNC edge-centric analysis setting (NetNC-FTI) employs positive false discovery rate [157] and an iterative minimum cut procedure [158] to derive clusters as follows:
- (a)
- Subgraphs with the same number of nodes as S are resampled from G, application of steps 1 and 2 to these subgraphs generates an empirical null distribution of neighbourhood clustering p-values (H0). This H0 accounts for the effect of the sample size and the structure of G on the Sij hypergeometric p-values (pij). Each NetNC run on TF_ALL in this study resampled 1000 subgraphs to derive H0.
- (b)
- Each edge in S is associated with a positive false discovery rate (q) estimated over pij using H1 and H0. The neighbourhood clustering subgraph C is induced by edges where the associated q ≤ Q. Therefore, Q is the NetNC-FTI threshold for false discovery rate (q).
- (c)
- An iterative minimum cut procedure [158] is applied to C until all components have density greater than or equal to a threshold Z. Edge weights in this procedure are taken as the negative log p-values from H1. Therefore, Z is the threshold for the density of network components output by NetNC-FTI.
- (d)
- As described below, thresholds Q and Z were chosen to optimise the performance of NetNC on the “Functional Target Identification” task using training data taken from KEGG. Connected components with less than three nodes are discarded, in line with common definitions of a “cluster”. Remaining nodes are taken as functionally coherent.
- 4.
- The node-centric, parameter-free approach (NetNC-FBT) proceeds by calculating degree-normalised node functional coherence scores (NFCS) from H1, then identifies statistical modes of the NFCS distribution using Gaussian Mixture Modelling (GMM) [159].
- (a)
- The node functional coherence score (NFCS) is calculated by summation of Sij p-values in H1 (pij) for fixed Si, normalised by the Si degree value in S (di) (Equation (2)):
- (b)
- GMM is applied to identify structure in the NFCS distribution. Expectation-maximization fits a mixture of Gaussians to the distribution using independent mean and standard deviation parameters for each Gaussian [159,160]. Models with 1..9 Gaussians are fitted and the final model selected using the Bayesian Information Criterion (BIC).
- (c)
- Nodes in high-scoring statistical mode(s) are predicted to be “Functionally Bound Targets” (FBTs) and retained. Firstly, any mode at NFCS < 0.05 is excluded because this typically represents nodes with no edges in S (where NFCS = 0). A second step eliminates the lowest scoring mode if >1 mode remains. Very rarely a unimodal model is returned, which may be due to a large non-Gaussian peak at NFCS = 0 confounding model fitting; if necessary, this is addressed by introducing a tiny Gaussian noise component (SD = 0.01) to the NFCS = 0 nodes to produce NFCS_GN0. GMM is performed on NFCS_GN0 and nodes eliminated according to the above procedure on the resulting model. This procedure was developed following manual inspection of results on training data from KEGG pathways with “synthetic neutral target genes” (STNGs) as nodes resampled from G (TRAIN-CL).
3.3. Estimating Positive False Discovery Rate for Hypergeometric Mutual Clustering p-Values
3.4. Estimating Local False Discovery Rate from Global False Discovery Rate
3.5. Median Difference and Correlation between Estimates of Functional Binding from NetNC Functional Target Identification and Local False Discovery Rate
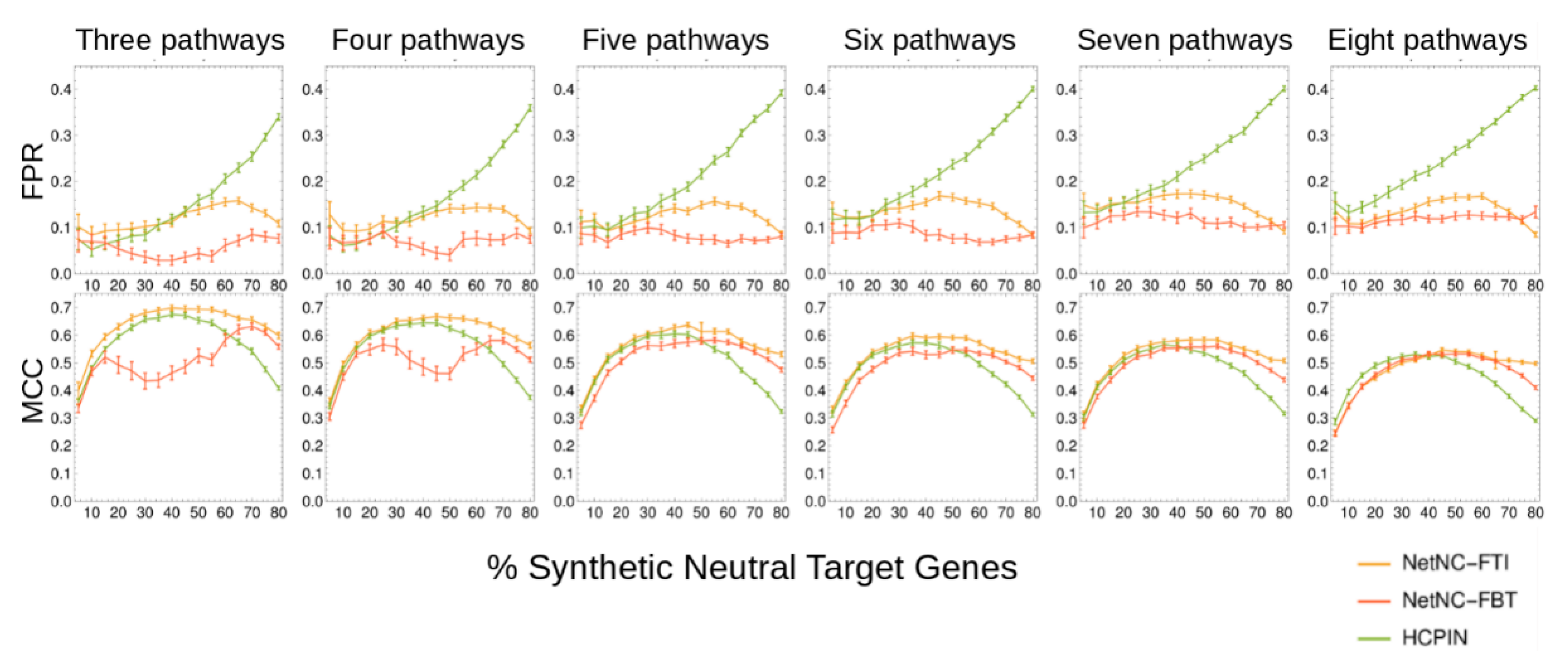
3.6. NetNC Benchmarking Data
3.7. Comparison of Synthetic NetNC Benchmark to Experimentally Determined TF Binding Data
3.8. NetNC-FTI Parameter Optimisation
3.9. Performance on Blind Test Data
3.10. Subsampling of Transcription Factor Binding Datasets and Statistical Testing
3.11. Transcription Factor Binding and Notch Modifier Datasets
3.12. Breast Cancer Transcriptome Datasets and Molecular Subtypes
3.13. Invasion Assays for Validation of Genes Selected from NetNC Results
3.14. Data and Software Availability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shlyueva, D.; Stampfel, G.; Stark, A. Transcriptional enhancers: From properties to genome-wide predictions. Nat. Rev. Genet. 2014, 15, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Stampfel, G.; Kazmar, T.; Frank, O.; Wienerroither, S.; Reiter, F.; Stark, A. Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature 2015, 528, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Rhee, D.Y.; Cho, D.-Y.; Zhai, B.; Slattery, M.; Ma, L.; Mintseris, J.; Wong, C.Y.; White, K.P.; Celniker, S.E.; Przytycka, T.M.; et al. Transcription factor networks in drosophila melanogaster. Cell Rep. 2014, 8, 2031–2043. [Google Scholar] [CrossRef] [PubMed]
- Zabidi, M.A.; Stark, A. Regulatory enhancer–core-promoter communication via transcription factors and cofactors. Trends Genet. 2016, 32, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Khoueiry, P.; Girardot, C.; Ciglar, L.; Peng, P.C.; Gustafson, E.H.; Sinha, S.; Furlong, E.E. Uncoupling evolutionary changes in DNA sequence, transcription factor occupancy and enhancer activity. eLife 2017, 6. [Google Scholar] [CrossRef]
- Wilczynski, B.; Furlong, E.E.M. Challenges for modeling global gene regulatory networks during development: Insights from Drosophila. Dev. Biol. 2010, 340, 161–169. [Google Scholar] [CrossRef]
- Li, X.; MacArthur, S.; Bourgon, R.; Nix, D.; Pollard, D.A.; Iyer, V.N.; Hechmer, A.; Simirenko, L.; Stapleton, M.; Hendriks, C.L.L.; et al. Transcription factors bind thousands of active and inactive regions in the drosophila blastoderm. PLoS Biol. 2008, 6, e27. [Google Scholar] [CrossRef]
- Ozdemir, A.; Fisher-Aylor, K.I.; Pepke, S.; Samanta, M.; Dunipace, L.; McCue, K.; Zeng, L.; Ogawa, N.; Wold, B.J.; Stathopoulos, A. High resolution mapping of twist to DNA in drosophila embryos: Efficient functional analysis and evolutionary conservation. Genome Res. 2011, 21, 566–577. [Google Scholar] [CrossRef]
- Biggin, M.D. Animal transcription networks as highly connected, quantitative continua. Dev. Cell 2011, 21, 611–626. [Google Scholar] [CrossRef]
- Roy, S.; Ernst, J.; Kharchenko, P.V.; Kheradpour, P.; Negre, N.; Eaton, M.L.; Landolin, J.M.; Bristow, C.A.; Ma, L.; Lin, M.F.; et al. Identification of functional elements and regulatory circuits by drosophila modENCODE. Science 2010, 330, 1787–1797. [Google Scholar] [CrossRef]
- Kvon, E.Z.; Stampfel, G.; Yáñez-Cuna, J.O.; Dickson, B.J.; Stark, A. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev. 2012, 26, 908–913. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, H.; Liu, F.; Ren, C.; Wang, S.; Bo, X.; Shu, W. Functional annotation of hot regions in the human genome: Implications for human disease and cancer. Sci. Rep. 2015, 5, 11633. [Google Scholar] [CrossRef] [PubMed]
- Moorman, C.; Sun, L.V.; Wang, J.; de Wit, E.; Talhout, W.; Ward, L.D.; Greil, F.; Lu, X.-J.; White, K.P.; Bussemaker, H.J.; et al. Hotspots of transcription factor colocalization in the genome of drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2006, 103, 12027–12032. [Google Scholar] [CrossRef]
- Montavon, T.; Soshnikova, N.; Mascrez, B.; Joye, E.; Thevenet, L.; Splinter, E.; de Laat, W.; Spitz, F.; Duboule, D. A regulatory archipelago controls hox genes transcription in digits. Cell 2011, 147, 1132–1145. [Google Scholar] [CrossRef]
- Teytelman, L.; Thurtle, D.M.; Rine, J.; van Oudenaarden, A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 18602–18607. [Google Scholar] [CrossRef]
- Cannavò, E.; Khoueiry, P.; Garfield, D.; Geeleher, G.; Zichner, T.; Gustafson, E.; Ciglar, L.; Korbel, J.; Furlong, E. Shadow enhancers are pervasive features of developmental regulatory networks. Curr. Biol. 2016, 26, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef]
- Keung, A.J.; Bashor, C.J.; Kiriakov, S.; Collins, J.J.; Khalil, A.S. Using targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation. Cell 2014, 158, 110–120. [Google Scholar] [CrossRef]
- Brown, J.B.; Celniker, S.E. Lessons from modENCODE. Annu. Rev. Genom. Hum. Genet. 2015, 16, 31–53. [Google Scholar] [CrossRef]
- Igual, J.C.; Johnson, A.L.; Johnston, L.H. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996, 15, 5001–5013. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Snyder, M.; Altman, R.B.; Tatonetti, N.P. Coherent functional modules improve transcription factor target identification, cooperativity prediction, and disease association. PLoS Genet. 2014, 10, e1004122. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, S.; Li, X.-Y.; Li, J.; Brown, J.B.; Chu, H.C.; Zeng, L.; Grondona, B.P.; Hechmer, A.; Simirenko, L.; Keränen, S.V.; et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009, 10, R80. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H.; Hopfield, J.J.; Leibler, S.; Murray, A.W. From molecular to modular cell biology. Nature 1999, 402, C47–C52. [Google Scholar] [CrossRef] [PubMed]
- Hooper, S.D.; Boué, S.; Krause, R.; Jensen, L.J.; Mason, C.E.; Ghanim, M.; White, K.P.; Furlong, E.E.; Bork, P. Identification of tightly regulated groups of genes during Drosophila melanogaster embryogenesis. Mol. Syst. Biol. 2007, 3, 72. [Google Scholar] [CrossRef] [PubMed]
- Ideker, T.; Ozier, O.; Schwikowski, B.; Siegel, A.F. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics 2002, 18, S233–S240. [Google Scholar] [CrossRef]
- Vidal, M.; Cusick, M.E.; Barabási, A.-L. Interactome networks and human disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Igea, A.; Arroyo, R.; Alcalde, V.; Canovas, B.; Orozco, M.; Nebreda, A.R.; Aloy, P. Quantification of pathway cross-talk reveals novel synergistic drug combinations for breast cancer. Cancer Res. 2017, 77, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.X.; Thomas, C.E.; Brunak, S. Network biology concepts in complex disease comorbidities. Nat. Rev. Genet. 2016, 17, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Greene, C.S.; Krishnan, A.; Wong, A.K.; Ricciotti, E.; Zelaya, R.A.; Himmelstein, D.S.; Zhang, R.; Hartmann, B.M.; Zaslavsky, E.; Sealfon, S.C.; et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015, 47, 569–576. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, M.; Chen, J.; Pan, Y. A fast hierarchical clustering algorithm for functional modules discovery in protein interaction networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2011, 8, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Pawson, T.; Nash, P. Assembly of cell regulatory systems through protein interaction domains. Science 2003, 300, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef]
- Lim, J.; Thiery, J.P. Epithelial-mesenchymal transitions: Insights from development. Development 2012, 139, 3471–3486. [Google Scholar] [CrossRef]
- Giampieri, S.; Manning, C.; Hooper, S.; Jones, L.; Hill, C.S.; Sahai, E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat. Cell Biol. 2009, 11, 1287–1296. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38, D355–D360. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Pacifico, S.; Liu, G.; Finley, R.L., Jr. DroID: The drosophila interactions database, a comprehensive resource for annotated gene and protein interactions. BMC Genom. 2008, 9, 461. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Guruharsha, K.G.; Rual, J.-F.; Zhai, B.; Mintseris, J.; Vaidya, P.; Vaidya, N.; Beekman, C.; Wong, C.; Rhee, D.Y.; Cenaj, O.; et al. A protein complex network of drosophila melanogaster. Cell 2011, 147, 690. [Google Scholar] [CrossRef] [PubMed]
- Vitali, F.; Li, Q.; Schissler, A.G.; Berghout, J.; Kenost, C.; Lussier, Y.A. Developing a ‘personalome’ for precision medicine: Emerging methods that compute interpretable effect sizes from single-subject transcriptomes. Brief. Bioinform. 2017, bbx149. [Google Scholar] [CrossRef]
- Goldberg, D.S.; Roth, F.P. Assessing experimentally derived interactions in a small world. Proc. Natl. Acad. Sci. USA 2003, 100, 4372–4376. [Google Scholar] [CrossRef]
- Leiserson, M.D.M.; Vandin, F.; Wu, H.-T.; Dobson, J.R.; Eldridge, J.V.; Thomas, J.L.; Papoutsaki, A.; Kim, Y.; Niu, B.; McLellan, M.; et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 2015, 47, 106–114. [Google Scholar] [CrossRef]
- Taşan, M.; Musso, G.; Hao, T.; Vidal, M.; MacRae, C.A.; Roth, F.P. Selecting causal genes from genome-wide association studies via functionally-coherent subnetworks. Nat. Methods 2015, 12, 154–159. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef]
- Simonis, M.; Klous, P.; Splinter, E.; Moshkin, Y.; Willemsen, R.; de Wit, E.; van Steensel, B.; de Laat, W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 2006, 38, 1348. [Google Scholar] [CrossRef]
- Dostie, J.; Richmond, T.A.; Arnaout, R.A.; Selzer, R.R.; Lee, W.L.; Honan, T.A.; Rubio, E.D.; Krumm, A.; Lamb, J.; Nusbaum, C.; et al. Chromosome conformation capture carbon copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006, 16, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Wang, H.; Li, W.; Zang, C.; Li, B.; Wong, Y.J.; Meyer, C.; Liu, J.S.; Aster, J.C.; Liu, S. Network analysis of gene essentiality in functional genomics experiments. Genome Biol. 2015, 16, 239. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, T.; Girardot, C.; Brehme, M.; Tongprasit, W.; Stolc, V.; Furlong, E.E.M. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007, 21, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Zeitlinger, J.; Zinzen, R.P.; Stark, A.; Kellis, M.; Zhang, H.; Young, R.A.; Levine, M. Whole-genome ChIP-chip analysis of dorsal, twist, and snail suggests integration of diverse patterning processes in the drosophila embryo. Genes Dev. 2007, 21, 385–390. [Google Scholar] [CrossRef]
- Chen, R.A.-J.; Stempor, P.; Down, T.A.; Zeiser, E.; Feuer, S.K.; Ahringer, J. Extreme HOT regions are CpG-dense promoters in C. elegans and humans. Genome Res. 2014, 24, 1138–1146. [Google Scholar] [CrossRef]
- Boyle, A.P.; Araya, C.L.; Brdlik, C.; Cayting, P.; Cheng, C.; Cheng, Y.; Gardner, K.; Hillier, L.; Janette, J.; Jiang, L.; et al. Comparative analysis of regulatory information and circuits across distant species. Nature 2014, 512, 453. [Google Scholar] [CrossRef]
- Jolma, A.; Yin, Y.; Nitta, K.R.; Dave, K.; Popov, A.; Taipale, M.; Enge, M.; Kivioja, T.; Morgunova, E.; Taipale, J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 2015, 527, 384–388. [Google Scholar] [CrossRef]
- Long, H.K.; Prescott, S.L.; Wysocka, J. Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell 2016, 167, 1170–1187. [Google Scholar] [CrossRef]
- Spitz, F.; Furlong, E.E.M. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef]
- Chen, J.; Hu, Z.; Phatak, M.; Reichard, J.; Freudenberg, J.M.; Sivaganesan, S.; Medvedovic, M. Genome-wide signatures of transcription factor activity: Connecting transcription factors, disease, and small molecules. PLoS Comput. Biol. 2013, 9, e1003198. [Google Scholar] [CrossRef]
- Aris-Brosou, S. Determinants of adaptive evolution at the molecular level: The extended complexity hypothesis. Mol. Biol. Evol. 2005, 22, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Wieschaus, E.; Nüsslein-Volhard, C. The heidelberg screen for pattern mutants of drosophila: A personal account. Annu. Rev. Cell Dev. Biol. 2016, 32, 1–46. [Google Scholar] [CrossRef]
- Gheisari, E.; Aakhte, M.; Müller, H.-A.J. Gastrulation in drosophila melanogaster: Genetic control, cellular basis and biomechanics. Mech. Dev. 2020, 103629. [Google Scholar] [CrossRef] [PubMed]
- Guruharsha, K.G.; Kankel, M.W.; Artavanis-Tsakonas, S. The Notch signalling system: Recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 2012, 13, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Ntziachristos, P.; Lim, J.S.; Sage, J.; Aifantis, I. From fly wings to targeted cancer therapies: A centennial for notch signaling. Cancer Cell 2014, 25, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Nowell, C.S.; Radtke, F. Notch as a tumour suppressor. Nat. Rev. Cancer 2017, 17, 145. [Google Scholar] [CrossRef]
- Bernard, F.; Krejci, A.; Housden, B.; Adryan, B.; Bray, S.J. Specificity of notch pathway activation: Twist controls the transcriptional output in adult muscle progenitors. Development 2010, 137, 2633–2642. [Google Scholar] [CrossRef]
- Sahlgren, C.; Gustafsson, M.V.; Jin, S.; Poellinger, L.; Lendahl, U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. USA 2008, 105, 6392–6397. [Google Scholar] [CrossRef]
- Baylies, M.K.; Bate, M. Twist: A myogenic switch in drosophila. Science 1996, 272, 1481–1484. [Google Scholar] [CrossRef]
- Xie, Y.; Li, X.; Deng, X.; Hou, Y.; O’Hara, K.; Urso, A.; Peng, Y.; Chen, L.; Zhu, S. The ets protein pointed prevents both premature differentiation and dedifferentiation of drosophila intermediate neural progenitors. Development 2016, 143, 3109–3118. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-M.; Freedman, J.A.; Bettler, D.R.; Manning, S.D.; Giep, S.N.; Steiner, J.; Ellis, H.M. Polychaetoid is required to restrict segregation of sensory organ precursors from proneural clusters in drosophila. Mech. Dev. 1996, 57, 215–227. [Google Scholar] [CrossRef]
- Lo, P.C.H.; Skeath, J.B.; Gajewski, K.; Schulz, R.A.; Frasch, M. Homeotic genes autonomously specify the anteroposterior subdivision of the drosophila dorsal vessel into aorta and heart. Dev. Biol. 2002, 251, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, G.V.; Nodal, D.H.; Lovato, C.V.; Hendren, J.D.; Helander, L.A.; Lovato, T.L.; Bodmer, R.; Cripps, R.M. The canonical wingless signaling pathway is required but not sufficient for inflow tract formation in the drosophila melanogaster heart. Dev. Biol. 2016, 413, 16–25. [Google Scholar] [CrossRef]
- Hammonds, A.S.; Bristow, C.A.; Fisher, W.W.; Weiszmann, R.; Wu, S.; Hartenstein, V.; Kellis, M.; Yu, B.; Frise, E.; Celniker, S.E. Spatial expression of transcription factors in Drosophila embryonic organ development. Genome Biol. 2013, 14, R140. [Google Scholar] [CrossRef]
- Tomancak, P.; Beaton, A.; Weiszmann, R.; Kwan, E.; Shu, S.; Lewis, S.E.; Richards, S.; Ashburner, M.; Hartenstein, V.; Celniker, S.E.; et al. Systematic determination of patterns of gene expression during drosophila embryogenesis. Genome Biol. 2002, 3, research0088. [Google Scholar] [CrossRef]
- Hartley, D.; Xu, T.; Artavanis-Tsakonas, S. The embryonic expression of the notch locus of drosophila melanogaster and the implications of point mutations in the extracellular EGF-like domain of the predicted protein., the embryonic expression of the notch locus of drosophila melanogaster and the implications of point mutations in the extracellular EGF-like domain of the predicted protein. EMBO J. 1987, 6, 3407–3417. [Google Scholar]
- Kusch, T.; Reuter, R. Functions for drosophila brachyenteron and forkhead in mesoderm specification and cell signalling. Development 1999, 126, 3991–4003. [Google Scholar]
- Millo, H.; Bownes, M. The expression pattern and cellular localisation of myosin VI during the drosophila melanogaster life cycle. Gene Expr. Patterns 2007, 7, 501–510. [Google Scholar] [CrossRef]
- Kuroda, M.I.; Kang, H.; De, S.; Kassis, J.A. Dynamic competition of polycomb and trithorax in transcriptional programming. Annu. Rev. Biochem. 2020, 89, 235–253. [Google Scholar] [CrossRef]
- Shao, Z.; Raible, F.; Mollaaghababa, R.; Guyon, J.R.; Wu, C.; Bender, W.; Kingston, R.E. Stabilization of chromatin structure by PRC1, a polycomb complex. Cell 1999, 98, 37–46. [Google Scholar] [CrossRef]
- Schotta, G.; Ebert, A.; Krauss, V.; Fischer, A.; Hoffmann, J.; Rea, S.; Jenuwein, T.; Dorn, R.; Reuter, G. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002, 21, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Lopez, A.; Higuet, D.; Rosset, R.; Deutsch, J.; Peronnet, F. Corto genetically interacts with Pc-G and trx-G genes and maintains the anterior boundary of Ultrabithorax expression in drosophila larvae. Mol. Gen. Genom. 2001, 266, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Mishra, K.; Chopra, V.S.; Srinivasan, A.; Mishra, R.K. Trl-GAGA directly interacts with lola like and both are part of the repressive complex of polycomb group of genes. Mech. Dev. 2003, 120, 681–689. [Google Scholar] [CrossRef]
- Schuster, K.J.; Smith-Bolton, R.K. Taranis protects regenerating tissue from fate changes induced by the wound response in drosophila. Dev. Cell 2015, 34, 119–128. [Google Scholar] [CrossRef]
- Tie, F.; Banerjee, R.; Saiakhova, A.R.; Howard, B.; Monteith, K.E.; Scacheri, P.C.; Cosgrove, M.S.; Harte, P.J. Trithorax monomethylates histone H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize Polycomb silencing. Development (Cambrige. Engl.) 2014, 141, 1129. [Google Scholar] [CrossRef]
- Gutierrez, L. The drosophila trithorax group gene tonalli(tna) interacts genetically with the Brahma remodeling complex and encodes an SP-RING finger protein. Development 2003, 130, 343–354. [Google Scholar] [CrossRef]
- Crosby, M.A.; Miller, C.; Alon, T.; Watson, K.L.; Verrijzer, C.P.; Goldman-Levi, R.; Zak, N.B. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in drosophila melanogaster. Mol. Cell. Biol. 1999, 19, 1159–1170. [Google Scholar] [CrossRef]
- Fanti, L.; Dorer, D.R.; Berloco, M.; Henikoff, S.; Pimpinelli, S. Heterochromatin protein 1 binds transgene arrays. Chromosoma 1998, 107, 286–292. [Google Scholar] [CrossRef]
- Pulikkan, J.A.; Hegde, M.; Ahmad, H.M.; Belaghzal, H.; Illendula, A.; Yu, J.; O’Hagan, K.; Ou, J.; Muller-Tidow, C.; Wolfe, S.A.; et al. CBFβ-SMMHC inhibition triggers apoptosis by disrupting MYC chromatin dynamics in acute myeloid leukemia. Cell 2018, 174, 172–186.e21. [Google Scholar] [CrossRef]
- Bao, X.; Deng, H.; Johansen, J.; Girton, J.; Johansen, K.M. Loss-of-function alleles of the JIL-1 histone H3S10 kinase enhance position-effect variegation at pericentric sites in drosophila heterochromatin. Genetics 2007, 176, 1355–1358. [Google Scholar] [CrossRef] [PubMed]
- Sparmann, A.; van Lohuizen, M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 2006, 6, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Koppens, M.; van Lohuizen, M. Context-dependent actions of polycomb repressors in cancer. Oncogene 2016, 35, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Herranz, N.; Pasini, D.; Díaz, V.M.; Francí, C.; Gutierrez, A.; Dave, N.; Escrivà, M.; Hernandez-Muñoz, I.; Croce, L.D.; Helin, K.; et al. Polycomb complex 2 is required for E-cadherin repression by the snail1 transcription factor. Mol. Cell. Biol. 2008, 28, 4772–4781. [Google Scholar] [CrossRef] [PubMed]
- Leptin, M. Twist and snail as positive and negative regulators during drosophila mesoderm development. Genes Dev. 1991, 5, 1568–1576. [Google Scholar] [CrossRef]
- Gilmour, D.; Rembold, M.; Leptin, M. From morphogen to morphogenesis and back. Nature 2017, 541, 311–320. [Google Scholar] [CrossRef]
- Ashraf, S.I.; Ip, Y.T. The snail protein family regulates neuroblast expression of inscuteable and string, genes involved in asymmetry and cell division in drosophila. Development 2001, 128, 4757–4767. [Google Scholar]
- Zander, M.A.; Burns, S.E.; Yang, G.; Kaplan, D.R.; Miller, F.D. Snail coordinately regulates downstream pathways to control multiple aspects of mammalian neural precursor development. J. Neurosci. 2014, 34, 5164–5175. [Google Scholar] [CrossRef]
- Nevil, M.; Bondra, E.R.; Schulz, K.N.; Kaplan, T.; Harrison, M.M. Stable binding of the conserved transcription factor grainy head to its target genes throughout drosophila melanogaster development. Genetics 2017, 205, 605–620. [Google Scholar] [CrossRef]
- Caron, S.J.C.; Ruta, V.; Abbott, L.F.; Axel, R. Random convergence of olfactory inputs in the drosophila mushroom body. Nature 2013, 497, 113–117. [Google Scholar] [CrossRef]
- Lin, S.; Ewen-Campen, B.; Ni, X.; Housden, B.E.; Perrimon, N. In vivo transcriptional activation using CRISPR/Cas9 in drosophila. Genetics 2015, 201, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Vesuna, F.; van Diest, P.; Chen, J.H.; Raman, V. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem. Biophys. Res. Commun. 2008, 367, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Mohr, S.E.; Hu, Y.; Kim, K.; Housden, B.E.; Perrimon, N. Resources for functional genomics studies in drosophila melanogaster. Genetics 2014, 197, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef]
- Cejalvo, J.M.; de Dueñas, E.M.; Galvan, P.; García-Recio, S.; Gasión, O.B.; Paré, L.; Antolin, S.; Martinello, R.; Blancas, I.; Adamo, B.; et al. Intrinsic subtypes and gene expression profiles in primary and metastatic breast cancer. Cancer Res. 2017, 77, 2213–2221. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346. [Google Scholar] [CrossRef]
- Stylianou, S.; Clarke, R.B.; Brennan, K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006, 66, 1517–1525. [Google Scholar] [CrossRef]
- Barnawi, R.; Al-Khaldi, S.; Majed Sleiman, G.; Sarkar, A.; Al-Dhfyan, A.; Al-Mohanna, F.; Ghebeh, H.; Al-Alwan, M. Fascin is critical for the maintenance of breast cancer stem cell pool predominantly via the activation of the notch self-renewal pathway. Stem Cells 2016, 34, 2799–2813. [Google Scholar] [CrossRef]
- Ingthorsson, S.; Briem, E.; Bergthorsson, J.T.; Gudjonsson, T. Epithelial plasticity during human breast morphogenesis and cancer progression. J. Mammary Gland. Biol. Neoplasia 2016, 21, 139–148. [Google Scholar] [CrossRef]
- Moleirinho, S.; Chang, N.; Sims, A.H.; Tilston-Lünel, A.M.; Angus, L.; Steele, A.; Boswell, V.; Barnett, S.C.; Ormandy, C.; Faratian, D.; et al. KIBRA exhibits MST-independent functional regulation of the hippo signaling pathway in mammals. Oncogene 2013, 32, 1821–1830. [Google Scholar] [CrossRef]
- Venet, D.; Dumont, J.E.; Detours, V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput. Biol. 2011, 7, e1002240. [Google Scholar] [CrossRef]
- Sarrió, D.; Rodriguez-Pinilla, S.M.; Hardisson, D.; Cano, A.; Moreno-Bueno, G.; Palacios, J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008, 68, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Beltran, A.S.; Graves, L.M.; Blancafort, P. Novel role of engrailed 1 as a prosurvival transcription factor in basal-like breast cancer and engineering of interference peptides block its oncogenic function. Oncogene 2014, 33, 4767–4777. [Google Scholar] [CrossRef]
- Adélaïde, J.; Finetti, P.; Bekhouche, I.; Repellini, L.; Geneix, J.; Sircoulomb, F.; Charafe-Jauffret, E.; Cervera, N.; Desplans, J.; Parzy, D.; et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007, 67, 11565–11575. [Google Scholar] [CrossRef] [PubMed]
- Letessier, A.; Ginestier, C.; Charafe-Jauffret, E.; Cervera, N.; Adélaïde, J.; Gelsi-Boyer, V.; Ahomadegbe, J.-C.; Benard, J.; Jacquemier, J.; Birnbaum, D.; et al. ETV6 gene rearrangements in invasive breast carcinoma. Genes Chromosomes Cancer 2005, 44, 103–108. [Google Scholar] [CrossRef]
- Chapellier, M.; Bachelard-Cascales, E.; Schmidt, X.; Clément, F.; Treilleux, I.; Delay, E.; Jammot, A.; Ménétrier-Caux, C.; Pochon, G.; Besançon, R.; et al. Disequilibrium of BMP2 levels in the breast stem cell niche launches epithelial transformation by overamplifying BMPR1B cell response. Stem Cell Rep. 2015, 4, 239–254. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Lu, M.-F.; Schwartz, R.J.; Martin, J.F. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development 2005, 132, 5601–5611. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kand, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Popovici, V.; Chen, W.; Gallas, B.G.; Hatzis, C.; Shi, W.; Samuelson, F.W.; Nikolsky, Y.; Tsyganova, M.; Ishkin, A.; Nikolskaya, T.; et al. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 2010, 12, R5. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.-J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- DiMeo, T.A.; Anderson, K.; Phadke, P.; Feng, C.; Perou, C.M.; Naber, S.; Kuperwasser, C. A novel lung metastasis signature links wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009, 69, 5364–5373. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.M.; Panzilius, E.; Bartsch, H.S.; Irmler, M.; Beckers, J.; Kari, V.; Linnemann, J.R.; Dragoi, D.; Hirschi, B.; Kloos, U.J.; et al. Stem-cell-like properties and epithelial plasticity arise as stable traits after transient twist1 activation. Cell Rep. 2015, 10, 131–139. [Google Scholar] [CrossRef]
- Sieuwerts, A.M.; Kraan, J.; Bolt, J.; van der Spoel, P.; Elstrodt, F.; Schutte, M.; Martens, J.W.M.; Gratama, J.-W.; Sleijfer, S.; Foekens, J.A. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J. Natl. Cancer Inst. 2009, 101, 61–66. [Google Scholar] [CrossRef]
- Lawson, D.A.; Bhakta, N.R.; Kessenbrock, K.; Prummel, K.D.; Yu, Y.; Takai, K.; Zhou, A.; Eyob, H.; Balakrishnan, S.; Wang, C.-Y.; et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015, 526, 131–135. [Google Scholar] [CrossRef]
- Guo, W.; Keckesova, Z.; Donaher, J.L.; Shibue, T.; Tischler, V.; Reinhardt, F.; Itzkovitz, S.; Noske, A.; Zürrer-Härdi, U.; Bell, G.; et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012, 148, 1015–1028. [Google Scholar] [CrossRef]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23. [Google Scholar] [CrossRef]
- Resende, L.P.F.; Truong, M.E.; Gomez, A.; Jones, D.L. Intestinal stem cell ablation reveals differential requirements for survival in response to chemical challenge. Dev. Biol. 2017, 424, 10–17. [Google Scholar] [CrossRef]
- Steneberg, P.; Englund, C.; Kronhamn, J.; Weaver, T.A.; Samakovlis, C. Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the drosophila trachea. Genes Dev. 1998, 12, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Amith, S.R.; Fliegel, L. Na+/H+ exchanger-mediated hydrogen ion extrusion as a carcinogenic signal in triple-negative breast cancer etiopathogenesis and prospects for its inhibition in therapeutics. Semin. Cancer Biol. 2017, 43, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Kimball, S.; Liu, H.; Holowatyj, A.; Yang, Z.-Q.; Liu, L.; Kimball, S.; Liu, H.; Holowatyj, A.; Yang, Z.-Q. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget 2014, 6, 2466–2482. [Google Scholar] [CrossRef] [PubMed]
- Cheong, J.K.; Gunaratnam, L.; Zang, Z.J.; Yang, C.M.; Sun, X.; Nasr, S.L.; Sim, K.G.; Peh, B.K.; Rashid, S.B.A.; Bonventre, J.V.; et al. TRIP-Br2 promotes oncogenesis in nude mice and is frequently overexpressed in multiple human tumors. J. Transl. Med. 2009, 7, 8. [Google Scholar] [CrossRef]
- García-Pedrero, J.M.; Kiskinis, E.; Parker, M.G.; Belandia, B. The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J. Biol. Chem. 2006, 281, 22656–22664. [Google Scholar] [CrossRef]
- Sethuraman, A.; Brown, M.; Seagroves, T.N.; Wu, Z.-H.; Pfeffer, L.M.; Fan, M. SMARCE1 regulates metastatic potential of breast cancer cells through the HIF1A/PTK2 pathway. Breast Cancer Res. 2016, 18, 81. [Google Scholar] [CrossRef]
- Sokol, E.S.; Feng, Y.-X.; Jin, D.X.; Tizabi, M.D.; Miller, D.H.; Cohen, M.A.; Sanduja, S.; Reinhardt, F.; Pandey, J.; Superville, D.A.; et al. SMARCE1 is required for the invasive progression of in situ cancers. Proc. Natl. Acad. Sci. USA 2017, 201703931. [Google Scholar] [CrossRef]
- Mohd-Sarip, A.; Teeuwssen, M.; Bot, A.G.; De Herdt, M.J.; Willems, S.M.; Baatenburg de Jong, R.J.; Looijenga, L.H.J.; Zatreanu, D.; Bezstarosti, K.; van Riet, J.; et al. DOC1-dependent recruitment of NURD reveals antagonism with SWI/SNF during epithelial-mesenchymal transition in oral cancer cells. Cell Rep. 2017, 20, 61–75. [Google Scholar] [CrossRef]
- Hemberger, M.; Dean, W.; Reik, W. Epigenetic dynamics of stem cells and cell lineage commitment: Digging waddington’s canal. Nat. Rev. Mol. Cell Biol. 2009, 10, 526–537. [Google Scholar] [CrossRef]
- Dhasarathy, A.; Kajita, M.; Wade, P.A. The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor alpha. Mol. Endocrinol. 2007, 21, 2907–2918. [Google Scholar] [CrossRef]
- Lacroix, M.; Leclercq, G. Relevance of breast cancer cell lines as models for breast tumours: An update. Breast Cancer Res. Treat. 2004, 83, 249–289. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.O.; Lalit, P.A.; Biermann, M.; Markandeya, Y.S.; Capes, D.L.; Addesso, L.; Patel, G.; Han, T.; John, M.C.; Powers, P.A.; et al. Irx4 marks a multipotent, ventricular-specific progenitor cell. Stem Cells 2016, 34, 2875–2888. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Hussain, W.M.; Vijai, J.; Offit, K.; Rubin, M.A.; Demichelis, F.; Klein, R.J. Variants at IRX4 as prostate cancer expression quantitative trait loci. Eur. J. Hum. Genet. 2014, 22, 558–563. [Google Scholar] [CrossRef]
- Marat, A.L.; Haucke, V. Phosphatidylinositol 3-phosphates-at the interface between cell signalling and membrane traffic. EMBO J. 2016, 35, 561–579. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Hu, Z.; Liu, J.; Gao, J.; Lin, B. Gene expression profile analysis identifies metastasis and chemoresistance-associated genes in epithelial ovarian carcinoma cells. Med. Oncol 2015, 32, 426. [Google Scholar] [CrossRef]
- Doherty, J.; Baehrecke, E.H. Life, death and autophagy. Nat. Cell Biol. 2018, 20, 1110. [Google Scholar] [CrossRef]
- Li, J.; Yang, B.; Zhou, Q.; Wu, Y.; Shang, D.; Guo, Y.; Song, Z.; Zheng, Q.; Xiong, J. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial–mesenchymal transition. Carcinogenesis 2013, 34, 1343–1351. [Google Scholar] [CrossRef]
- Mohler, J.; Weiss, N.; Murli, S.; Mohammadi, S.; Vani, K.; Vasilakis, G.; Song, C.H.; Epstein, A.; Kuang, T.; English, J. The embryonically active gene, unkempt, of Drosophila encodes a Cys3His finger protein. Genetics 1992, 131, 377–388. [Google Scholar]
- Murn, J.; Zarnack, K.; Yang, Y.J.; Durak, O.; Murphy, E.A.; Cheloufi, S.; Gonzalez, D.M.; Teplova, M.; Curk, T.; Zuber, J.; et al. Control of a neuronal morphology program by an RNA-binding zinc finger protein, unkempt. Genes Dev. 2015, 29, 501–512. [Google Scholar] [CrossRef]
- Murn, J.; Teplova, M.; Zarnack, K.; Shi, Y.; Patel, D.J. Recognition of distinct RNA motifs by the clustered CCCH zinc fingers of neuronal protein unkempt. Nat. Struct. Mol. Biol. 2016, 23, 16–23. [Google Scholar] [CrossRef]
- Parkinson, H.; Kapushesky, M.; Kolesnikov, N.; Rustici, G.; Shojatalab, M.; Abeygunawardena, N.; Berube, H.; Dylag, M.; Emam, I.; Farne, A.; et al. Array express update-from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009, 37, D868–D872. [Google Scholar] [CrossRef] [PubMed]
- Overton, I.M.; Graham, S.; Gould, K.A.; Hinds, J.; Botting, C.H.; Shirran, S.; Barton, G.J.; Coote, P.J. Global network analysis of drug tolerance, mode of action and virulence in methicillin-resistant S. aureus. BMC Syst. Biol. 2011, 5, 68. [Google Scholar] [CrossRef] [PubMed]
- Ben-Hur, A.; Noble, W.S. Kernel methods for predicting protein-protein interactions. Bioinformatics 2005, 21, i38–i46. [Google Scholar] [CrossRef]
- Van Rijsbergen, C.J. Information Retrieval; Butterworths: London, UK, 1979. [Google Scholar]
- Zhou, N.; Jiang, Y.; Bergquist, T.R.; Lee, A.J.; Kacsoh, B.Z.; Crocker, A.W.; Lewis, K.A.; Georghiou, G.; Nguyen, H.N.; Hamid, M.N.; et al. The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome Biol. 2019, 20, 244. [Google Scholar] [CrossRef]
- Park, Y.; Marcotte, E.M. Flaws in evaluation schemes for pair-input computational predictions. Nat. Methods 2012, 9, 1134–1136. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2002, 64, 479–498. [Google Scholar] [CrossRef]
- Ford, L.R.; Fulkerson, D.R. Maximal flow through a network. Can. J. Math. 1956, 8, 399–404. [Google Scholar] [CrossRef]
- Lubbock, A.L.R.; Katz, E.; Harrison, D.J.; Overton, I.M. TMA navigator: Network inference, patient stratification and survival analysis with tissue microarray data. Nucleic Acids Res. 2013, 41, W562–W568. [Google Scholar] [CrossRef]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B 1977, 39, 1–38. [Google Scholar] [CrossRef]
- Yamada, T.; Bork, P. Evolution of biomolecular networks-lessons from metabolic and protein interactions. Nat. Rev. Mol. Cell Biol. 2009, 10, 791–803. [Google Scholar] [CrossRef]
- Fitzgibbon, M.; Li, Q.; McIntosh, M. Modes of inference for evaluating the confidence of peptide identifications. J. Proteome Res. 2008, 7, 35–39. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sennels, L.; Bukowski-Wills, J.-C.; Rappsilber, J. Improved results in proteomics by use of local and peptide-class specific false discovery rates. BMC Bioinform. 2009, 10, 179. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; van Oudenaarden, A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 2008, 135, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Rifkin, S.A.; Andersen, E.; van Oudenaarden, A. Variability in gene expression underlies incomplete penetrance. Nature 2010, 463, 913–918. [Google Scholar] [CrossRef]
- Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323–334. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R.; Storey, J.D.; Tusher, V. Empirical bayes analysis of a microarray experiment. J. Am. Stat. Assoc. 2001, 96, 1151–1160. [Google Scholar] [CrossRef]
- López-Schier, H.; St Johnston, D. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 2001, 15, 1393–1405. [Google Scholar] [CrossRef]
- Schmitt, A.; Nebreda, A.R. Signalling pathways in oocyte meiotic maturation. J. Cell Sci 2002, 115, 2457–2459. [Google Scholar]
- Acharya, U.; Patel, S.; Koundakjian, E.; Nagashima, K.; Han, X.; Acharya, J.K. Modulating sphingolipid biosynthetic pathway rescues photoreceptor degeneration. Science 2003, 299, 1740–1743. [Google Scholar] [CrossRef]
- Dasgupta, U.; Bamba, T.; Chiantia, S.; Karim, P.; Tayoun, A.N.A.; Yonamine, I.; Rawat, S.S.; Rao, R.P.; Nagashima, K.; Fukusaki, E.; et al. Ceramide kinase regulates phospholipase C and phosphatidylinositol 4, 5, bisphosphate in phototransduction. Proc. Natl. Acad. Sci. USA 2009, 106, 20063–20068. [Google Scholar] [CrossRef]
- Yonamine, I.; Bamba, T.; Nirala, N.K.; Jesmin, N.; Kosakowska-Cholody, T.; Nagashima, K.; Fukusaki, E.; Acharya, J.K.; Acharya, U. Sphingosine kinases and their metabolites modulate endolysosomal trafficking in photoreceptors. J. Cell Biol 2011, 192, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Stathopoulos, A.; Van Drenth, M.; Erives, A.; Markstein, M.; Levine, M. Whole-genome analysis of dorsal-ventral patterning in the drosophila embryo. Cell 2002, 111, 687–701. [Google Scholar] [CrossRef]
- Campos-Ortega, J.A.; Hartenstein, V. The Embryonic Development of Drosophila Melanogaster, 2nd ed.; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1997; ISBN 978-3-662-22489-2. [Google Scholar]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Gramates, L.S.; Marygold, S.J.; dos Santos, G.; Urbano, J.-M.; Antonazzo, G.; Matthews, B.B.; Rey, A.J.; Tabone, C.J.; Crosby, M.A.; Emmert, D.B.; et al. Fly base at 25: Looking to the future. Nucleic Acids Res. 2017, 45, D663–D671. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Wang, P.; Boyd, A.D.; Kostov, G.; Athey, B.; Jones, E.G.; Bunney, W.E.; Myers, R.M.; Speed, T.P.; Akil, H.; et al. Evolving gene/transcript definitions significantly alter the interpretation of gene chip data. Nucleic Acids Res. 2005, 33, e175. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Bolstad, B.M.; Collin, F.; Cope, L.M.; Hobbs, B.; Speed, T.P. Summaries of affymetrix gene chip probe level data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Sims, A.H.; Smethurst, G.J.; Hey, Y.; Okoniewski, M.J.; Pepper, S.D.; Howell, A.; Miller, C.J.; Clarke, R.B. The removal of multiplicative, systematic bias allows integration of breast cancer gene expression datasets-improving meta-analysis and prediction of prognosis. BMC Med. Genom. 2008, 1, 42. [Google Scholar] [CrossRef]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef]
- Östlund, G.; Schmitt, T.; Forslund, K.; Köstler, T.; Messina, D.N.; Roopra, S.; Frings, O.; Sonnhammer, E.L.L. In paranoid 7: New algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2009, 38, D196–D203. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jiang, G.; Liu, S.; Liu, Z.; Pan, H.; Yao, R.; Liang, J. Lentivirus-delivered short hairpin RNA targeting SNAIL inhibits HepG2 cell growth. Oncol. Rep. 2013, 30, 1483–1487. [Google Scholar] [CrossRef] [PubMed]
- Peluso, S.; Douglas, A.; Hill, A.; Angelis, C.D.; Moore, B.L.; Grimes, G.; Petrovich, G.; Essafi, A.; Hill, R.E. Fibroblast growth factors (FGFs) prime the limb specific Shh enhancer for chromatin changes that balance histone acetylation mediated by E26 transformation-specific (ETS) factors. eLife 2017, 6, e28590. [Google Scholar] [CrossRef] [PubMed]
- Essafi, A.; Webb, A.; Berry, R.L.; Slight, J.; Burn, S.F.; Spraggon, L.; Velecela, V.; Martinez-Estrada, O.M.; Wiltshire, J.H.; Roberts, S.G.E.; et al. A Wt1-controlled chromatin switching mechanism underpins tissue-specific Wnt4 activation and repression. Dev. Cell 2011, 21, 559–574. [Google Scholar] [CrossRef]
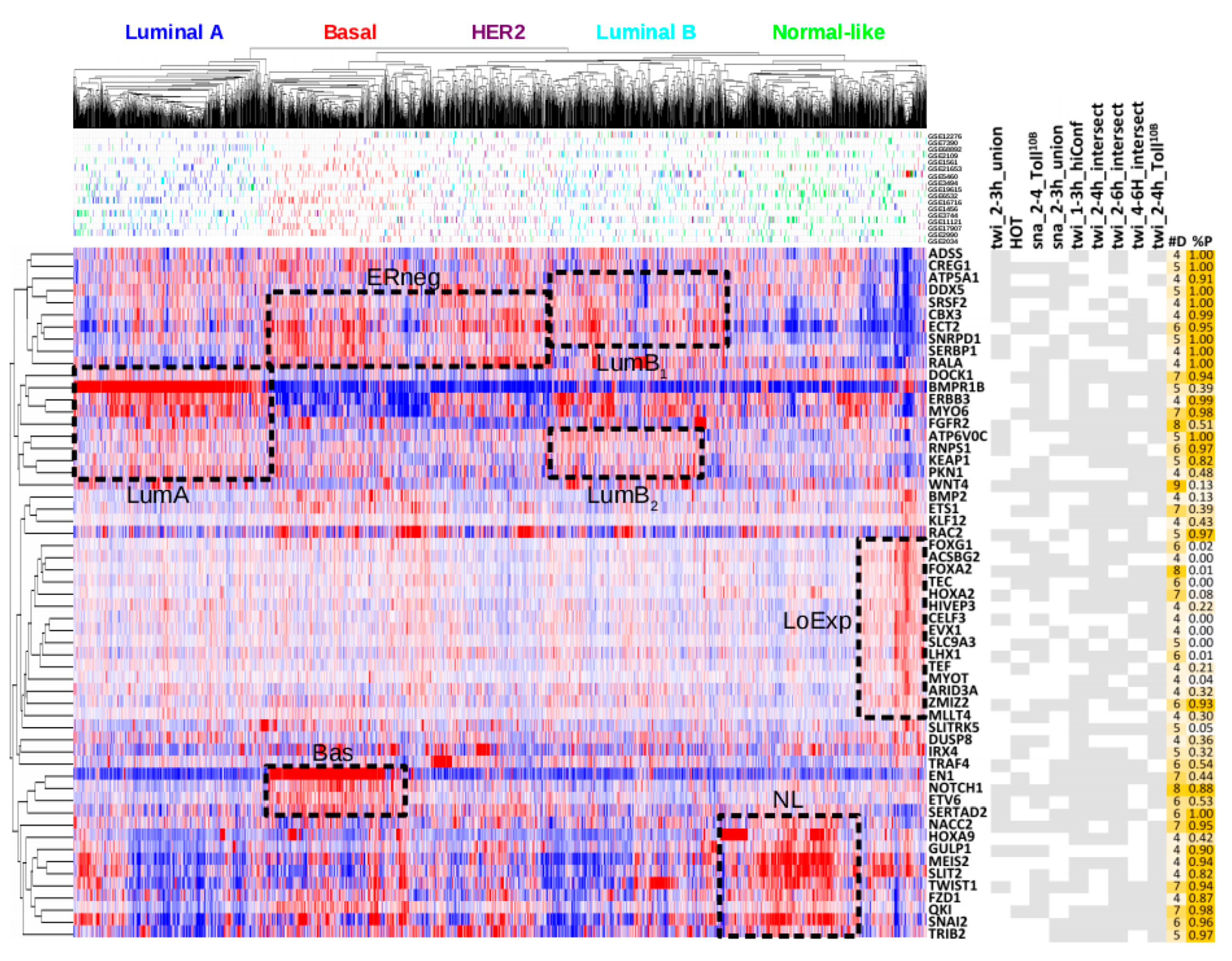
| Dataset | Predicted Functional Targets ‡ | ||||
|---|---|---|---|---|---|
| Name | Developmental Time Period(s) | Total Candidate Target Genes * | Candidate Target Genes in DroFN | NetNC-FTI | NetNC-lcFDR (95% CI) |
| twi_1–3h_hiConf | 1–3 h | 755 | 664 | 202 (30%) | 37% (32–39%) |
| twi_2–6h_intersect | 2–4 h and 4–6 h | 743 | 615 | 241 (39%) | 31% (25–33%) |
| twi_2–4h_intersect | 2–4 h only (not 4–6 h) | 1028 | 801 | 182 (23%) | 19% (14–21%) |
| twi_4–6h_intersect | 4–6 h only (not 2–4 h) | 1026 | 818 | 126 (15%) | 20% (14–22%) |
| HOT | 0–12 h + | 1648 | 677 | 174 (26%) | 27% (19–28%) |
| twi_2–3h_union | 2–3 h | 2285 | 1848 | 424 (23%) | 21% (17–22%) |
| sna_2–3h_union | 2–3 h | 1424 | 1158 | 226 (20%) | 20% (15–21%) |
| twi_2–4h_Toll10b | 2–4 h | 1578 | 1238 | 279 (23%) | 25% (20–25%) |
| sna_2–4h_Toll10b | 2–4 h | 1822 | 1488 | 211 (14%) | 13% (5–14%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Overton, I.M.; Sims, A.H.; Owen, J.A.; Heale, B.S.E.; Ford, M.J.; Lubbock, A.L.R.; Pairo-Castineira, E.; Essafi, A. Functional Transcription Factor Target Networks Illuminate Control of Epithelial Remodelling. Cancers 2020, 12, 2823. https://doi.org/10.3390/cancers12102823
Overton IM, Sims AH, Owen JA, Heale BSE, Ford MJ, Lubbock ALR, Pairo-Castineira E, Essafi A. Functional Transcription Factor Target Networks Illuminate Control of Epithelial Remodelling. Cancers. 2020; 12(10):2823. https://doi.org/10.3390/cancers12102823
Chicago/Turabian StyleOverton, Ian M., Andrew H. Sims, Jeremy A. Owen, Bret S. E. Heale, Matthew J. Ford, Alexander L. R. Lubbock, Erola Pairo-Castineira, and Abdelkader Essafi. 2020. "Functional Transcription Factor Target Networks Illuminate Control of Epithelial Remodelling" Cancers 12, no. 10: 2823. https://doi.org/10.3390/cancers12102823
APA StyleOverton, I. M., Sims, A. H., Owen, J. A., Heale, B. S. E., Ford, M. J., Lubbock, A. L. R., Pairo-Castineira, E., & Essafi, A. (2020). Functional Transcription Factor Target Networks Illuminate Control of Epithelial Remodelling. Cancers, 12(10), 2823. https://doi.org/10.3390/cancers12102823







