VE1 Immunohistochemistry Improves the Limit of Genotyping for Detecting BRAFV600E Mutation in Papillary Thyroid Cancer
Abstract
1. Introduction
2. Results
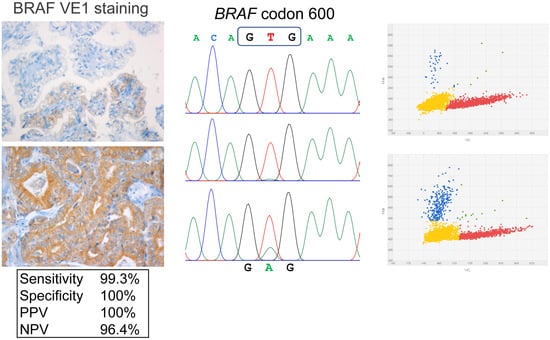
2.1. VE1 Immunohistochemical Staining vs. Sanger Sequencing
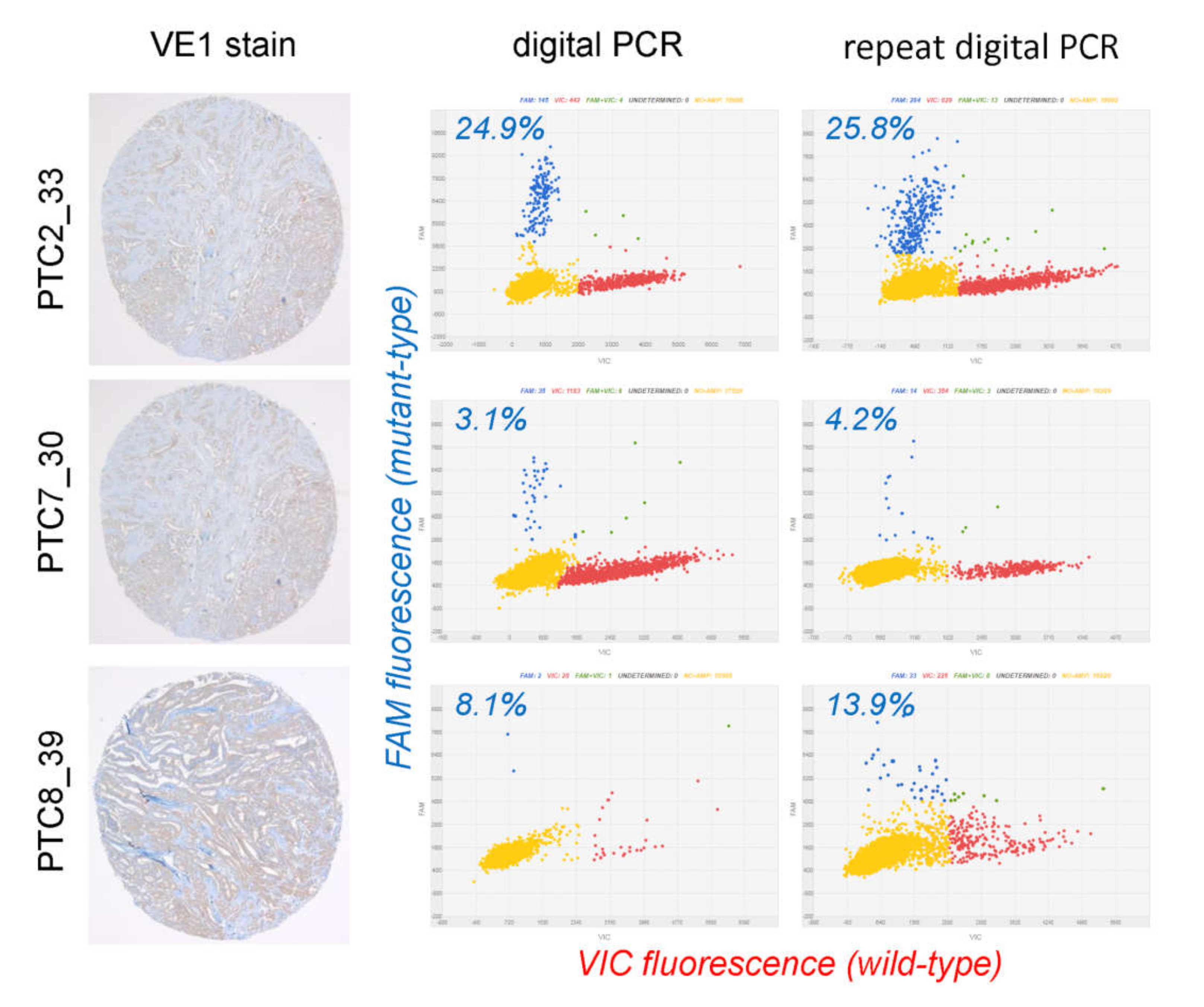
2.2. Evaluation of Discordant Cases
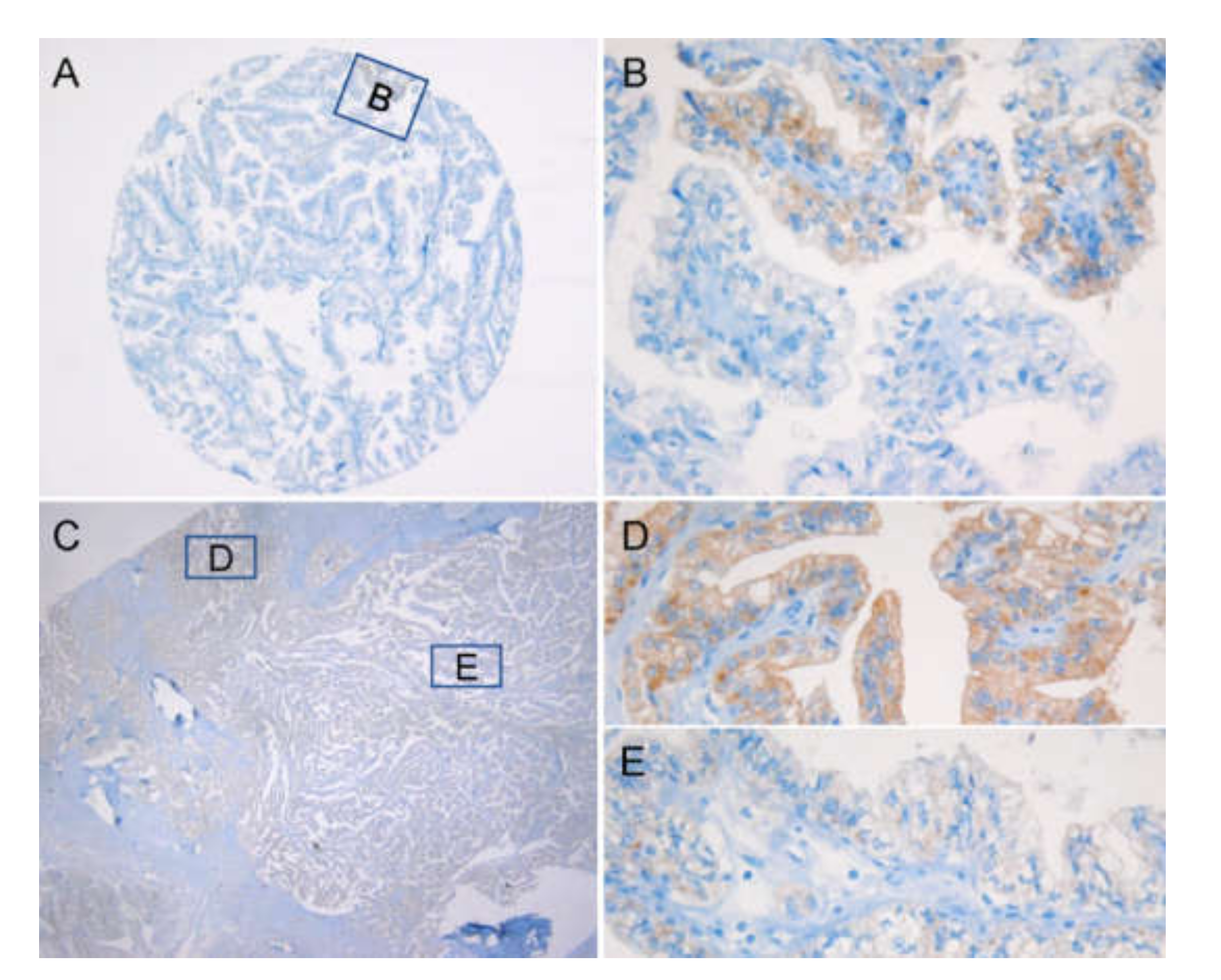
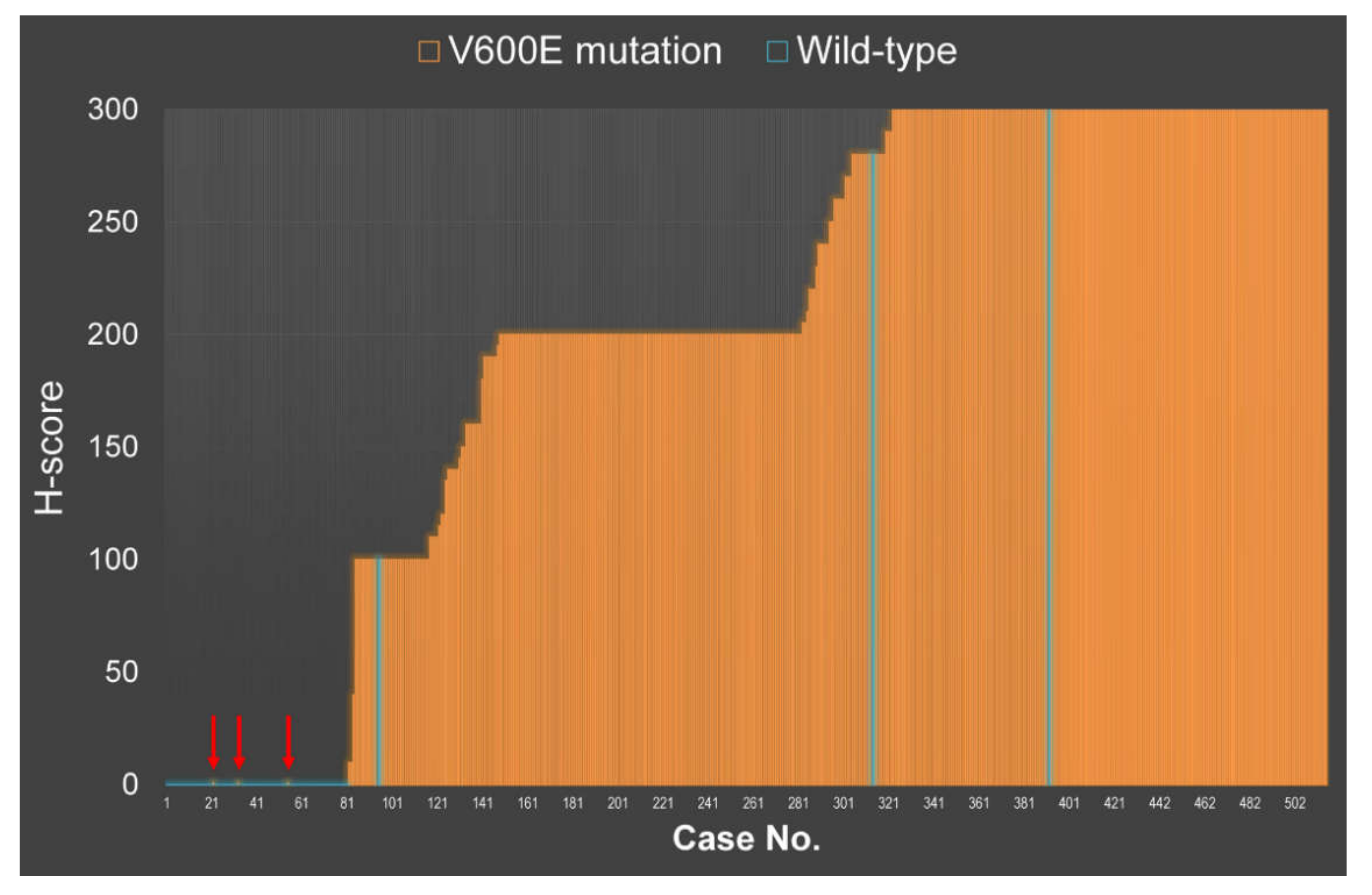
2.3. VE1 Performance in PTC Tissue Microarray (TMA)
2.4. Clinical Correlation
3. Discussion
4. Materials and Methods
4.1. Case Selection
4.2. Tissue Microarray Construction
4.3. VE1 Immunohistochemistry
4.4. BRAF Sanger Sequencing
4.5. Digital PCR Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Vaccarella, S.; Franceschi, S.; Bray, F.; Wild, C.P.; Plummer, M.; Dal Maso, L. Worldwide Thyroid-Cancer Epidemic? The Increasing Impact of Overdiagnosis. N. Engl. J. Med. 2016, 375, 614–617. [Google Scholar] [CrossRef] [PubMed]
- Pellegriti, G.; Frasca, F.; Regalbuto, C.; Squatrito, S.; Vigneri, R. Worldwide increasing incidence of thyroid cancer: Update on epidemiology and risk factors. J. Cancer Epidemiol. 2013, 2013, 965212. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Miyauchi, A.; Kihara, M.; Fukushima, M.; Higashiyama, T.; Miya, A. Overall Survival of Papillary Thyroid Carcinoma Patients: A Single-Institution Long-Term Follow-Up of 5897 Patients. World J. Surg. 2018, 42, 615–622. [Google Scholar] [CrossRef]
- Fagin, J.A.; Wells, S.A., Jr. Biologic and Clinical Perspectives on Thyroid Cancer. N. Engl. J. Med. 2016, 375, 1054–1067. [Google Scholar] [CrossRef]
- Bychkov, A.; Jung, C.K.; Liu, Z.; Kakudo, K. Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features in Asian Practice: Perspectives for Surgical Pathology and Cytopathology. Endocr. Pathol. 2018, 29, 276–288. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Xing, M. BRAF mutation in papillary thyroid cancer: Pathogenic role, molecular bases, and clinical implications. Endocr. Rev. 2007, 28, 742–762. [Google Scholar] [CrossRef]
- Xing, M. Prognostic utility of BRAF mutation in papillary thyroid cancer. Mol. Cell Endocrinol. 2010, 321, 86–93. [Google Scholar] [CrossRef]
- Xing, M.; Alzahrani, A.S.; Carson, K.A.; Viola, D.; Elisei, R.; Bendlova, B.; Yip, L.; Mian, C.; Vianello, F.; Tuttle, R.M.; et al. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA 2013, 309, 1493–1501. [Google Scholar] [CrossRef]
- Liu, C.; Chen, T.; Liu, Z. Associations between BRAF(V600E) and prognostic factors and poor outcomes in papillary thyroid carcinoma: A meta-analysis. World J. Surg. Oncol. 2016, 14, 241. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Qu, S.; Zhu, G.; Wang, F.; Liu, R.; Shen, X.; Viola, D.; Elisei, R.; Puxeddu, E.; Fugazzola, L.; et al. BRAF V600E Mutation-Assisted Risk Stratification of Solitary Intrathyroidal Papillary Thyroid Cancer for Precision Treatment. J. Natl. Cancer Inst. 2018, 110, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Bible, K.C.; Ryder, M. Mutated BRAF and personalised medicine in differentiated thyroid cancer. Lancet Oncol. 2016, 17, 1181–1183. [Google Scholar] [CrossRef]
- Matrone, A.; Valerio, L.; Pieruzzi, L.; Giani, C.; Cappagli, V.; Lorusso, L.; Agate, L.; Puleo, L.; Viola, D.; Bottici, V.; et al. Protein kinase inhibitors for the treatment of advanced and progressive radiorefractory thyroid tumors: From the clinical trials to the real life. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.S.; Lim, J.A.; Park, Y.J. Mutation Profile of Well-Differentiated Thyroid Cancer in Asians. Endocrinol. Metab. (Seoul) 2015, 30, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Bychkov, A. Prevalence of BRAF(V600E) mutation in Asian patients with thyroid cancer. Malays. J. Pathol. 2017, 39, 95–96. [Google Scholar] [PubMed]
- Fisher, K.E.; Neill, S.G.; Ehsani, L.; Caltharp, S.A.; Siddiqui, M.T.; Cohen, C. Immunohistochemical Investigation of BRAF p.V600E mutations in thyroid carcinoma using 2 separate BRAF antibodies. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 562–567. [Google Scholar] [CrossRef]
- Zimmermann, A.K.; Camenisch, U.; Rechsteiner, M.P.; Bode-Lesniewska, B.; Rossle, M. Value of immunohistochemistry in the detection of BRAF(V600E) mutations in fine-needle aspiration biopsies of papillary thyroid carcinoma. Cancer Cytopathol. 2014, 122, 48–58. [Google Scholar] [CrossRef]
- Martinuzzi, C.; Pastorino, L.; Andreotti, V.; Garuti, A.; Minuto, M.; Fiocca, R.; Bianchi-Scarra, G.; Ghiorzo, P.; Grillo, F.; Mastracci, L. A combination of immunohistochemistry and molecular approaches improves highly sensitive detection of BRAF mutations in papillary thyroid cancer. Endocrine 2016, 53, 672–680. [Google Scholar] [CrossRef]
- Oh, H.S.; Kwon, H.; Park, S.; Kim, M.; Jeon, M.J.; Kim, T.Y.; Shong, Y.K.; Kim, W.B.; Choi, J.; Kim, W.G.; et al. Comparison of Immunohistochemistry and Direct Sanger Sequencing for Detection of the BRAF(V600E) Mutation in Thyroid Neoplasm. Endocrinol. Metab. (Seoul) 2018, 33, 62–69. [Google Scholar] [CrossRef]
- Rossi, E.D.; Martini, M.; Capodimonti, S.; Cenci, T.; Straccia, P.; Angrisani, B.; Ricci, C.; Lanza, P.; Lombardi, C.P.; Pontecorvi, A.; et al. Analysis of immunocytochemical and molecular BRAF expression in thyroid carcinomas: A cytohistologic institutional experience. Cancer Cytopathol. 2014, 122, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Lu, H.; Guo, L.; Huang, W.; Ling, Y.; Shan, L.; Li, W.; Ying, J.; Lv, N. Detection of BRAF mutation in Chinese tumor patients using a highly sensitive antibody immunohistochemistry assay. Sci. Rep. 2015, 5, 9211. [Google Scholar] [CrossRef] [PubMed]
- Szymonek, M.; Kowalik, A.; Kopczynski, J.; Gasior-Perczak, D.; Palyga, I.; Walczyk, A.; Gadawska-Juszczyk, K.; Plusa, A.; Mezyk, R.; Chrapek, M.; et al. Immunohistochemistry cannot replace DNA analysis for evaluation of BRAF V600E mutations in papillary thyroid carcinoma. Oncotarget 2017, 8, 74897–74909. [Google Scholar] [CrossRef] [PubMed]
- Routhier, C.A.; Mochel, M.C.; Lynch, K.; Dias-Santagata, D.; Louis, D.N.; Hoang, M.P. Comparison of 2 monoclonal antibodies for immunohistochemical detection of BRAF V600E mutation in malignant melanoma, pulmonary carcinoma, gastrointestinal carcinoma, thyroid carcinoma, and gliomas. Hum. Pathol. 2013, 44, 2563–2570. [Google Scholar] [CrossRef]
- Zagzag, J.; Pollack, A.; Dultz, L.; Dhar, S.; Ogilvie, J.B.; Heller, K.S.; Deng, F.M.; Patel, K.N. Clinical utility of immunohistochemistry for the detection of the BRAF v600e mutation in papillary thyroid carcinoma. Surgery 2013, 154, 1199–1205. [Google Scholar] [CrossRef]
- Andrici, J.; Gill, A.J.; Hornick, J.L. Next generation immunohistochemistry: Emerging substitutes to genetic testing? Semin. Diagn. Pathol. 2018, 35, 161–169. [Google Scholar] [CrossRef]
- Lin, F.; Prichard, J. Handbook of practical immunohistochemistry, 2nd ed.; Springer: New York, NY, USA, 2015; pp. 2014–2246. [Google Scholar]
- Trier, N.H.; Houen, G. Peptide Antibodies in Clinical Laboratory Diagnostics. Adv. Clin. Chem. 2017, 81, 43–96. [Google Scholar] [CrossRef]
- Trier, N.; Hansen, P.; Houen, G. Peptides, Antibodies, Peptide Antibodies and More. Int. J. Mol. Sci. 2019, 20, e6289. [Google Scholar] [CrossRef]
- Capper, D.; Preusser, M.; Habel, A.; Sahm, F.; Ackermann, U.; Schindler, G.; Pusch, S.; Mechtersheimer, G.; Zentgraf, H.; von Deimling, A. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011, 122, 11–19. [Google Scholar] [CrossRef]
- Bullock, M.; O’Neill, C.; Chou, A.; Clarkson, A.; Dodds, T.; Toon, C.; Sywak, M.; Sidhu, S.B.; Delbridge, L.W.; Robinson, B.G.; et al. Utilization of a MAB for BRAF(V600E) detection in papillary thyroid carcinoma. Endocr. Relat. Cancer 2012, 19, 779–784. [Google Scholar] [CrossRef]
- Ilie, M.I.; Lassalle, S.; Long-Mira, E.; Bonnetaud, C.; Bordone, O.; Lespinet, V.; Lamy, A.; Sabourin, J.C.; Haudebourg, J.; Butori, C.; et al. Diagnostic value of immunohistochemistry for the detection of the BRAF(V600E) mutation in papillary thyroid carcinoma: Comparative analysis with three DNA-based assays. Thyroid 2014, 24, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Na, J.I.; Kim, J.H.; Kim, H.J.; Kim, H.K.; Moon, K.S.; Lee, J.S.; Lee, J.H.; Lee, K.H.; Park, J.T. VE1 immunohistochemical detection of the BRAF V600E mutation in thyroid carcinoma: A review of its usefulness and limitations. Virchows Arch. 2015, 467, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Pyo, J.S.; Sohn, J.H.; Kang, G. BRAF Immunohistochemistry Using Clone VE1 is Strongly Concordant with BRAF(V600E) Mutation Test in Papillary Thyroid Carcinoma. Endocr. Pathol. 2015, 26, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Luo, Y.; Bai, Q.; Lu, Y.; Lu, Y.; Wu, L.; Zhou, X. Specific immunohistochemical detection of the BRAF V600E mutation in primary and metastatic papillary thyroid carcinoma. Exp. Mol. Pathol. 2016, 100, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Ritterhouse, L.L.; Barletta, J.A. BRAF V600E mutation-specific antibody: A review. Semin. Diagn. Pathol. 2015, 32, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Singarayer, R.; Mete, O.; Perrier, L.; Thabane, L.; Asa, S.L.; Van Uum, S.; Ezzat, S.; Goldstein, D.P.; Sawka, A.M. A Systematic Review and Meta-Analysis of the Diagnostic Performance of BRAF V600E Immunohistochemistry in Thyroid Histopathology. Endocr. Pathol. 2019, 30, 201–218. [Google Scholar] [CrossRef]
- Dvorak, K.; Aggeler, B.; Palting, J.; McKelvie, P.; Ruszkiewicz, A.; Waring, P. Immunohistochemistry with the anti-BRAF V600E (VE1) antibody: Impact of pre-analytical conditions and concordance with DNA sequencing in colorectal and papillary thyroid carcinoma. Pathology 2014, 46, 509–517. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, J.; Lu, J.; Gao, J.; Lu, T.; Ren, X.; Duan, H.; Liang, Z. Immunohistochemistry is highly sensitive and specific for detecting the BRAF V600E mutation in papillary thyroid carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 15072–15078. [Google Scholar]
- Kim, J.K.; Seong, C.Y.; Bae, I.E.; Yi, J.W.; Yu, H.W.; Kim, S.J.; Won, J.K.; Chai, Y.J.; Choi, J.Y.; Lee, K.E. Comparison of Immunohistochemistry and Direct Sequencing Methods for Identification of the BRAF(V600E) Mutation in Papillary Thyroid Carcinoma. Ann. Surg. Oncol. 2018, 25, 1775–1781. [Google Scholar] [CrossRef]
- Jung, Y.Y.; Yoo, J.H.; Park, E.S.; Kim, M.K.; Lee, T.J.; Cho, B.Y.; Chung, Y.J.; Kang, K.H.; Ahn, H.Y.; Kim, H.S. Clinicopathologic correlations of the BRAFV600E mutation, BRAF V600E immunohistochemistry, and BRAF RNA in situ hybridization in papillary thyroid carcinoma. Pathol. Res. Pract. 2015, 211, 162–170. [Google Scholar] [CrossRef]
- Abd Elmageed, Z.Y.; Sholl, A.B.; Tsumagari, K.; Al-Qurayshi, Z.; Basolo, F.; Moroz, K.; Boulares, A.H.; Friedlander, P.; Miccoli, P.; Kandil, E. Immunohistochemistry as an accurate tool for evaluating BRAF-V600E mutation in 130 samples of papillary thyroid cancer. Surgery 2017, 161, 1122–1128. [Google Scholar] [CrossRef] [PubMed]
- Paja Fano, M.; Ugalde Olano, A.; Fuertes Thomas, E.; Oleaga Alday, A. Immunohistochemical detection of the BRAF V600E mutation in papillary thyroid carcinoma. Evaluation against real-time polymerase chain reaction. Endocrinol. Diabetes Nutr. 2017, 64, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, L.; Wang, J.; Zhao, H.; Wu, J.; Liu, S.; Zhang, L.; Li, Y.; Xing, X. Immunohistochemistry is a feasible method to screen BRAF V600E mutation in colorectal and papillary thyroid carcinoma. Exp. Mol. Pathol. 2018, 105, 153–159. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, P.; Yu, Y.; Zhi, J.; Zheng, X.; Yu, J.; Gao, M. Comparison of diagnostic methods for the detection of a BRAF mutation in papillary thyroid cancer. Oncol. Lett. 2019, 17, 4661–4666. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Choi, S.E.; Yoon, S.O.; Hong, S.W. A testing algorithm for detection of the B-type Raf kinase V600E mutation in papillary thyroid carcinoma. Hum. Pathol. 2014, 45, 1483–1488. [Google Scholar] [CrossRef] [PubMed]
- McKelvie, P.A.; Chan, F.; Yu, Y.; Waring, P.; Gresshoff, I.; Farrell, S.; Williams, R.A. The prognostic significance of the BRAF V600E mutation in papillary thyroid carcinoma detected by mutation-specific immunohistochemistry. Pathology 2013, 45, 637–644. [Google Scholar] [CrossRef]
- Capper, D.; Berghoff, A.S.; Magerle, M.; Ilhan, A.; Wohrer, A.; Hackl, M.; Pichler, J.; Pusch, S.; Meyer, J.; Habel, A.; et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol. 2012, 123, 223–233. [Google Scholar] [CrossRef]
- Crescenzi, A.; Guidobaldi, L.; Nasrollah, N.; Taccogna, S.; Cicciarella Modica, D.D.; Turrini, L.; Nigri, G.; Romanelli, F.; Valabrega, S.; Giovanella, L.; et al. Immunohistochemistry for BRAF(V600E) antibody VE1 performed in core needle biopsy samples identifies mutated papillary thyroid cancers. Horm. Metab. Res. 2014, 46, 370–374. [Google Scholar] [CrossRef]
- Jung, C.K.; Baek, J.H.; Na, D.G.; Oh, Y.L.; Yi, K.H.; Kang, H.C. 2019 Practice guidelines for thyroid core needle biopsy: A report of the Clinical Practice Guidelines Development Committee of the Korean Thyroid Association. J. Pathol. Transl. Med. 2020, 54, 64–86. [Google Scholar] [CrossRef]
- Cohen, Y.; Xing, M.; Mambo, E.; Guo, Z.; Wu, G.; Trink, B.; Beller, U.; Westra, W.H.; Ladenson, P.W.; Sidransky, D. BRAF mutation in papillary thyroid carcinoma. J. Natl. Cancer Inst. 2003, 95, 625–627. [Google Scholar] [CrossRef]
- Lloyd, R.V.; Osamura, R.Y.; Kloppel, G.; Rosai, J. WHO Classification of Tumors of Endocrine Organs, 4th ed.; International Agency for Research on Cancer (IARC): Lyon, France, 2017; Volume 10, pp. 65–91. [Google Scholar]
- Jung, C.K.; Kim, Y.; Jeon, S.; Jo, K.; Lee, S.; Bae, J.S. Clinical utility of EZH1 mutations in the diagnosis of follicular-patterned thyroid tumors. Hum. Pathol. 2018, 81, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, M.H.; Jeon, S.; Kim, J.; Kim, C.; Bae, J.S.; Jung, C.K. Prognostic implication of histological features associated with EHD2 expression in papillary thyroid carcinoma. PLoS ONE 2017, 12, e0174737. [Google Scholar] [CrossRef] [PubMed]
- Cho, U.; Mete, O.; Kim, M.H.; Bae, J.S.; Jung, C.K. Molecular correlates and rate of lymph node metastasis of non-invasive follicular thyroid neoplasm with papillary-like nuclear features and invasive follicular variant papillary thyroid carcinoma: The impact of rigid criteria to distinguish non-invasive follicular thyroid neoplasm with papillary-like nuclear features. Mod. Pathol. 2017, 30, 810–825. [Google Scholar] [CrossRef] [PubMed]


| ## | ID | TMA | BRAF (Sanger 1st) | First Interpretation | WTS | BRAF (Sanger 2nd) | Digital PCR * (Ground Truth) | Conclusion | ||
|---|---|---|---|---|---|---|---|---|---|---|
| VE1 Score | VE1 Result | VE1 Score | VE1 Result | |||||||
| 1 | PTC2_33 | 100 | VE1+ | wt | VE1-FP | 300 | VE1+ | wt | 24.9%, 25.8% | concordant |
| 2 | PTC4_22 | 0 | VE1− | V600E | VE1-FN | 0 | VE1− | V600E | n/a | VE1-FN |
| 3 | PTC5_41 | 0 | VE1− | V600E | VE1-FN | 0 | VE1− | V600E | n/a | VE1-FN |
| 4 | PTC7_30 | 300 | VE1+ | wt | VE1-FP | 300 | VE1+ | wt | 3.1%, 4.2% | concordant |
| 5 | PTC8_31 | 0 | VE1− | V600E | VE1-FN | 0 | VE1− | V600E | n/a | VE1-FN |
| 6 | PTC8_39 | 280 | VE1+ | wt | VE1-FP | 250 | VE1+ | wt | 8.1%; 13.9% | concordant |
| Source | Year | Molecular Test 1 | Discordant, n | Molecular Test 2 | VE1-FP after Test 1 | VE1-FP after Test 2 | VE1-FN after Test 1 | VE1-FN after Test 2 | Remained Discordant |
|---|---|---|---|---|---|---|---|---|---|
| Bullock [31] | 2012 | Sseq | 11/96 (11%) | re-Sseq, MPSeq | 10 | 3 | 1 | 0 | 3 |
| McKelvie [47] | 2013 | C-PCR | 9/71 (13%) | SNaPshot | 9 | 1 | 0 | 0 | 1 |
| Dvorak [38] | 2014 | Sseq | 7/73 (10%) | re-Sseq, SNaPshot, NGS | 4 | 0 * | 3 | 2 | 2 |
| Fisher [17] | 2014 | Pyroseq | 4/29 (14%) | re-Pyroseq | 4 | 4 | 0 | 0 | 4 |
| Na [33] | 2015 | real-time PCR | 8/104 (8%) | Sseq, nested PCR | 8 | 4 ** | 0 | 0 | 4 |
| Martinuzzi [19] | 2016 | Sseq, qPCR | 7/86 (8%) | NGS | 2 | 2 | 5 | 5 *** | 7 |
| Zhu [35] | 2016 | Sseq | 8/118 (7%) | ARMS | 8 | 7 | 0 | 0 | 7 |
| Szymonek [23] | 2017 | real-time PCR | 8/137 (6%) | NGS | 6 | 6 | 2 | 2 | 8 |
| Zhang [44] | 2018 | real-time PCR | 7/132 (5%) | Sseq | 7 | 5 | 0 | 0 | 5 |
| current study | 2019 | Sseq | 6/514 (1.2%) | digital PCR | 3 | 0 | 3 | 3 | 3 |
| TOTAL | 75/1360 (5.5%) | 61 | 32 | 14 | 12 | 44/75 (59%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choden, S.; Keelawat, S.; Jung, C.K.; Bychkov, A. VE1 Immunohistochemistry Improves the Limit of Genotyping for Detecting BRAFV600E Mutation in Papillary Thyroid Cancer. Cancers 2020, 12, 596. https://doi.org/10.3390/cancers12030596
Choden S, Keelawat S, Jung CK, Bychkov A. VE1 Immunohistochemistry Improves the Limit of Genotyping for Detecting BRAFV600E Mutation in Papillary Thyroid Cancer. Cancers. 2020; 12(3):596. https://doi.org/10.3390/cancers12030596
Chicago/Turabian StyleChoden, Sonam, Somboon Keelawat, Chan Kwon Jung, and Andrey Bychkov. 2020. "VE1 Immunohistochemistry Improves the Limit of Genotyping for Detecting BRAFV600E Mutation in Papillary Thyroid Cancer" Cancers 12, no. 3: 596. https://doi.org/10.3390/cancers12030596
APA StyleChoden, S., Keelawat, S., Jung, C. K., & Bychkov, A. (2020). VE1 Immunohistochemistry Improves the Limit of Genotyping for Detecting BRAFV600E Mutation in Papillary Thyroid Cancer. Cancers, 12(3), 596. https://doi.org/10.3390/cancers12030596