Non-Coding RNAs as Regulators and Markers for Targeting of Breast Cancer and Cancer Stem Cells
Abstract
1. Introduction
2. BCSCs and Their Regulation
3. BCSCs and Tumor Microenvironment
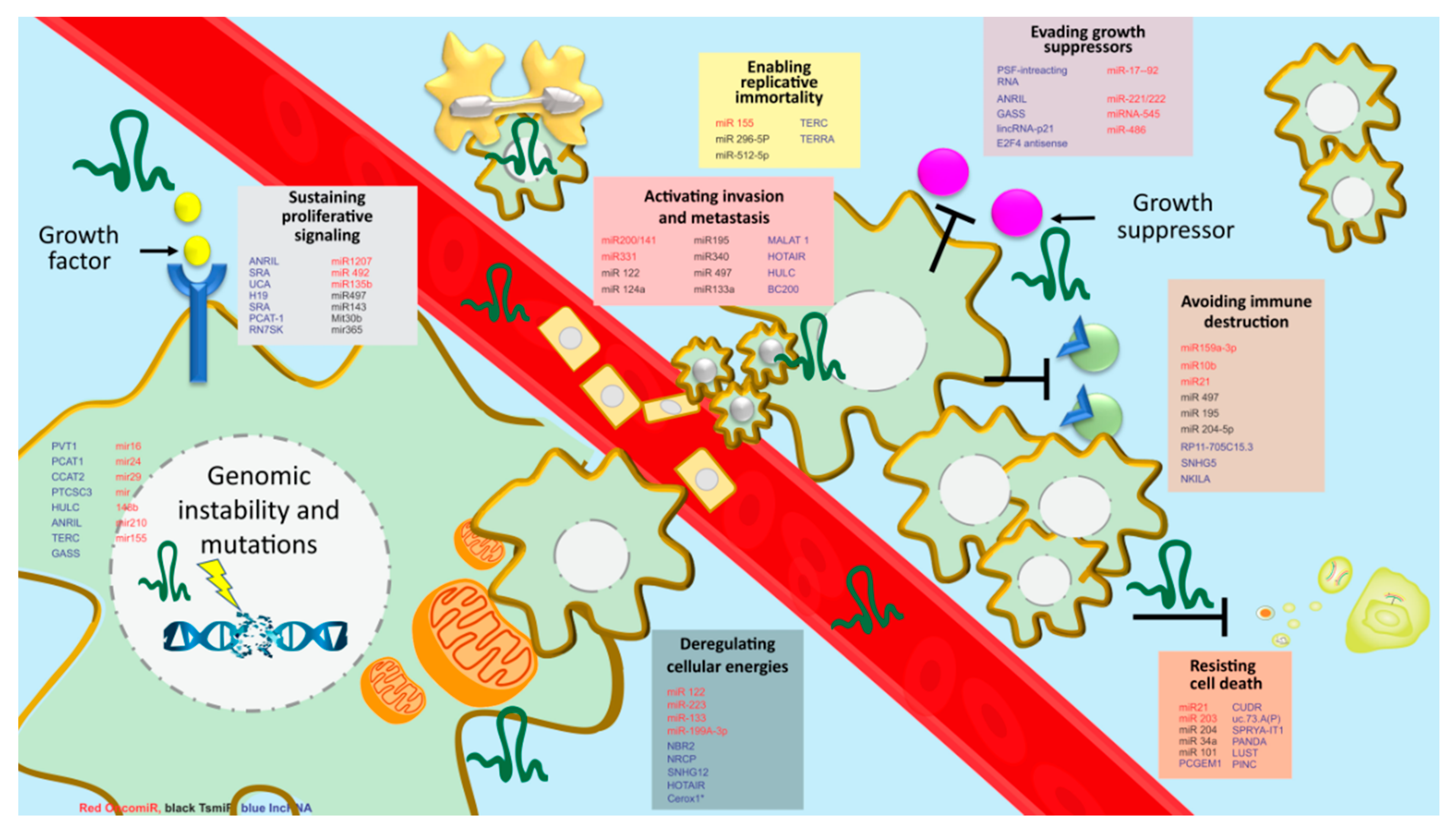
4. Regulatory Pathways Associated with BCSC
5. Role of MicroRNAs and LncRNA in BCSCs
6. Exosomal miRNAs: A Future Tool for Prognosis, Drug Discovery and As Therapeutic Targets
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALDH1A3 | Aldehyde Dehydrogenase 1A3 |
| ANRIL | antisense to the CDKN2B locus |
| ASCO | American Society of Clinical Oncology |
| BC | Breast cancer |
| BCL-2 | B-cell lymphoma 2 |
| BCSC | Breast Cancer Stem Cells |
| CAF | cancer-associated fibroblast |
| CAFs | cancer-associated fibroblasts |
| CCAT2 | Colon Cancer Associated Transcript 2 |
| CCL2 | monocyte chemotactic protein-1 |
| CCL7 | monocyte chemotactic protein-7 |
| CCND1 | cyclin D1 |
| CDK6 | Cyclin-Dependent Kinase 6 |
| Cernan | Competing endogenous RNA |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CRNDE | colorectal neoplasia differentially expressed |
| CSC | Cancer stem cells |
| CTNNB1 | b-catenin |
| E2F1 | E2F transcription factor 1 |
| E2F3 | E2F transcription factor 3 |
| E-BCSC | Epithelial |
| ECM | extracellular matrix |
| EIF4EBP1 | Eukaryotic Translation Initiation Factor 4E Binding Protein 1 |
| EMT | Epithelial-to-Mesenchymal Transition |
| ENCODE | Encyclopedia of DNA Elements project |
| ER | Estrogen receptor |
| ERBB2 | Receptor tyrosine-protein kinase erbB-2 |
| ErbB2/3 | Receptor tyrosine-protein kinase 2/3 |
| EZH2 | Enhancer of Zester Homolog 2 |
| FGF13-AS1 | fibroblast growth factor 13-antisense RNA 1 |
| FGF5 | fibroblast growth factor 5 |
| FOXC1 | Forehead box C1 |
| GAS5 | growth arrest specific 5 |
| HAS2 | hyaluronic synthase |
| HIF 1α | Hypoxia-inducible factor-1 |
| HOTAIR | HOX Transcript Antisense Intergenic RNA |
| HOX | Homeobox |
| KLF4 | Rappel-Like Factor 4 |
| lncRNA | Long intervening/intergenic noncoding RNAs |
| LINK-A | Long intergenic non-coding RNA for kinase activation |
| LncRNA | long non-coding RNA |
| LUCAT1 | Lung Cancer Associated Transcript 1 |
| MALAT1 | metastasis-associated lung adenocarcinoma transcript 1 |
| MAPK | Microtubule Associated Protein Kinase |
| Mass | mammary stem cells |
| M-BCSC | Mesenchymal |
| MEG3 | Maternally Expressed Gene 3 |
| MIAT | myocardial infarction associated transcript |
| miRNA | Micro RNA |
| mRNA | Messenger RNAs |
| MSCs | mesenchymal stem cells |
| MYC | MYC Proto-Oncogene |
| MYT-1 | Myelin Transcription Factor 1 |
| NCCN | National Comprehensive Cancer Network |
| NCID | Notch intracellular domain |
| ncRNAs | noncoding RNAs |
| NEAT1 | nuclear Para speckle assembly transcript 1 |
| NEDD4L | Neural precursor cell expressed developmentally down-regulated gene 4-like |
| NRAD1 | non-coding RNA in the aldehyde dehydrogenase 1A pathway |
| OCT4 | octamer-binding transcription factor 4 |
| p53 | protein p53 |
| PARP | Poly ADP ribose polymerase |
| PDGF-BB | platelet derived growth factor BB |
| PDK1 | Phosphoinositide-dependent kinase 1 |
| PHLDA1 | Pleckstrin homology-like domain, family A member 1 |
| pine | Piwi-interacting RNA |
| PLAGL2 | pleomorphic gene like-2 |
| PR | Progesterone receptor |
| RMRP | RNA component of mitochondrial RNA processing endoribonuclease |
| RMST | Rhabdomyosarcoma 2-associated transcript |
| ROR | receptor tyrosine kinase-like orphan receptor |
| RUNX1 | Chr. Runt-related transcription factor 1 |
| S1P | sphingosine-1-phosphate |
| siRNA | small interfering Ribonucleic Acid |
| SIRT1 | silent mating type information regulation 2 homolog |
| SnaR | steroid receptor RNA activator |
| sncRNAs | short noncoding RNAs |
| SNHG12 | Small nucleolar RNA host gene 12 |
| snoRNA | Small nucleolar RNAs |
| snRNA | Small nuclear RNA |
| SOX 2 | Sry-related high mobility group box 2 |
| Sox9 | Sry-related HMG box 9 |
| STAT3-CPTIB-FAO | JAK/STAT3-Regulated Fatty Acid β-Oxidation I |
| STXBP5-AS1 | STXBP5 Antisense RNA 1 |
| SUZ12 | suppressor of zeste 12 |
| TAM | Tumor-Associated Macrophages |
| TCF/LEF | T cell factor/lymphoid enhancing factor |
| TERC | Telomerase RNA component |
| TERRA | telomeric repeat-containing RNA |
| TET | ten-eleven translocation |
| TGF-β | Transforming growth factor beta |
| TME | tumor microenvironment |
| TNBC | Triple Negative Breast Cancer |
| Tregs | T regulatory cells |
| tRFs | tRNA-derived fragments |
| UCA1 | urothelial carcinoma associated 1 |
| XIST | X inactive specific transcript |
| ZBTB10 | Zinc finger and BTB domain containing 10 |
| ZEB 2 | Zinc finger E-box binding homeobox 2 |
| ZEB1 | zinc finger E-box-binding homeobox 1 |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. (Lausanne) 2017, 4, 227. [Google Scholar] [CrossRef] [PubMed]
- Fragomeni, S.M.; Sciallis, A.; Jeruss, J.S. Molecular Subtypes and Local-Regional Control of Breast Cancer. Surg. Oncol. Clin. N. Am. 2018, 27, 95–120. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Reis-Filho, J.S.; Pusztai, L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet 2011, 378, 1812–1823. [Google Scholar] [CrossRef]
- Gradishar, W.J.; Anderson, B.O.; Balassanian, R.; Blair, S.L.; Burstein, H.J.; Cyr, A.; Elias, A.D.; Farrar, W.B.; Forero, A.; Giordano, S.H.; et al. NCCN Guidelines Insights: Breast Cancer, Version 1.2017. J. Natl. Compr. Canc. Netw. 2017, 15, 433–451. [Google Scholar] [CrossRef]
- Breast Cancer (ASCO). Available online: https://ascopubs.org.doi/10.1200/EDBK_237715 (accessed on 3 February 2020).
- Chan, C.W.H.; Law, B.M.H.; So, W.K.W.; Chow, K.M.; Waye, M.M.Y. Novel Strategies on Personalized Medicine for Breast Cancer Treatment: An Update. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Prensner, J.R.; Iyer, M.K.; Balbin, O.A.; Dhanasekaran, S.M.; Cao, Q.; Brenner, J.C.; Laxman, B.; Asangani, I.A.; Grasso, C.S.; Kominsky, H.D.; et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011, 29, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, E. Genomics. ENCODE project writes eulogy for junk DNA. Science 2012, 337, 1159–1161. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef] [PubMed]
- O’Day, E.; Lal, A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010, 12, 201. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Liu, Y.; Sharma, S.; Watabe, K. Roles of lncRNA in breast cancer. Front. Biosci. (Schol. Ed.) 2015, 7, 94–108. [Google Scholar]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinf. 2017, 15, 177–186. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, L.; Li, J.; Le, T.D. LncmiRSRN: identification and analysis of long non-coding RNA related miRNA sponge regulatory network in human cancer. Bioinformatics 2018, 34, 4232–4240. [Google Scholar] [CrossRef]
- Xue, M.; Zhuo, Y.; Shan, B. MicroRNAs, Long Noncoding RNAs, and Their Functions in Human Disease. Methods Mol. Biol. 2017, 1617, 1–25. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2017, 9, 852. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome. Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.C.; Kean, M.J.; Xu, J.; Chua, N.H. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome. Res. 2014, 24, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Yu, K.; Sun, S.; Xie, C.; Skogerbo, G.; Miao, R.; Xiao, H.; Liao, Q.; Luo, H.; Zhao, G.; et al. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic. Acids. Res. 2012, 40, D210–D215. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Sharp, P.A. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol. Cell. Biol. 2003, 23, 1961–1967. [Google Scholar] [CrossRef]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007, 8, 39. [Google Scholar] [CrossRef]
- Kishore, S.; Gruber, A.R.; Jedlinski, D.J.; Syed, A.P.; Jorjani, H.; Zavolan, M. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome. Biol. 2013, 14, R45. [Google Scholar] [CrossRef]
- Vicens, Q.; Westhof, E. Biogenesis of Circular RNAs. Cell 2014, 159, 13–14. [Google Scholar] [CrossRef]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Hirose, T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013, 10, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.H.; Nakagawa, S.; Hirose, T.; Bond, C.S. Paraspeckles: Where Long Noncoding RNA Meets Phase Separation. Trends Biochem. Sci. 2018, 43, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Tragante, V.; Moore, J.H.; Asselbergs, F.W. The ENCODE project and perspectives on pathways. Genet. Epidemiol. 2014, 38, 275–280. [Google Scholar] [CrossRef]
- Tang, Q.; Hann, S.S. HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell Physiol. Biochem. 2018, 47, 893–913. [Google Scholar] [CrossRef]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef]
- DiStefano, J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. Methods Mol. Biol. 2018, 1706, 91–110. [Google Scholar] [CrossRef]
- Cipolla, G.A.; de Oliveira, J.C.; Salviano-Silva, A.; Lobo-Alves, S.C.; Lemos, D.S.; Oliveira, L.C.; Jucoski, T.S.; Mathias, C.; Pedroso, G.A.; Zambalde, E.P.; et al. Long Non-Coding RNAs in Multifactorial Diseases: Another Layer of Complexity. Noncoding RNA 2018, 4. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, H.; Yan, G.; Wu, T.; Liu, S.; Chen, W.; Ning, Y.; Lu, Z. Long Non-Coding RNA and Breast Cancer. Technol. Cancer Res. Treat. 2019, 18. [Google Scholar] [CrossRef]
- Gasch, C.; Ffrench, B.; O’Leary, J.J.; Gallagher, M.F. Catching moving targets: cancer stem cell hierarchies, therapy-resistance & considerations for clinical intervention. Mol. Cancer 2017, 16, 43. [Google Scholar] [CrossRef]
- Yu, Z.; Pestell, T.G.; Lisanti, M.P.; Pestell, R.G. Cancer stem cells. Int. J. Biochem. Cell Biol. 2012, 44, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Palomeras, S.; Ruiz-Martinez, S.; Puig, T. Targeting Breast Cancer Stem Cells to Overcome Treatment Resistance. Molecules 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, Q.; Zou, Y.; Chen, H.; Qi, L.; Chen, Y. Stem Cells and Cellular Origins of Breast Cancer: Updates in the Rationale, Controversies, and Therapeutic Implications. Front. Oncol. 2019, 9, 820. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem. Cell Reports 2014, 2, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef]
- LaBarge, M.A.; Petersen, O.W.; Bissell, M.J. Of microenvironments and mammary stem cells. Stem Cell Rev. 2007, 3, 137–146. [Google Scholar] [CrossRef]
- Wiseman, B.S.; Werb, Z. Stromal effects on mammary gland development and breast cancer. Science 2002, 296, 1046–1049. [Google Scholar] [CrossRef]
- Silberstein, G.B. Tumour-stromal interactions. Role of the stroma in mammary development. Breast Cancer Res. 2001, 3, 218–223. [Google Scholar] [CrossRef]
- Parmar, H.; Cunha, G.R. Epithelial-stromal interactions in the mouse and human mammary gland in vivo. Endocr. Relat. Cancer 2004, 11, 437–458. [Google Scholar] [CrossRef]
- Bocci, F.; Gearhart-Serna, L.; Boareto, M.; Ribeiro, M.; Ben-Jacob, E.; Devi, G.R.; Levine, H.; Onuchic, J.N.; Jolly, M.K. Toward understanding cancer stem cell heterogeneity in the tumor microenvironment. Proc. Natl. Acad. Sci. USA 2019, 116, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Bhat, V.; Allan, A.L.; Raouf, A. Role of the Microenvironment in Regulating Normal and Cancer Stem Cell Activity: Implications for Breast Cancer Progression and Therapy Response. Cancers (Basel) 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Liubomirski, Y.; Lerrer, S.; Meshel, T.; Rubinstein-Achiasaf, L.; Morein, D.; Wiemann, S.; Korner, C.; Ben-Baruch, A. Tumor-Stroma-Inflammation Networks Promote Pro-metastatic Chemokines and Aggressiveness Characteristics in Triple-Negative Breast Cancer. Front. Immunol. 2019, 10, 757. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Liu, S.; Wicha, M.S. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J. Clin. Invest. 2011, 121, 3804–3809. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Basak, P.; Buchel, E.; Safneck, J.; Murphy, L.C.; Mowat, M.; Kung, S.K.; Eirew, P.; Eaves, C.J.; Raouf, A. Breast Cancers Activate Stromal Fibroblast-Induced Suppression of Progenitors in Adjacent Normal Tissue. Stem Cell Reports 2018, 10, 196–211. [Google Scholar] [CrossRef]
- Tsuyada, A.; Chow, A.; Wu, J.; Somlo, G.; Chu, P.; Loera, S.; Luu, T.; Li, A.X.; Wu, X.; Ye, W.; et al. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer Res. 2012, 72, 2768–2779. [Google Scholar] [CrossRef]
- Ohlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef]
- Sugimoto, H.; Mundel, T.M.; Kieran, M.W.; Kalluri, R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol. Ther. 2006, 5, 1640–1646. [Google Scholar] [CrossRef]
- Cazet, A.S.; Hui, M.N.; Elsworth, B.L.; Wu, S.Z.; Roden, D.; Chan, C.L.; Skhinas, J.N.; Collot, R.; Yang, J.; Harvey, K.; et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat. Commun. 2018, 9, 2897. [Google Scholar] [CrossRef]
- Valenti, G.; Quinn, H.M.; Heynen, G.; Lan, L.; Holland, J.D.; Vogel, R.; Wulf-Goldenberg, A.; Birchmeier, W. Cancer Stem Cells Regulate Cancer-Associated Fibroblasts via Activation of Hedgehog Signaling in Mammary Gland Tumors. Cancer Res. 2017, 77, 2134–2147. [Google Scholar] [CrossRef]
- Al-Khalaf, H.H.; Ghebeh, H.; Inass, R.; Aboussekhra, A. Senescent Breast Luminal Cells Promote Carcinogenesis through Interleukin-8-Dependent Activation of Stromal Fibroblasts. Mol. Cell Biol. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Chen, J.; Yao, H.; Liu, J.; Yu, S.; Lao, L.; Wang, M.; Luo, M.; Xing, Y.; Chen, F.; et al. CD10(+)GPR77(+) Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 2018, 172, 841–856.e16. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ginestier, C.; Ou, S.J.; Clouthier, S.G.; Patel, S.H.; Monville, F.; Korkaya, H.; Heath, A.; Dutcher, J.; Kleer, C.G.; et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011, 71, 614–624. [Google Scholar] [CrossRef]
- Ma, X.J.; Dahiya, S.; Richardson, E.; Erlander, M.; Sgroi, D.C. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009, 11, R7. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liao, D.; Chen, C.; Liu, Y.; Chuang, T.H.; Xiang, R.; Markowitz, D.; Reisfeld, R.A.; Luo, Y. Tumor-associated macrophages regulate murine breast cancer stem cells through a novel paracrine EGFR/Stat3/Sox-2 signaling pathway. Stem Cells 2013, 31, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Okuda, H.; Kobayashi, A.; Xia, B.; Watabe, M.; Pai, S.K.; Hirota, S.; Xing, F.; Liu, W.; Pandey, P.R.; Fukuda, K.; et al. Hyaluronan synthase HAS2 promotes tumor progression in bone by stimulating the interaction of breast cancer stem-like cells with macrophages and stromal cells. Cancer Res. 2012, 72, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Clauser, K.R.; Tam, W.L.; Frose, J.; Ye, X.; Eaton, E.N.; Reinhardt, F.; Donnenberg, V.S.; Bhargava, R.; Carr, S.A.; et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat. Cell Biol. 2014, 16, 1105–1117. [Google Scholar] [CrossRef]
- Nalla, L.V.; Kalia, K.; Khairnar, A. Self-renewal signaling pathways in breast cancer stem cells. Int J Biochem. Cell Biol. 2019, 107, 140–153. [Google Scholar] [CrossRef]
- Al-Hussaini, H.; Subramanyam, D.; Reedijk, M.; Sridhar, S.S. Notch signaling pathway as a therapeutic target in breast cancer. Mol. Cancer Ther. 2011, 10, 9–15. [Google Scholar] [CrossRef]
- Habib, J.G.; O’Shaughnessy, J.A. The hedgehog pathway in triple-negative breast cancer. Cancer Med. 2016, 5, 2989–3006. [Google Scholar] [CrossRef]
- King, T.D.; Suto, M.J.; Li, Y. The Wnt/beta-catenin signaling pathway: A potential therapeutic target in the treatment of triple negative breast cancer. J. Cell Biochem. 2012, 113, 13–18. [Google Scholar] [CrossRef]
- Borah, A.; Raveendran, S.; Rochani, A.; Maekawa, T.; Kumar, D.S. Targeting self-renewal pathways in cancer stem cells: clinical implications for cancer therapy. Oncogenesis 2015, 4, e177. [Google Scholar] [CrossRef] [PubMed]
- Matsui, W.H. Cancer stem cell signaling pathways. Medicine (Baltimore) 2016, 95, S8–S19. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Tanikawa, T.; Li, W.; Zhao, L.; Vatan, L.; Szeliga, W.; Wan, S.; Wei, S.; Wang, Y.; Liu, Y.; et al. Myeloid-Derived Suppressor Cells Endow Stem-like Qualities to Breast Cancer Cells through IL6/STAT3 and NO/NOTCH Cross-talk Signaling. Cancer Res. 2016, 76, 3156–3165. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Cai, Y.; Liu, J.; Wang, Z.; Wu, Q.; Zhang, Z.; Yang, C.J.; Yuan, L.; Ouyang, G. Twist2 contributes to breast cancer progression by promoting an epithelial-mesenchymal transition and cancer stem-like cell self-renewal. Oncogene 2011, 30, 4707–4720. [Google Scholar] [CrossRef]
- Thiagarajan, P.S.; Zheng, Q.; Bhagrath, M.; Mulkearns-Hubert, E.E.; Myers, M.G.; Lathia, J.D.; Reizes, O. STAT3 activation by leptin receptor is essential for TNBC stem cell maintenance. Endocr. Relat. Cancer 2017, 24, 415–426. [Google Scholar] [CrossRef]
- Wang, T.; Fahrmann, J.F.; Lee, H.; Li, Y.J.; Tripathi, S.C.; Yue, C.; Zhang, C.; Lifshitz, V.; Song, J.; Yuan, Y.; et al. JAK/STAT3-Regulated Fatty Acid beta-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab. 2018, 27, 136–150.e5. [Google Scholar] [CrossRef]
- Zhong, Y.; Shen, S.; Zhou, Y.; Mao, F.; Lin, Y.; Guan, J.; Xu, Y.; Zhang, S.; Liu, X.; Sun, Q. NOTCH1 is a poor prognostic factor for breast cancer and is associated with breast cancer stem cells. Onco. Targets Ther. 2016, 9, 6865–6871. [Google Scholar] [CrossRef]
- Gonzalez, M.E.; Moore, H.M.; Li, X.; Toy, K.A.; Huang, W.; Sabel, M.S.; Kidwell, K.M.; Kleer, C.G. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 3098–3103. [Google Scholar] [CrossRef]
- Hirata, N.; Yamada, S.; Shoda, T.; Kurihara, M.; Sekino, Y.; Kanda, Y. Sphingosine-1-phosphate promotes expansion of cancer stem cells via S1PR3 by a ligand-independent Notch activation. Nat. Commun. 2014, 5, 4806. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.K.; Shao, C.; Wang, J.; Wei, Q.; Wang, X.; Collier, Z.; Tang, S.; Liu, H.; Zhang, F.; Huang, J.; et al. Wnt/beta-catenin signaling plays an ever-expanding role in stem cell self-renewal, tumorigenesis and cancer chemoresistance. Genes Dis. 2016, 3, 11–40. [Google Scholar] [CrossRef] [PubMed]
- Domenici, G.; Aurrekoetxea-Rodriguez, I.; Simoes, B.M.; Rabano, M.; Lee, S.Y.; Millan, J.S.; Comaills, V.; Oliemuller, E.; Lopez-Ruiz, J.A.; Zabalza, I.; et al. A Sox2-Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene 2019, 38, 3151–3169. [Google Scholar] [CrossRef]
- Protecting Workers’ Health. Available online: https://www.who.int/news-room/fact-sheets/detail/protecting-workers’-health (accessed on 3 February 2020).
- Wang, L.; Duan, W.; Kang, L.; Mao, J.; Yu, X.; Fan, S.; Li, L.; Tao, Y. Smoothened activates breast cancer stem-like cell and promotes tumorigenesis and metastasis of breast cancer. Biomed. Pharmacother. 2014, 68, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Qu, Y.; Jin, Y.; Yu, Y.; Deng, N.; Wawrowsky, K.; Zhang, X.; Li, N.; Bose, S.; Wang, Q.; et al. FOXC1 Activates Smoothened-Independent Hedgehog Signaling in Basal-like Breast Cancer. Cell Rep. 2015, 13, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- Loh, H.Y.; Norman, B.P.; Lai, K.S.; Rahman, N.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Luo, Q.; Li, X.; Gao, Y.; Long, Y.; Chen, L.; Huang, Y.; Fang, L. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int. 2013, 13, 95. [Google Scholar] [CrossRef]
- Guo, X.; Connick, M.C.; Vanderhoof, J.; Ishak, M.A.; Hartley, R.S. MicroRNA-16 modulates HuR regulation of cyclin E1 in breast cancer cells. Int. J. Mol. Sci. 2015, 16, 7112–7132. [Google Scholar] [CrossRef]
- Shukla, K.; Sharma, A.K.; Ward, A.; Will, R.; Hielscher, T.; Balwierz, A.; Breunig, C.; Munstermann, E.; Konig, R.; Keklikoglou, I.; et al. MicroRNA-30c-2-3p negatively regulates NF-kappaB signaling and cell cycle progression through downregulation of TRADD and CCNE1 in breast cancer. Mol. Oncol. 2015, 9, 1106–1119. [Google Scholar] [CrossRef]
- Huang, X.; Lyu, J. Tumor suppressor function of miR-483-3p on breast cancer via targeting of the cyclin E1 gene. Exp. Ther. Med. 2018, 16, 2615–2620. [Google Scholar] [CrossRef]
- Yan, C.; Chen, Y.; Kong, W.; Fu, L.; Liu, Y.; Yao, Q.; Yuan, Y. PVT1-derived miR-1207-5p promotes breast cancer cell growth by targeting STAT6. Cancer Sci. 2017, 108, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; He, M.; Ma, M.T.; Wu, H.Z.; Yu, Z.J.; Guan, S.; Jiang, L.Y.; Wang, Y.; Zheng, D.D.; Jin, F.; et al. MicroRNA-148a inhibits breast cancer migration and invasion by directly targeting WNT-1. Oncol. Rep. 2016, 35, 1425–1432. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi-Yeganeh, S.; Paryan, M.; Arefian, E.; Vasei, M.; Ghanbarian, H.; Mahdian, R.; Karimipoor, M.; Soleimani, M. MicroRNA-340 inhibits the migration, invasion, and metastasis of breast cancer cells by targeting Wnt pathway. Tumour Biol 2016, 37, 8993–9000. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Furukawa, T.; Tsukagoshi, H. Motor paresis improved by sympathetic block. A motor form of reflex sympathetic dystrophy? Arch Neurol 1989, 46, 683–687. [Google Scholar] [CrossRef]
- Pan, Y.; Jiao, G.; Wang, C.; Yang, J.; Yang, W. MicroRNA-421 inhibits breast cancer metastasis by targeting metastasis associated 1. Biomed. Pharmacother. 2016, 83, 1398–1406. [Google Scholar] [CrossRef]
- Xie, F.; Hosany, S.; Zhong, S.; Jiang, Y.; Zhang, F.; Lin, L.; Wang, X.; Gao, S.; Hu, X. MicroRNA-193a inhibits breast cancer proliferation and metastasis by downregulating WT1. PLoS ONE 2017, 12, e0185565. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Z.; Li, X.; Tang, X.; He, J.; Lu, S. MicroRNA-1297 contributes to tumor growth of human breast cancer by targeting PTEN/PI3K/AKT signaling. Oncol. Rep. 2017, 38, 2435–2443. [Google Scholar] [CrossRef]
- Miao, Y.; Zheng, W.; Li, N.; Su, Z.; Zhao, L.; Zhou, H.; Jia, L. MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci. Rep. 2017, 7, 41942. [Google Scholar] [CrossRef]
- Hong, B.S.; Ryu, H.S.; Kim, N.; Kim, J.; Lee, E.; Moon, H.; Kim, K.H.; Jin, M.S.; Kwon, N.H.; Kim, S.; et al. Tumor Suppressor miRNA-204-5p Regulates Growth, Metastasis, and Immune Microenvironment Remodeling in Breast Cancer. Cancer Res. 2019, 79, 1520–1534. [Google Scholar] [CrossRef]
- Khan, A.Q.; Ahmed, E.I.; Elareer, N.R.; Junejo, K.; Steinhoff, M.; Uddin, S. Role of miRNA-Regulated Cancer Stem Cells in the Pathogenesis of Human Malignancies. Cells 2019, 8. [Google Scholar] [CrossRef]
- Fan, X.; Chen, W.; Fu, Z.; Zeng, L.; Yin, Y.; Yuan, H. MicroRNAs, a subpopulation of regulators, are involved in breast cancer progression through regulating breast cancer stem cells. Oncol. Lett. 2017, 14, 5069–5076. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lim, Y.Y.; Wright, J.A.; Attema, J.L.; Gregory, P.A.; Bert, A.G.; Smith, E.; Thomas, D.; Lopez, A.F.; Drew, P.A.; Khew-Goodall, Y.; et al. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J. Cell Sci. 2013, 126, 2256–2266. [Google Scholar] [CrossRef] [PubMed]
- Polytarchou, C.; Iliopoulos, D.; Struhl, K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc. Natl. Acad. Sci. USA 2012, 109, 14470–14475. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Lindahl-Allen, M.; Polytarchou, C.; Hirsch, H.A.; Tsichlis, P.N.; Struhl, K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 2010, 39, 761–772. [Google Scholar] [CrossRef]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell. Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef]
- Dykxhoorn, D.M.; Wu, Y.; Xie, H.; Yu, F.; Lal, A.; Petrocca, F.; Martinvalet, D.; Song, E.; Lim, B.; Lieberman, J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS ONE 2009, 4, e7181. [Google Scholar] [CrossRef]
- Knezevic, J.; Pfefferle, A.D.; Petrovic, I.; Greene, S.B.; Perou, C.M.; Rosen, J.M. Expression of miR-200c in claudin-low breast cancer alters stem cell functionality, enhances chemosensitivity and reduces metastatic potential. Oncogene 2015, 34, 5997–6006. [Google Scholar] [CrossRef]
- van den Beucken, T.; Koch, E.; Chu, K.; Rupaimoole, R.; Prickaerts, P.; Adriaens, M.; Voncken, J.W.; Harris, A.L.; Buffa, F.M.; Haider, S.; et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat. Commun. 2014, 5, 5203. [Google Scholar] [CrossRef]
- Song, S.J.; Poliseno, L.; Song, M.S.; Ala, U.; Webster, K.; Ng, C.; Beringer, G.; Brikbak, N.J.; Yuan, X.; Cantley, L.C.; et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family dependent chromatin remodeling. Cell 2013, 154, 311–324. [Google Scholar] [CrossRef]
- Valastyan, S.; Chang, A.; Benaich, N.; Reinhardt, F.; Weinberg, R.A. Concurrent suppression of integrin alpha5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res. 2010, 70, 5147–5154. [Google Scholar] [CrossRef]
- Valastyan, S.; Reinhardt, F.; Benaich, N.; Calogrias, D.; Szasz, A.M.; Wang, Z.C.; Brock, J.E.; Richardson, A.L.; Weinberg, R.A. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 2009, 137, 1032–1046. [Google Scholar] [CrossRef]
- Sachdeva, M.; Mo, Y.Y. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010, 70, 378–387. [Google Scholar] [CrossRef]
- Spizzo, R.; Nicoloso, M.S.; Lupini, L.; Lu, Y.; Fogarty, J.; Rossi, S.; Zagatti, B.; Fabbri, M.; Veronese, A.; Liu, X.; et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death. Differ. 2010, 17, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Bian, C.; Yang, Z.; Bo, Y.; Li, J.; Zeng, L.; Zhou, H.; Zhao, R.C. miR-145 inhibits breast cancer cell growth through RTKN. Int. J. Oncol. 2009, 34, 1461–1466. [Google Scholar] [PubMed]
- Jiang, S.; Zhang, H.W.; Lu, M.H.; He, X.H.; Li, Y.; Gu, H.; Liu, M.F.; Wang, E.D. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010, 70, 3119–3127. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; He, L.; Coppola, M.; Guo, J.; Esposito, N.N.; Coppola, D.; Cheng, J.Q. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J. Biol. Chem. 2010, 285, 17869–17879. [Google Scholar] [CrossRef]
- Kong, W.; Yang, H.; He, L.; Zhao, J.J.; Coppola, D.; Dalton, W.S.; Cheng, J.Q. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell Biol. 2008, 28, 6773–6784. [Google Scholar] [CrossRef]
- Carpenter, R.L.; Paw, I.; Dewhirst, M.W.; Lo, H.W. Akt phosphorylates and activates HSF-1 independent of heat shock, leading to Slug overexpression and epithelial-mesenchymal transition (EMT) of HER2-overexpressing breast cancer cells. Oncogene 2015, 34, 546–557. [Google Scholar] [CrossRef]
- Song, B.; Wang, C.; Liu, J.; Wang, X.; Lv, L.; Wei, L.; Xie, L.; Zheng, Y.; Song, X. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J. Exp. Clin. Cancer Res. 2010, 29, 29. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Bart, J.; Tan, L.P.; Platteel, I.; Sluis, T.; Huitema, S.; Harms, G.; Fu, L.; Hollema, H.; Berg, A. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 2009, 9, 163. [Google Scholar] [CrossRef]
- Huang, G.L.; Zhang, X.H.; Guo, G.L.; Huang, K.T.; Yang, K.Y.; Hu, X.Q. Expression of microRNA-21 in invasive ductal carcinoma of the breast and its association with phosphatase and tensin homolog deleted from chromosome expression and clinicopathologic features. Chinese Med. J. 2008, 88, 2833–2837. [Google Scholar]
- Qian, B.; Katsaros, D.; Lu, L.; Preti, M.; Durando, A.; Arisio, R.; Mu, L.; Yu, H. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res. Treat. 2009, 117, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.K.; Goga, A.; Bhaumik, D.; Berger, C.E.; Sullivan, C.S.; Benz, C.C. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 2007, 282, 1479–1486. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, Z.; Zhao, Y.; Ding, Y.; Liu, H.; Xi, Y.; Xiong, W.; Li, G.; Lu, J.; Fodstad, O.; et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 2010, 285, 21496–21507. [Google Scholar] [CrossRef]
- Hofmann, M.H.; Heinrich, J.; Radziwill, G.; Moelling, K. A short hairpin DNA analogous to miR-125b inhibits C-Raf expression, proliferation, and survival of breast cancer cells. Mol. Cancer Res. 2009, 7, 1635–1644. [Google Scholar] [CrossRef]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; Weinberg, R.A. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347. [Google Scholar] [CrossRef]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef]
- Ahmad, A.; Ginnebaugh, K.R.; Yin, S.; Bollig-Fischer, A.; Reddy, K.B.; Sarkar, F.H. Functional role of miR-10b in tamoxifen resistance of ER-positive breast cancer cells through down-regulation of HDAC4. BMC Cancer 2015, 15, 540. [Google Scholar] [CrossRef]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef]
- Wu, H.; Zhu, S.; Mo, Y.Y. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009, 19, 439–448. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial-to-mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Camps, C.; Buffa, F.M.; Colella, S.; Moore, J.; Sotiriou, C.; Sheldon, H.; Harris, A.L.; Gleadle, J.M.; Ragoussis, J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin. Cancer Res. 2008, 14, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sun, H.; Dai, H.; Walsh, R.M.; Imakura, M.; Schelter, J.; Burchard, J.; Dai, X.; Chang, A.N.; Diaz, R.L.; et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle 2009, 8, 2756–2768. [Google Scholar] [CrossRef] [PubMed]
- Luthra, R.; Singh, R.R.; Luthra, M.G.; Li, Y.X.; Hannah, C.; Romans, A.M.; Barkoh, B.A.; Chen, S.S.; Ensor, J.; Maru, D.M.; et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene 2008, 27, 6667–6678. [Google Scholar] [CrossRef]
- He, H.; Tian, W.; Chen, H.; Jiang, K. MiR-944 functions as a novel oncogene and regulates the chemoresistance in breast cancer. Tumour. Biol. 2016, 37, 1599–1607. [Google Scholar] [CrossRef]
- Shen, H.; Wang, D.; Li, L.; Yang, S.; Chen, X.; Zhou, S.; Zhong, S.; Zhao, J.; Tang, J. MiR-222 promotes drug-resistance of breast cancer cells to adriamycin via modulation of PTEN/Akt/FOXO1 pathway. Gene 2017, 596, 110–118. [Google Scholar] [CrossRef]
- Zhang, X.; Zhong, S.; Xu, Y.; Yu, D.; Ma, T.; Chen, L.; Zhao, Y.; Chen, X.; Yang, S.; Wu, Y.; et al. MicroRNA-3646 Contributes to Docetaxel Resistance in Human Breast Cancer Cells by GSK-3beta/beta-Catenin Signaling Pathway. PLoS ONE 2016, 11, e0153194. [Google Scholar] [CrossRef]
- Kastl, L.; Brown, I.; Schofield, A.C. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res. Treat. 2012, 131, 445–454. [Google Scholar] [CrossRef]
- Yao, Y.S.; Qiu, W.S.; Yao, R.Y.; Zhang, Q.; Zhuang, L.K.; Zhou, F.; Sun, L.B.; Yue, L. miR-141 confers docetaxel chemoresistance of breast cancer cells via regulation of EIF4E expression. Oncol. Rep. 2015, 33, 2504–2512. [Google Scholar] [CrossRef]
- Su, C.M.; Wang, M.Y.; Hong, C.C.; Chen, H.A.; Su, Y.H.; Wu, C.H.; Huang, M.T.; Chang, Y.W.; Jiang, S.S.; Sung, S.Y.; et al. miR-520h is crucial for DAPK2 regulation and breast cancer progression. Oncogene 2016, 35, 1134–1142. [Google Scholar] [CrossRef]
- Kato, M.; Paranjape, T.; Muller, R.U.; Nallur, S.; Gillespie, E.; Keane, K.; Esquela-Kerscher, A.; Weidhaas, J.B.; Slack, F.J. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 2009, 28, 2419–2424. [Google Scholar] [CrossRef] [PubMed]
- Bhaumik, D.; Scott, G.K.; Schokrpur, S.; Patil, C.K.; Campisi, J.; Benz, C.C. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene 2008, 27, 5643–5647. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.D.; Ohshiro, K.; Rayala, S.K.; Kumar, R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res 2008, 68, 8195–8200. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.P.; Picard, D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol Cell Biol 2009, 29, 3783–3790. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Di Leva, G.; Li, M.; Fang, F.; Devlin, C.; Hartman-Frey, C.; Burow, M.E.; Ivan, M.; Croce, C.M.; Nephew, K.P. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 2011, 30, 1082–1097. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, N.; Ahmad, H.M.; Molparia, B.; Kulshreshtha, R. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis 2013, 34, 1889–1899. [Google Scholar] [CrossRef]
- Tavazoie, S.F.; Alarcon, C.; Oskarsson, T.; Padua, D.; Wang, Q.; Bos, P.D.; Gerald, W.L.; Massague, J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008, 451, 147–152. [Google Scholar] [CrossRef]
- Trompeter, H.I.; Abbad, H.; Iwaniuk, K.M.; Hafner, M.; Renwick, N.; Tuschl, T.; Schira, J.; Muller, H.W.; Wernet, P. MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS ONE 2011, 6, e16138. [Google Scholar] [CrossRef]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef]
- Xia, P.; Wang, Z.; Liu, X.; Wu, B.; Wang, J.; Ward, T.; Zhang, L.; Ding, X.; Gibbons, G.; Shi, Y.; et al. EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl. Acad. Sci. USA 2012, 109, 16564–16569. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Xie, H.; Zhou, Z.; Chen, H.; Hu, T.; Bai, Y.; Shen, Y.; Yuan, W.; Jing, Q.; et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol. Cell Biochem. 2011, 351, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Siragam, V.; Rutnam, Z.J.; Yang, W.; Fang, L.; Luo, L.; Yang, X.; Li, M.; Deng, Z.; Qian, J.; Peng, C.; et al. MicroRNA miR-98 inhibits tumor angiogenesis and invasion by targeting activin receptor-like kinase-4 and matrix metalloproteinase-11. Oncotarget 2012, 3, 1370–1385. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.T.; Xu, Q.; Li, W.; Lu, B.; et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2013, 5, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.T.; Chen, P.S.; Johansson, G.; Chu, C.Y.; Wang, M.Y.; Jeng, Y.M.; Yu, S.L.; Chen, J.S.; Chang, K.J.; Jee, S.H.; et al. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 2010, 70, 2675–2685. [Google Scholar] [CrossRef]
- Plummer, P.N.; Freeman, R.; Taft, R.J.; Vider, J.; Sax, M.; Umer, B.A.; Gao, D.; Johns, C.; Mattick, J.S.; Wilton, S.D.; et al. MicroRNAs regulate tumor angiogenesis modulated by endothelial progenitor cells. Cancer Res. 2013, 73, 341–352. [Google Scholar] [CrossRef]
- Lu, Y.; Qin, T.; Li, J.; Wang, L.; Zhang, Q.; Jiang, Z.; Mao, J. MicroRNA-140-5p inhibits invasion and angiogenesis through targeting VEGF-A in breast cancer. Cancer Gene Ther. 2017, 24, 386–392. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, L.; Chen, Q.; Song, Y.; Xu, S.; Ma, F.; Wang, X.; Wang, J.; Yu, H.; Cao, X.; et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J. Immunol. 2012, 188, 5500–5510. [Google Scholar] [CrossRef]
- Liang, Z.; Bian, X.; Shim, H. Downregulation of microRNA-206 promotes invasion and angiogenesis of triple negative breast cancer. Biochem. Biophys. Res. Commun. 2016, 477, 461–466. [Google Scholar] [CrossRef]
- Anfossi, S.; Giordano, A.; Gao, H.; Cohen, E.N.; Tin, S.; Wu, Q.; Garza, R.J.; Debeb, B.G.; Alvarez, R.H.; Valero, V.; et al. High serum miR-19a levels are associated with inflammatory breast cancer and are predictive of favorable clinical outcome in patients with metastatic HER2+ inflammatory breast cancer. PLoS ONE 2014, 9, e83113. [Google Scholar] [CrossRef]
- Taguchi, A.; Yanagisawa, K.; Tanaka, M.; Cao, K.; Matsuyama, Y.; Goto, H.; Takahashi, T. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008, 68, 5540–5545. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Sul, K.; Krukovets, I.; Nestor, C.; Li, J.; Adognravi, O.S. Novel tissue-specific mechanism of regulation of angiogenesis and cancer growth in response to hyperglycemia. J. Am. Heart Assoc. 2012, 1, e005967. [Google Scholar] [CrossRef] [PubMed]
- Bishnoi, V.; Kumar, B.; Bhagat, H.; Salunke, P.; Bishnoi, S. Comparison of Dexmedetomidine Versus Midazolam-Fentanyl Combination for Monitored Anesthesia Care During Burr-Hole Surgery for Chronic Subdural Hematoma. J. Neurosurg. Anesthesiol. 2016, 28, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Tomar, D.; Yadav, A.S.; Kumar, D.; Bhadauriya, G.; Kundu, G.C. Non-coding RNAs as potential therapeutic targets in breast cancer. Biochim. Biophys. Acta. Gene. Regul. Mech. 2019. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.E.; Tu, G.; Yang, G.; Li, G.; Yang, D.; Lang, L.; Xi, L.; Sun, K.; Chen, Y.; Shu, K.; et al. MiR-205/YAP1 in Activated Fibroblasts of Breast Tumor Promotes VEGF-independent Angiogenesis through STAT3 Signaling. Theranostics 2017, 7, 3972–3988. [Google Scholar] [CrossRef]
- Jiang, L.; Yu, L.; Zhang, X.; Lei, F.; Wang, L.; Liu, X.; Wu, S.; Zhu, J.; Wu, G.; Cao, L.; et al. miR-892b Silencing Activates NF-kappaB and Promotes Aggressiveness in Breast Cancer. Cancer Res. 2016, 76, 1101–1111. [Google Scholar] [CrossRef]
- Jung, E.J.; Santarpia, L.; Kim, J.; Esteva, F.J.; Moretti, E.; Buzdar, A.U.; Di Leo, A.; Le, X.F.; Bast, R.C., Jr.; Park, S.T.; et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer 2012, 118, 2603–2614. [Google Scholar] [CrossRef]
- Rothe, F.; Ignatiadis, M.; Chaboteaux, C.; Haibe-Kains, B.; Kheddoumi, N.; Majjaj, S.; Badran, B.; Fayyad-Kazan, H.; Desmedt, C.; Harris, A.L.; et al. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS ONE 2011, 6, e20980. [Google Scholar] [CrossRef]
- Gu, X.; Li, J.Y.; Guo, J.; Li, P.S.; Zhang, W.H. Influence of MiR-451 on Drug Resistances of Paclitaxel-Resistant Breast Cancer Cell Line. Med. Sci. Monit. 2015, 21, 3291–3297. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, R.; He, Y.; Fu, X.; Fu, L.; Zhu, Z.; Fu, L.; Dong, J.T. MicroRNA 100 sensitizes luminal A breast cancer cells to paclitaxel treatment in part by targeting mTOR. Oncotarget 2016, 7, 5702–5714. [Google Scholar] [CrossRef]
- Zhang, H.D.; Sun, D.W.; Mao, L.; Zhang, J.; Jiang, L.H.; Li, J.; Wu, Y.; Ji, H.; Chen, W.; Wang, J.; et al. MiR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem. Biophys. Res. Commun. 2015, 465, 702–713. [Google Scholar] [CrossRef]
- Yu, X.; Luo, A.; Liu, Y.; Wang, S.; Li, Y.; Shi, W.; Liu, Z.; Qu, X. MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol. Cancer 2015, 14, 208. [Google Scholar] [CrossRef] [PubMed]
- Esteva, F.J.; Yu, D.; Hung, M.C.; Hortobagyi, G.N. Molecular predictors of response to trastuzumab and lapatinib in breast cancer. Nat. Rev. Clin. Oncol. 2010, 7, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhou, S.; Zheng, M.; Deng, X.; Yi, Y.; Huang, T. MiR-199a-3p enhances breast cancer cell sensitivity to cisplatin by downregulating TFAM (TFAM). Biomed. Pharmacother. 2017, 88, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, A.; Cheung, D.G.; Balsari, A.; Tagliabue, E.; Coppola, V.; Iorio, M.V.; Palmieri, D.; Croce, C.M. miR-302b enhances breast cancer cell sensitivity to cisplatin by regulating E2F1 and the cellular DNA damage response. Oncotarget 2016, 7, 786–797. [Google Scholar] [CrossRef]
- He, X.; Xiao, X.; Dong, L.; Wan, N.; Zhou, Z.; Deng, H.; Zhang, X. MiR-218 regulates cisplatin chemosensitivity in breast cancer by targeting BRCA1. Tumour. Biol. 2015, 36, 2065–2075. [Google Scholar] [CrossRef]
- Tan, X.; Peng, J.; Fu, Y.; An, S.; Rezaei, K.; Tabbara, S.; Teal, C.B.; Man, Y.G.; Brem, R.F.; Fu, S.W. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple-negative breast cancer. Breast Cancer Res. 2014, 16, 435. [Google Scholar] [CrossRef]
- Zhong, S.; Li, W.; Chen, Z.; Xu, J.; Zhao, J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene 2013, 531, 8–14. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wei, Y.; Li, M.; Yu, S.; Ye, M.; Zhang, H.; Chen, S.; Liu, W.; Zhang, J. MiR-129-3p promotes docetaxel resistance of breast cancer cells via CP110 inhibition. Sci. Rep. 2015, 5, 15424. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, H.; Lou, J.R.; Zheng, J.; Zhu, H.; Popescu, N.I.; Lupu, F.; Lind, S.E.; Ding, W.Q. MicroRNA-19 (miR-19) regulates tissue factor expression in breast cancer cells. J. Biol. Chem. 2011, 286, 1429–1435. [Google Scholar] [CrossRef]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef]
- Yu, F.; Jiao, Y.; Zhu, Y.; Wang, Y.; Zhu, J.; Cui, X.; Liu, Y.; He, Y.; Park, E.Y.; Zhang, H.; et al. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J Biol Chem 2012, 287, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Chen, W.; Wei, F.; Zhou, B.P.; Hung, M.C.; Xie, X. Nanoparticle Delivery of miR-34a Eradicates Long-term-cultured Breast Cancer Stem Cells via Targeting C22ORF28 Directly. Theranostics 2017, 7, 4805–4824. [Google Scholar] [CrossRef] [PubMed]
- Christoffersen, N.R.; Shalgi, R.; Frankel, L.B.; Leucci, E.; Lees, M.; Klausen, M.; Pilpel, Y.; Nielsen, F.C.; Oren, M.; Lund, A.H. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death. Differ. 2010, 17, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Welch, C.; Chen, Y.; Stallings, R.L. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 2007, 26, 5017–5022. [Google Scholar] [CrossRef]
- Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13421–13426. [Google Scholar] [CrossRef]
- Kang, L.; Mao, J.; Tao, Y.; Song, B.; Ma, W.; Lu, Y.; Zhao, L.; Li, J.; Yang, B.; Li, L. MicroRNA-34a suppresses the breast cancer stem cell-like characteristics by downregulating Notch1 pathway. Cancer Sci. 2015, 106, 700–708. [Google Scholar] [CrossRef]
- Guarnieri, A.L.; Towers, C.G.; Drasin, D.J.; Oliphant, M.U.J.; Andrysik, Z.; Hotz, T.J.; Vartuli, R.L.; Linklater, E.S.; Pandey, A.; Khanal, S.; et al. The miR-106b-25 cluster mediates breast tumor initiation through activation of NOTCH1 via direct repression of NEDD4L. Oncogene 2018, 37, 3879–3893. [Google Scholar] [CrossRef]
- Wang, H.J.; Guo, Y.Q.; Tan, G.; Dong, L.; Cheng, L.; Li, K.J.; Wang, Z.Y.; Luo, H.F. miR-125b regulates side population in breast cancer and confers a chemoresistant phenotype. J. Cell. Biochem. 2013, 114, 2248–2257. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Tsuyada, A.; Ren, X.; Wu, X.; Stubblefield, K.; Rankin-Gee, E.K.; Wang, S.E. Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene 2011, 30, 1470–1480. [Google Scholar] [CrossRef]
- Niu, J.; Xue, A.; Chi, Y.; Xue, J.; Wang, W.; Zhao, Z.; Fan, M.; Yang, C.H.; Shao, Z.M.; Pfeffer, L.M.; et al. Induction of miRNA-181a by genotoxic treatments promotes chemotherapeutic resistance and metastasis in breast cancer. Oncogene 2016, 35, 1302–1313. [Google Scholar] [CrossRef]
- Kastrati, I.; Canestrari, E.; Frasor, J. PHLDA1 expression is controlled by an estrogen receptor-NFkappaB-miR-181 regulatory loop and is essential for formation of ER+ mammospheres. Oncogene 2015, 34, 2309–2316. [Google Scholar] [CrossRef] [PubMed]
- Min, S.; Li, L.; Zhang, M.; Zhang, Y.; Liang, X.; Xie, Y.; He, Q.; Li, Y.; Sun, J.; Liu, Q.; et al. TGF-beta-associated miR-27a inhibits dendritic cell-mediated differentiation of Th1 and Th17 cells by TAB3, p38 MAPK, MAP2K4 and MAP2K7. Genes. Immun. 2012, 13, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Chandran, P.A.; Keller, A.; Weinmann, L.; Seida, A.A.; Braun, M.; Andreev, K.; Fischer, B.; Horn, E.; Schwinn, S.; Junker, M.; et al. The TGF-beta-inducible miR-23a cluster attenuates IFN-gamma levels and antigen-specific cytotoxicity in human CD8(+) T cells. J. Leukoc. Biol. 2014, 96, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Cui, H.; Banerjee, S.; Tan, Z.; Salomao, R.; Fu, M.; Abraham, E.; Thannickal, V.J.; Liu, G. miR-27a regulates inflammatory response of macrophages by targeting IL-10. J. Immunol. 2014, 193, 327–334. [Google Scholar] [CrossRef]
- Tang, W.; Yu, F.; Yao, H.; Cui, X.; Jiao, Y.; Lin, L.; Chen, J.; Yin, D.; Song, E.; Liu, Q. miR-27a regulates endothelial differentiation of breast cancer stem like cells. Oncogene 2014, 33, 2629–2638. [Google Scholar] [CrossRef]
- GENECODE. Available online: https://www.gencodegenes.org (accessed on 3 February 2020).
- Klinge, C.M. Non-Coding RNAs in Breast Cancer: Intracellular and Intercellular Communication. Noncoding RNA 2018, 4. [Google Scholar] [CrossRef]
- Kong, X.; Liu, W.; Kong, Y. Roles and expression profiles of long non-coding RNAs in triple-negative breast cancers. J. Cell Mol. Med. 2018, 22, 390–394. [Google Scholar] [CrossRef]
- Balas, M.M.; Johnson, A.M. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. 2018, 3, 108–117. [Google Scholar] [CrossRef]
- Pecero, M.L.; Salvador-Bofill, J.; Molina-Pinelo, S. Long non-coding RNAs as monitoring tools and therapeutic targets in breast cancer. Cell Oncol. (Dordr.) 2019, 42, 1–12. [Google Scholar] [CrossRef]
- Chen, S.; Zhu, J.; Wang, F.; Guan, Z.; Ge, Y.; Yang, X.; Cai, J. LncRNAs and their role in cancer stem cells. Oncotarget 2017, 8, 110685–110692. [Google Scholar] [CrossRef]
- Huan, J.; Xing, L.; Lin, Q.; Xui, H.; Qin, X. Long noncoding RNA CRNDE activates Wnt/beta-catenin signaling pathway through acting as a molecular sponge of microRNA-136 in human breast cancer. Am. J. Transl. Res. 2017, 9, 1977–1989. [Google Scholar] [PubMed]
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014, 32, 2858–2868. [Google Scholar] [CrossRef]
- Deng, J.; Yang, M.; Jiang, R.; An, N.; Wang, X.; Liu, B. Long Non-Coding RNA HOTAIR Regulates the Proliferation, Self-Renewal Capacity, Tumor Formation and Migration of the Cancer Stem-Like Cell (CSC) Subpopulation Enriched from Breast Cancer Cells. PLoS ONE 2017, 12, e0170860. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Li, T.T.; Wang, K.L.; Xiao, G.Q.; Wang, J.H.; Zhao, H.D.; Kang, Z.J.; Fan, W.J.; Zhu, L.L.; Li, M.; et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017, 8, e2569. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Wang, J.H.; Fan, W.J.; Meng, Y.T.; Li, M.M.; Li, T.T.; Cui, B.; Wang, H.F.; Zhao, Y.; An, F.; et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 2018, 37, 1119. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Li, Y.; Ma, Y.; Lu, J.; Chen, Y.; Jiang, Q.; Qin, Q.; Zhao, L.; Huang, Q.; Luo, Z.; et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J. Exp. Clin. Cancer Res. 2018, 37, 289. [Google Scholar] [CrossRef]
- Tu, Z.; Schmollerl, J.; Cuiffo, B.G.; Karnoub, A.E. Microenvironmental Regulation of Long Noncoding RNA LINC01133 Promotes Cancer Stem Cell-Like Phenotypic Traits in Triple-Negative Breast Cancers. Stem Cells 2019, 37, 1281–1292. [Google Scholar] [CrossRef]
- Vidovic, D.; Huynh, T.T.; Konda, P.; Dean, C.; Cruickshank, B.M.; Sultan, M.; Coyle, K.M.; Gujar, S.; Marcato, P. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 2019. [Google Scholar] [CrossRef]
- Loewer, S.; Cabili, M.N.; Guttman, M.; Loh, Y.H.; Thomas, K.; Park, I.H.; Garber, M.; Curran, M.; Onder, T.; Agarwal, S.; et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010, 42, 1113–1117. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 2013, 25, 69–80. [Google Scholar] [CrossRef]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Wang, X.; Zhang, Y.; Gao, Z.; Wang, T.; Liu, R. LincRNA-ROR promotes metastasis and invasion of esophageal squamous cell carcinoma by regulating miR-145/FSCN1. Onco. Targets Ther. 2018, 11, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.W.; Li, L.; Wang, F.; Gu, Y.T. Inhibitory role of large intergenic noncoding RNA-ROR on tamoxifen resistance in the endocrine therapy of breast cancer by regulating the PI3K/Akt/mTOR signaling pathway. J. Cell Physiol. 2019, 234, 1904–1912. [Google Scholar] [CrossRef] [PubMed]
- Eades, G.; Wolfson, B.; Zhang, Y.; Li, Q.; Yao, Y.; Zhou, Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol. Cancer Res. 2015, 13, 330–338. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef]
- Chen, Y.M.; Liu, Y.; Wei, H.Y.; Lv, K.Z.; Fu, P. Linc-ROR induces epithelial-mesenchymal transition and contributes to drug resistance and invasion of breast cancer cells. Tumour. Biol. 2016, 37, 10861–10870. [Google Scholar] [CrossRef]
- Hou, L.; Tu, J.; Cheng, F.; Yang, H.; Yu, F.; Wang, M.; Liu, J.; Fan, J.; Zhou, G. Long noncoding RNA ROR promotes breast cancer by regulating the TGF-beta pathway. Cancer Cell Int. 2018, 18, 142. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Liang, F.; Zhang, J.W.; Wang, F.; Wang, L.; Kang, X.G. Effects of long noncoding RNA-ROR on tamoxifen resistance of breast cancer cells by regulating microRNA-205. Cancer Chemother. Pharmacol. 2017, 79, 327–337. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, B.; Zhu, H.; Qu, X.; Zhao, L.; Tan, Y.; Jiang, Y.; Liao, M.; Wu, X. Inhibition of long non-coding RNA ROR reverses resistance to Tamoxifen by inducing autophagy in breast cancer. Tumour. Biol. 2017, 39, 1010428317705790. [Google Scholar] [CrossRef]
- Zheng, A.; Song, X.; Zhang, L.; Zhao, L.; Mao, X.; Wei, M.; Jin, F. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/beta-catenin pathway. J. Exp. Clin. Cancer Res. 2019, 38, 305. [Google Scholar] [CrossRef]
- Zhou, M.; Hou, Y.; Yang, G.; Zhang, H.; Tu, G.; Du, Y.E.; Wen, S.; Xu, L.; Tang, X.; Tang, S.; et al. LncRNA-Hh Strengthen Cancer Stem Cells Generation in Twist-Positive Breast Cancer via Activation of Hedgehog Signaling Pathway. Stem Cells 2016, 34, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Liu, X.; Zhou, S.; Li, W.; Liu, C.; Chadwick, M.; Qian, C. Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 2019, 450, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, M.; Asadi, M.H. Long non-coding RNA ES1 controls the proliferation of breast cancer cells by regulating the Oct4/Sox2/miR-302 axis. FEBS J. 2019, 286, 2611–2623. [Google Scholar] [CrossRef] [PubMed]
- Shin, V.Y.; Chen, J.; Cheuk, I.W.; Siu, M.T.; Ho, C.W.; Wang, X.; Jin, H.; Kwong, A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019, 10, 270. [Google Scholar] [CrossRef]
- Youness, R.A.; Gad, M.Z. Long non-coding RNAs: Functional regulatory players in breast cancer. Noncoding RNA Res. 2019, 4, 36–44. [Google Scholar] [CrossRef]
- Hansji, H.; Leung, E.Y.; Baguley, B.C.; Finlay, G.J.; Askarian-Amiri, M.E. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front Genet. 2014, 5, 379. [Google Scholar] [CrossRef]
- Li, Z.; Hou, P.; Fan, D.; Dong, M.; Ma, M.; Li, H.; Yao, R.; Li, Y.; Wang, G.; Geng, P.; et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017, 24, 59–71. [Google Scholar] [CrossRef]
- Wu, W.; Chen, F.; Cui, X.; Yang, L.; Chen, J.; Zhao, J.; Huang, D.; Liu, J.; Yang, L.; Zeng, J.; et al. LncRNA NKILA suppresses TGF-beta-induced epithelial-mesenchymal transition by blocking NF-kappaB signaling in breast cancer. Int. J. Cancer 2018, 143, 2213–2224. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, B.; Zhang, M.; Guo, W.; Wu, Z.; Wang, Y.; Jia, L.; Li, S.; The Cancer Genome Atlas Research Network; Xie, W.; et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell 2018, 33, 706–720.e9. [Google Scholar] [CrossRef]
- Barsyte-Lovejoy, D.; Lau, S.K.; Boutros, P.C.; Khosravi, F.; Jurisica, I.; Andrulis, I.L.; Tsao, M.S.; Penn, L.Z. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006, 66, 5330–5337. [Google Scholar] [CrossRef]
- Si, X.; Zang, R.; Zhang, E.; Liu, Y.; Shi, X.; Zhang, E.; Shao, L.; Li, A.; Yang, N.; Han, X.; et al. LncRNA H19 confers chemoresistance in ERalpha-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget 2016, 7, 81452–81462. [Google Scholar] [CrossRef] [PubMed]
- Meseure, D.; Vacher, S.; Lallemand, F.; Alsibai, K.D.; Hatem, R.; Chemlali, W.; Nicolas, A.; De Koning, L.; Pasmant, E.; Callens, C.; et al. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br. J. Cancer 2016, 114, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; He, J.; Zhang, D. Suppression of long non-coding RNA CCAT2 improves tamoxifen-resistant breast cancer cells’ response to tamoxifen. Mol. Biol. (Mosk.) 2016, 50, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Luo, J. Long Non-Coding RNA (lncRNA) Urothelial Carcinoma-Associated 1 (UCA1) Enhances Tamoxifen Resistance in Breast Cancer Cells via Inhibiting mTOR Signaling Pathway. Med. Sci. Monit. 2016, 22, 3860–3867. [Google Scholar] [CrossRef] [PubMed]
- Saunders-Hastings, P.; Reisman, J.; Krewski, D. Assessing the State of Knowledge Regarding the Effectiveness of Interventions to Contain Pandemic Influenza Transmission: A Systematic Review and Narrative Synthesis. PLoS ONE 2016, 11, e0168262. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhai, L.; Wang, H.; Liu, C.; Zhang, J.; Chen, W.; Wei, Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 2016, 7, 27778–27786. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Wei, Y.; Lai, X.; Yu, S.; Chen, S.; Ma, Y.; Zhang, Y.; Li, H.; Zhu, X.; Yao, L.; Zhang, J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 2014, 147, 423–431. [Google Scholar] [CrossRef]
- Yu, D.D.; Wu, Y.; Zhang, X.H.; Lv, M.M.; Chen, W.X.; Chen, X.; Yang, S.J.; Shen, H.; Zhong, S.L.; Tang, J.H.; et al. Exosomes from adriamycin-resistant breast cancer cells transmit drug resistance partly by delivering miR-222. Tumour. Biol. 2016, 37, 3227–3235. [Google Scholar] [CrossRef]
- Chen, W.X.; Cai, Y.Q.; Lv, M.M.; Chen, L.; Zhong, S.L.; Ma, T.F.; Zhao, J.H.; Tang, J.H. Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumour. Biol. 2014, 35, 9649–9659. [Google Scholar] [CrossRef]
- Chen, W.X.; Liu, X.M.; Lv, M.M.; Chen, L.; Zhao, J.H.; Zhong, S.L.; Ji, M.H.; Hu, Q.; Luo, Z.; Wu, J.Z.; et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS ONE 2014, 9, e95240. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Li, J.; Chen, W.X.; Cai, Y.Q.; Yu, D.D.; Zhong, S.L.; Zhao, J.H.; Zhou, J.W.; Tang, J.H. Exosomes decrease sensitivity of breast cancer cells to adriamycin by delivering microRNAs. Tumour. Biol. 2016, 37, 5247–5256. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Peng, F.; Chen, J. The Role of Exosomal MicroRNAs in the Tumor Microenvironment of Breast Cancer. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef]
- Vaupel, P.; Mayer, A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef]
- Hashimoto, K.; Ochi, H.; Sunamura, S.; Kosaka, N.; Mabuchi, Y.; Fukuda, T.; Yao, K.; Kanda, H.; Ae, K.; Okawa, A.; et al. Cancer-secreted hsa-miR-940 induces an osteoblastic phenotype in the bone metastatic microenvironment via targeting ARHGAP1 and FAM134A. Proc. Natl. Acad Sci. USA 2018, 115, 2204–2209. [Google Scholar] [CrossRef]
- Turchinovich, A.; Samatov, T.R.; Tonevitsky, A.G.; Burwinkel, B. Circulating miRNAs: cell-cell communication function? Front Genet. 2013, 4, 119. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Y.; Xia, T.; Zhou, X.; Huang, Z.; Zhang, H.; Zhu, W.; Ding, Q.; Wang, S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat. 2018, 170, 257–270. [Google Scholar] [CrossRef]
- Eichelser, C.; Stuckrath, I.; Muller, V.; Milde-Langosch, K.; Wikman, H.; Pantel, K.; Schwarzenbach, H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014, 5, 9650–9663. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, J.; Li, J.; Shao, J.; Fang, L. MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem. Biophys. Res. Commun. 2018, 501, 486–493. [Google Scholar] [CrossRef]
- Chen, W.X.; Cheng, L.; Pan, M.; Qian, Q.; Zhu, Y.L.; Xu, L.Y.; Ding, Q. D Rhamnose beta-Hederin against human breast cancer by reducing tumor-derived exosomes. Oncol Lett. 2018, 16, 5172–5178. [Google Scholar] [CrossRef]
- Jang, J.Y.; Lee, J.K.; Jeon, Y.K.; Kim, C.W. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer 2013, 13, 421. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.D.; Yao, Y.F.; Zhong, S.L.; Zhao, J.H.; Tang, J.H. beta-Elemene Reverses Chemoresistance of Breast Cancer Cells by Reducing Resistance Transmission via Exosomes. Cell Physiol. Biochem. 2015, 36, 2274–2286. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Li, M.; Cui, S.; Wang, D.; Zhang, C.Y.; Zen, K.; Li, L. Shikonin Inhibits the Proliferation of Human Breast Cancer Cells by Reducing Tumor-Derived Exosomes. Molecules 2016, 21. [Google Scholar] [CrossRef]
- Hannafon, B.N.; Carpenter, K.J.; Berry, W.L.; Janknecht, R.; Dooley, W.C.; Ding, W.Q. Exosome-mediated microRNA signaling from breast cancer cells is altered by the anti-angiogenesis agent docosahexaenoic acid (DHA). Mol. Cancer 2015, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.P.; Khan, S.; Gilligan, K.E.; Zafar, H.; Lalor, P.; Glynn, C.; O’Flatharta, C.; Ingoldsby, H.; Dockery, P.; De Bhulbh, A.; et al. Employing mesenchymal stem cells to support tumor-targeted delivery of extracellular vesicle (EV)-encapsulated microRNA-379. Oncogene 2018, 37, 2137–2149. [Google Scholar] [CrossRef]
- Bliss, S.A.; Sinha, G.; Sandiford, O.A.; Williams, L.M.; Engelberth, D.J.; Guiro, K.; Isenalumhe, L.L.; Greco, S.J.; Ayer, S.; Bryan, M.; et al. Mesenchymal Stem Cell-Derived Exosomes Stimulate Cycling Quiescence and Early Breast Cancer Dormancy in Bone Marrow. Cancer Res. 2016, 76, 5832–5844. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Pereira, F.; Alves de Matos, A.P.; Fernandes, M.; Baptista, P.V.; Fernandes, A.R. Smuggling gold nanoparticles across cell types—A new role for exosomes in gene silencing. Nanomedicine 2017, 13, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Naseri, Z.; Oskuee, R.K.; Jaafari, M.R.; Forouzandeh Moghadam, M. Exosome-mediated delivery of functionally active miRNA-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. Int. J. Nanomedicine 2018, 13, 7727–7747. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Yu, Y.; Chrisler, W.B.; Xiong, Y.; Hu, D.; Lei, C. Delivery of MicroRNA-10b with Polylysine Nanoparticles for Inhibition of Breast Cancer Cell Wound Healing. Breast Cancer (Auckl.) 2012, 6, 9–19. [Google Scholar] [CrossRef]
- Devulapally, R.; Sekar, N.M.; Sekar, T.V.; Foygel, K.; Massoud, T.F.; Willmann, J.K.; Paulmurugan, R. Polymer nanoparticles mediated codelivery of antimiR-10b and antimiR-21 for achieving triple negative breast cancer therapy. ACS Nano. 2015, 9, 2290–2302. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Cao, M.; Zhang, J.; Hu, K.; Yin, Z.; Zhou, Z.; Xiao, X.; Yang, Y.; Sheng, W.; Wu, Y.; et al. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials 2014, 35, 4333–4344. [Google Scholar] [CrossRef] [PubMed]
- Ekin, A.; Karatas, O.F.; Culha, M.; Ozen, M. Designing a gold nanoparticle-based nanocarrier for microRNA transfection into the prostate and breast cancer cells. J. Gene Med. 2014, 16, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Zhi, F.; Dong, H.; Jia, X.; Guo, W.; Lu, H.; Yang, Y.; Ju, H.; Zhang, X.; Hu, Y. Functionalized graphene oxide mediated adriamycin delivery and miR-21 gene silencing to overcome tumor multidrug resistance in vitro. PLoS ONE 2013, 8, e60034. [Google Scholar] [CrossRef]
- Hydbring, P.; Wang, Y.; Fassl, A.; Li, X.; Matia, V.; Otto, T.; Choi, Y.J.; Sweeney, K.E.; Suski, J.M.; Yin, H.; et al. Cell-Cycle-Targeting MicroRNAs as Therapeutic Tools against Refractory Cancers. Cancer Cell 2017, 31, 576–590.e8. [Google Scholar] [CrossRef]
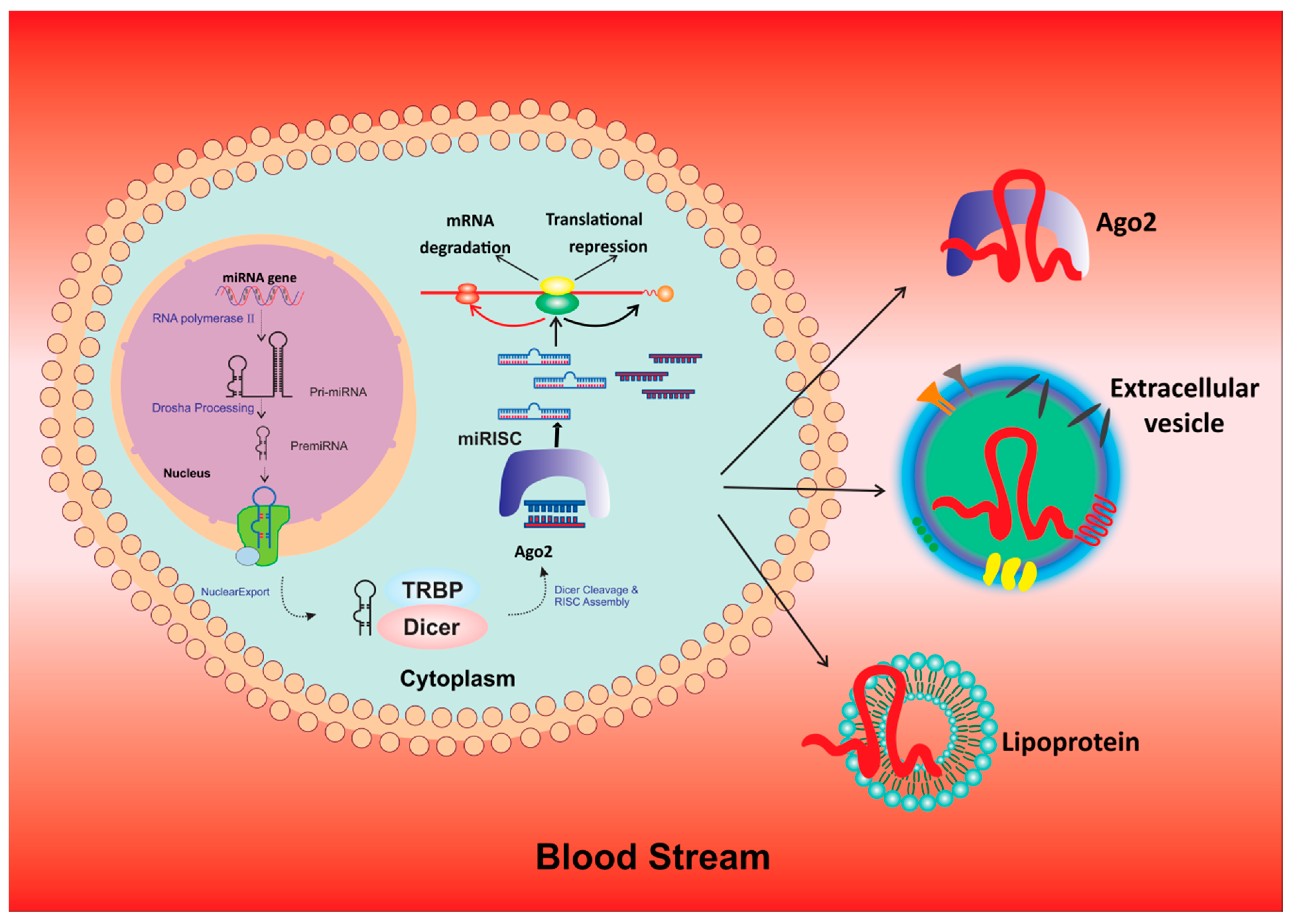
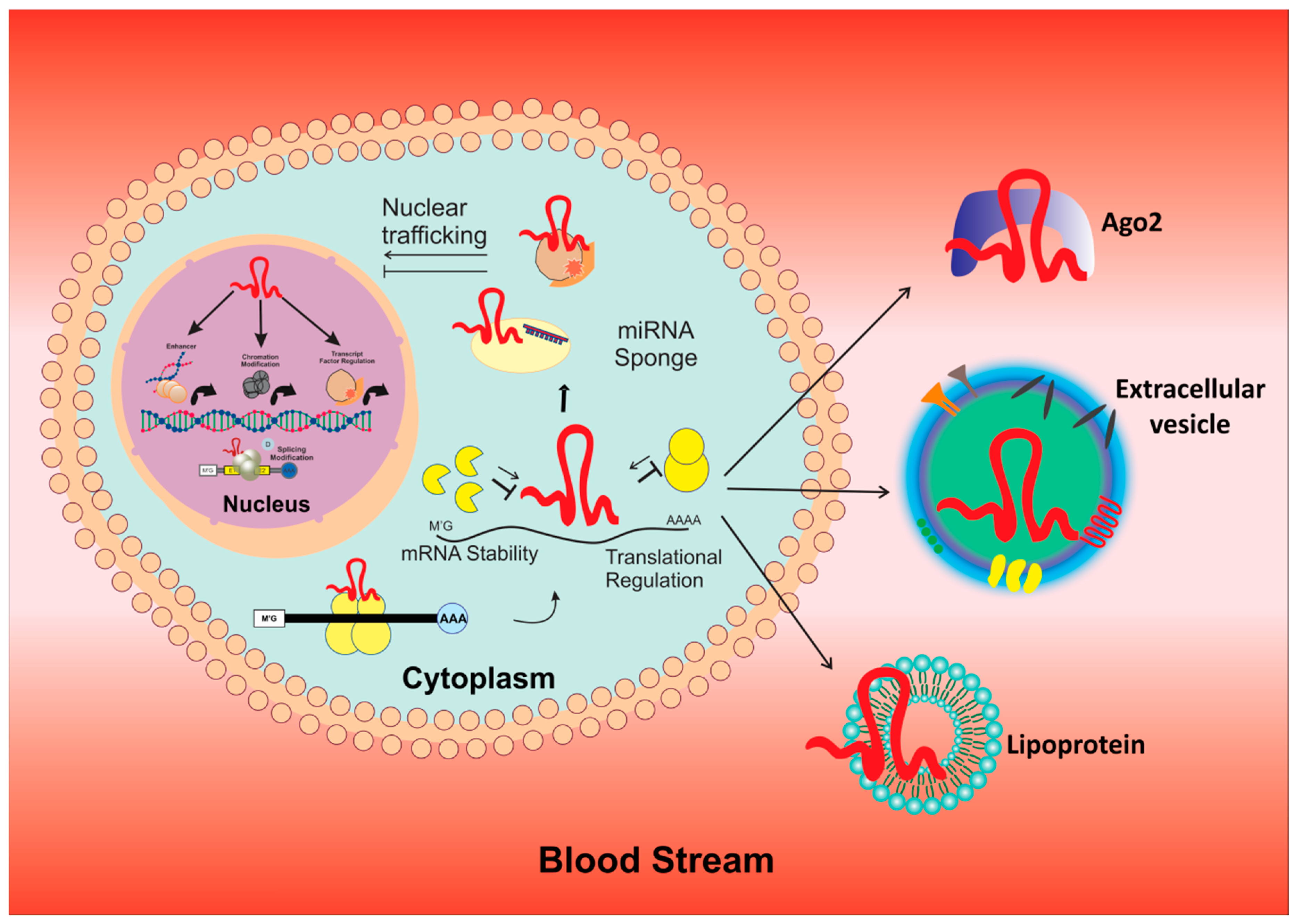
| miRNA | Type | Expression Level | Targets | Pathways | Reference |
|---|---|---|---|---|---|
| miR-31 | TsmiR | ↑/↓ | ITGA5, RDX, RHOA | Metastasis | [112,113] |
| miR-145 | TsmiR | ↓ | MUC1, ERA, RTKN | Proliferation, Apoptosis, Invasion | [114,115,116] |
| miR-155 | TsmiR | ↑ | FOXO3A, RHOA, SOCS1 | STAT3, Proliferation, TGFβ Signaling | [117,118,119] |
| miR-21 | OncomiR | ↑ | BCL2, PTEN, MMP3, TPM1, MASPIN, PDCD4, RHOB | EMT, Apoptosis, Invasion, Migration, Inflammatory Signals | [120,121,122,123,124] |
| miR-125b | TsmiR | ↑/↓ | BAK1, ERA, HER2, CRAF, RTKN, MUC1 | Migration, Proliferation, Apoptosis | [125,126,127] |
| miR-10b | OncomiR | ↑/↓ | HDAC4, TIAM, HOXD10, EMT | EMT, Metastasis, Invasion | [128,129,130] |
| miR-205 | TsmiR | ↓ | HER3, VEGFA, EMT | Proliferation, Invasion | [131,132,133] |
| miR-210 | OncomiR | ↑ | MNT, RAD52 | Hypoxia | [134,135] |
| miR-196A | OncomiR | ↑ | ANXA1 | Proliferation, Apoptosis, | [136] |
| miR-944 | OncomiR | ↑ | BNIP3 | Cell Proliferation, Migration, Invasion | [137] |
| miR-222 | OncomiR | ↑ | PTEN | PTEN, Akt/FOXP1 | [138] |
| miR-3646 | OncomiR | ↑ | GSK-3β | β Catenin | [139] |
| miR-34A | OncomiR | ↑ | BCL2, CCND1 | Apoptosis | [140] |
| miR-141 | OncomiR | ↑ | EIF4E | Apoptosis | [141] |
| miR-520h | OncomiR | ↑ | DAPK2 | PI3K/Akt | [142] |
| miR-34 | TsmiR | ↓ | BCL2, NOTCH | Apoptosis, NOTCH | [143] |
| miR-146 | TsmiR | ↓ | NFkB | Inflammatory Signals | [144] |
| miR-7 | TsmiR | ↓ | EGFR | EGFR | [145] |
| miR-22 | TsmiR | ↓ | HER3, CDK6, ERα, CDC25C, SP1 | Estrogen Signaling | [146] |
| miR-221 | TsmiR | ↑ | P27, P57 | Wnt/β-catenin | [147] |
| miR-191 | OncomiR | ↑ | SATB1, CDK6, BDNF | Estrogen Signaling | [148] |
| miR-196A | OncomiR | ↑ | ANXA1 | Apoptosis | [136] |
| miR-335 | TsmiR | ↑ | SOX4, TNC, PTPRN2, MERTK | Metastasis | [149] |
| miR-20 | OncomiR | ↑ | E2F | Proliferation | [150] |
| miR-9 | TsmiR | ↑ | LIFR, E-CADHERIN | EMT, Hippo-YAP | [151,152] |
| miR-126 | TsmiR | ↓ | VEGFA and PIK3R2 | VEGF/PI3K/AKT | [153] |
| miR-98 | TsmiR | ↑ | ALK4 and MMP11 | Angiogenesis, Invasion | [154] |
| miR-148a/152 | TsmiR | ↓ | DNMT1, IGF-IR and IRS1 | IGF-IR/PKM2 | [155] |
| miR-519c | TsmiR | ↓ | HIF-1α | Hypoxia | [156] |
| miR-10b | OncomiR | ↑ | HOXD10 | Hox pathway | [157] |
| miR-140-5p | TsmiR | ↓ | VEGFA | Metastasis, Angiogenesis | [158] |
| miR-494 | TsmiR | ↑ | PTEN | Akt, NF-kB, mTOR | [159] |
| miR-206 | TsmiR | ↓ | VEGF, MAPK3, and SOX9 | Invasion, Angiogenesis | [160] |
| miR-19a | OncomiR | ↑ | PTEN | Oncogenic PTEN Cell proliferation, Th1 immune response | [161] |
| miR-17-92 | TsmiR | ↓ | HIF-1α | Hypoxia, Angiogenesis. | [162] |
| miR-467 | OncomiR | ↑ | TSP-1 | Angiogenesis | [163,164] |
| miR-18 | OncomiR | ↑ | SMAD7 | EMT, Metastasis | [165] |
| miR-143 | OncomiR | ↑ | FOSL2 | EMT, Metastasis | [165] |
| miR-196B | OncomiR | ↑ | HOXD10 | Hox pathway | [157] |
| miR-200 | OncomiR | ↑ | ZEB1, ZEB2 | EMT | [165] |
| miR-205 | TsmiR | ↓ | YAP1 | miR-205/YAP1, Angiogenesis, Metastasis | [166] |
| miR-892b | TsmiR | ↑ | TRAF2, TAK1, and TAB3 | NF-kB | [167] |
| miR-210 RAD52 | OncomiR | ↑ | RAD52 | Invasion, Proliferation, Migration | [168] |
| mirR-155 | OncomiR | ↑ | SOC6 | STAT3 signaling | [169] |
| miR-451 | OncomiR | ↑ | Bcl-2 | Apoptosis | [170] |
| miR-100 | OncomiR | ↑ | mTOR | Cell proliferation, Survival | [171] |
| miR-139-5p | OncomiR | ↑ | Notch1 | Cell growth, Apoptosis | [172] |
| miR-214 | OncomiR | ↑ | UCP2 | Autophagy | [173] |
| miR-16 | OncomiR | ↑ | CCNJ, FUBP1 | PI3K/Akt | [174] |
| miR-199a-3p | TsmiR | ↑ | TFAM | Mitochondrial Biogenesis | [175] |
| miR-302b | TsmiR | ↑ | E2F1 | E2f1-ATM axis | [176] |
| miR-218 | TsmiR | ↑ | BRCA1 | DNA repair, Cell proliferation, Invasion | [177] |
| miR-638 | TsmiR | ↑ | BRCA1 | DNA repair, Cell proliferation, Invasion | [178] |
| miR-29A | OncomiR | ↑ | PTEN | Apoptosis | [179] |
| miR-129-3p | OncomiR | ↑ | CP110 | Apoptosis \, Cell Cycle, Cell Proliferation | [180] |
| miR-19 | OncomiR | ↓ | Tissue factor | Angiogenesis, Metastasis | [181] |
| lncRNA | Type | Expression Level | Targets | Pathways | Reference PMID |
|---|---|---|---|---|---|
| MEG 3 | Tumor suppressor | ↓ | p53 | p53 | [228] |
| HOTAIR | Oncogene | ↑ | BRCA1, PTEN | PI3K/AKT-BAD pathway, HOXD10 | [229] |
| ACNR | Tumor suppressor | ↓ | TGF-β | Metastasis, Invasion | [230] |
| PTENP1 | Tumor suppressor | ↓ | PTEN | Apoptosis | [228] |
| NKILA | Oncogene | ↓ | NF-kB | EMT | [231] |
| EPIC 1 | Oncogene | ↑ | Myc | Cell Cycle | [232] |
| PLNCRNA-1 | Oncogene | ↓ | TGF-β | Apoptosis, Metastasis, Invasion | [228] |
| H19 | Oncogene | ↑ | C-myc | AKT, BIK | [233,234] |
| MALAT-1 | Oncogene | ↑/↓ | AKT, p53 | APOPTOSIS | [235] |
| LINK-A | Oncogene | ↑ | HIF-1α | Hypoxia Pathway | [228] |
| CCAT2 | Oncogene | ↑ | ERK | MAPK | [236] |
| PVT-1 | Oncogene | ↑ | KLF-5,β-Catenin | WNT/β-Catenin | [228] |
| UCA1 | Oncogene | ↑ | mTOR,β-Catenin | mTOR, WNT/ β-Catenin | [237,238] |
| GAS5 | Tumor suppressor | ↓ | PTEN | Apoptosis | [239] |
| BCAR4 | Oncogene | ↑ | SNIP1, PNUTS | Hedgehog /GLI 2 Signaling Transduction | [228] |
| NEAT | Oncogene | ↑ | ZEB1, RAS | RAS, MAPK, RSF1 | [227] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prabhu, K.S.; Raza, A.; Karedath, T.; Raza, S.S.; Fathima, H.; Ahmed, E.I.; Kuttikrishnan, S.; Therachiyil, L.; Kulinski, M.; Dermime, S.; et al. Non-Coding RNAs as Regulators and Markers for Targeting of Breast Cancer and Cancer Stem Cells. Cancers 2020, 12, 351. https://doi.org/10.3390/cancers12020351
Prabhu KS, Raza A, Karedath T, Raza SS, Fathima H, Ahmed EI, Kuttikrishnan S, Therachiyil L, Kulinski M, Dermime S, et al. Non-Coding RNAs as Regulators and Markers for Targeting of Breast Cancer and Cancer Stem Cells. Cancers. 2020; 12(2):351. https://doi.org/10.3390/cancers12020351
Chicago/Turabian StylePrabhu, Kirti S., Afsheen Raza, Thasni Karedath, Syed Shadab Raza, Hamna Fathima, Eiman I. Ahmed, Shilpa Kuttikrishnan, Lubna Therachiyil, Michal Kulinski, Said Dermime, and et al. 2020. "Non-Coding RNAs as Regulators and Markers for Targeting of Breast Cancer and Cancer Stem Cells" Cancers 12, no. 2: 351. https://doi.org/10.3390/cancers12020351
APA StylePrabhu, K. S., Raza, A., Karedath, T., Raza, S. S., Fathima, H., Ahmed, E. I., Kuttikrishnan, S., Therachiyil, L., Kulinski, M., Dermime, S., Junejo, K., Steinhoff, M., & Uddin, S. (2020). Non-Coding RNAs as Regulators and Markers for Targeting of Breast Cancer and Cancer Stem Cells. Cancers, 12(2), 351. https://doi.org/10.3390/cancers12020351









