Angiogenesis-Related Gene Expression Signatures Predicting Prognosis in Gastric Cancer Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
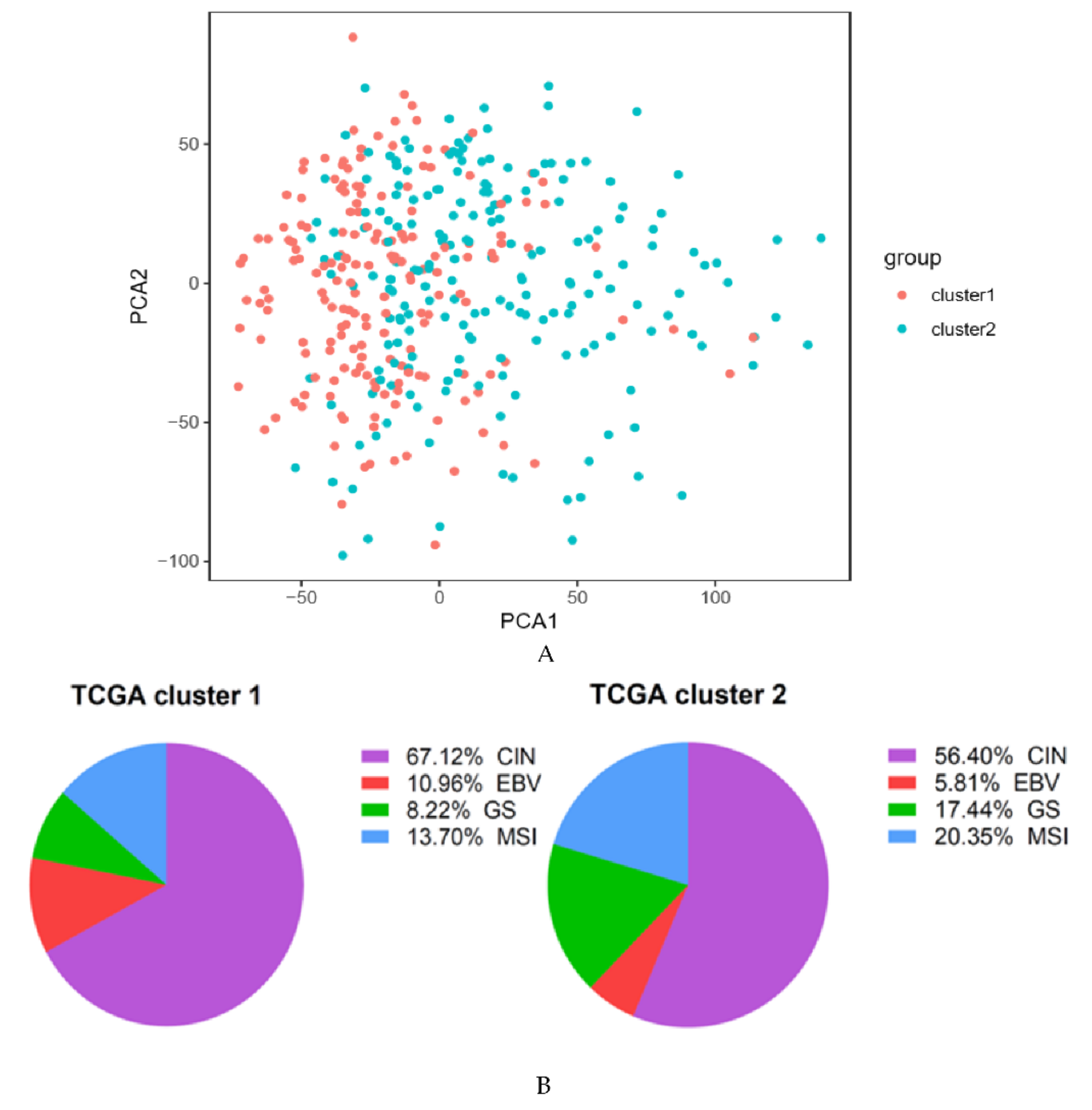
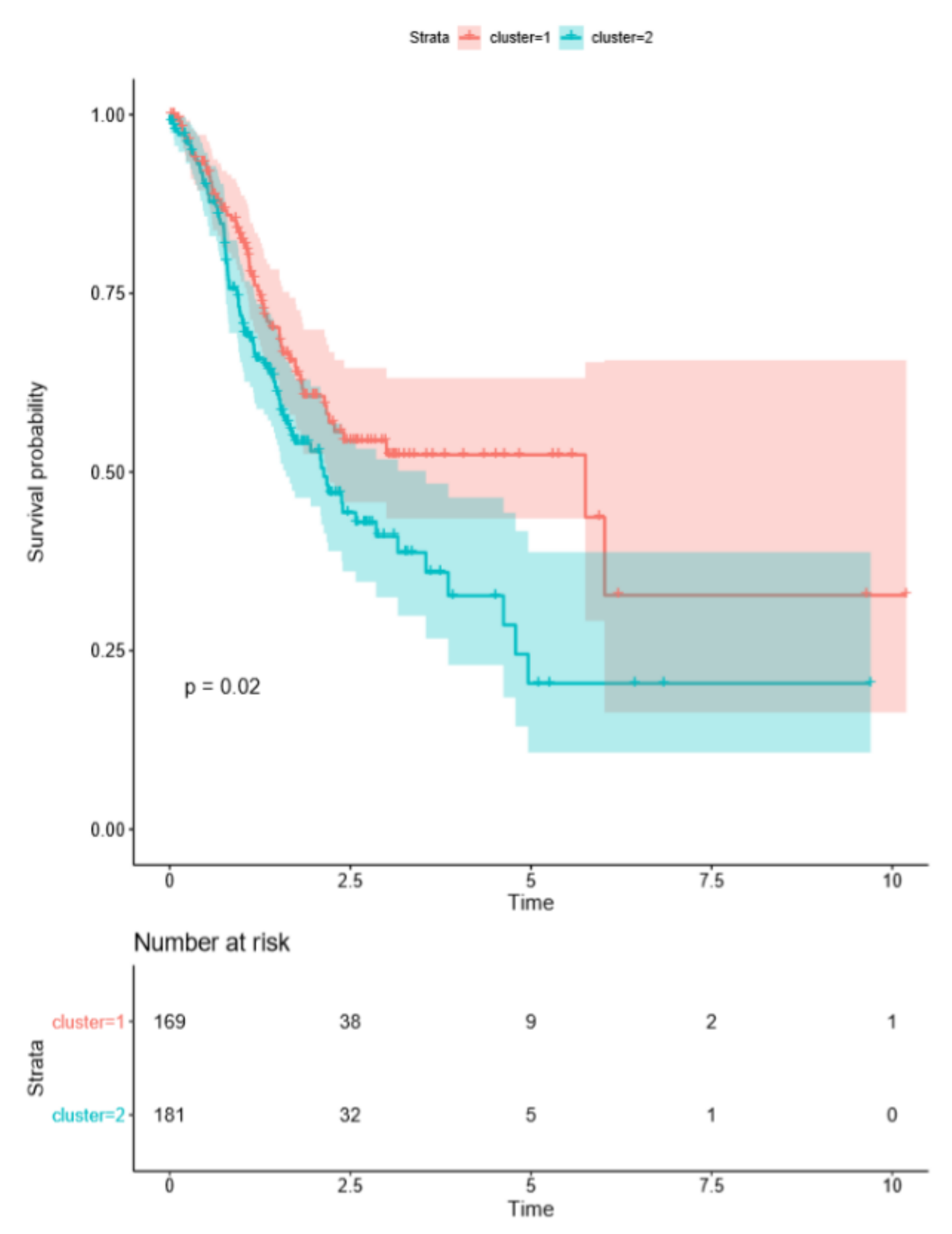
2.1. Cluster Analysis Based on ARG Expression Profiles
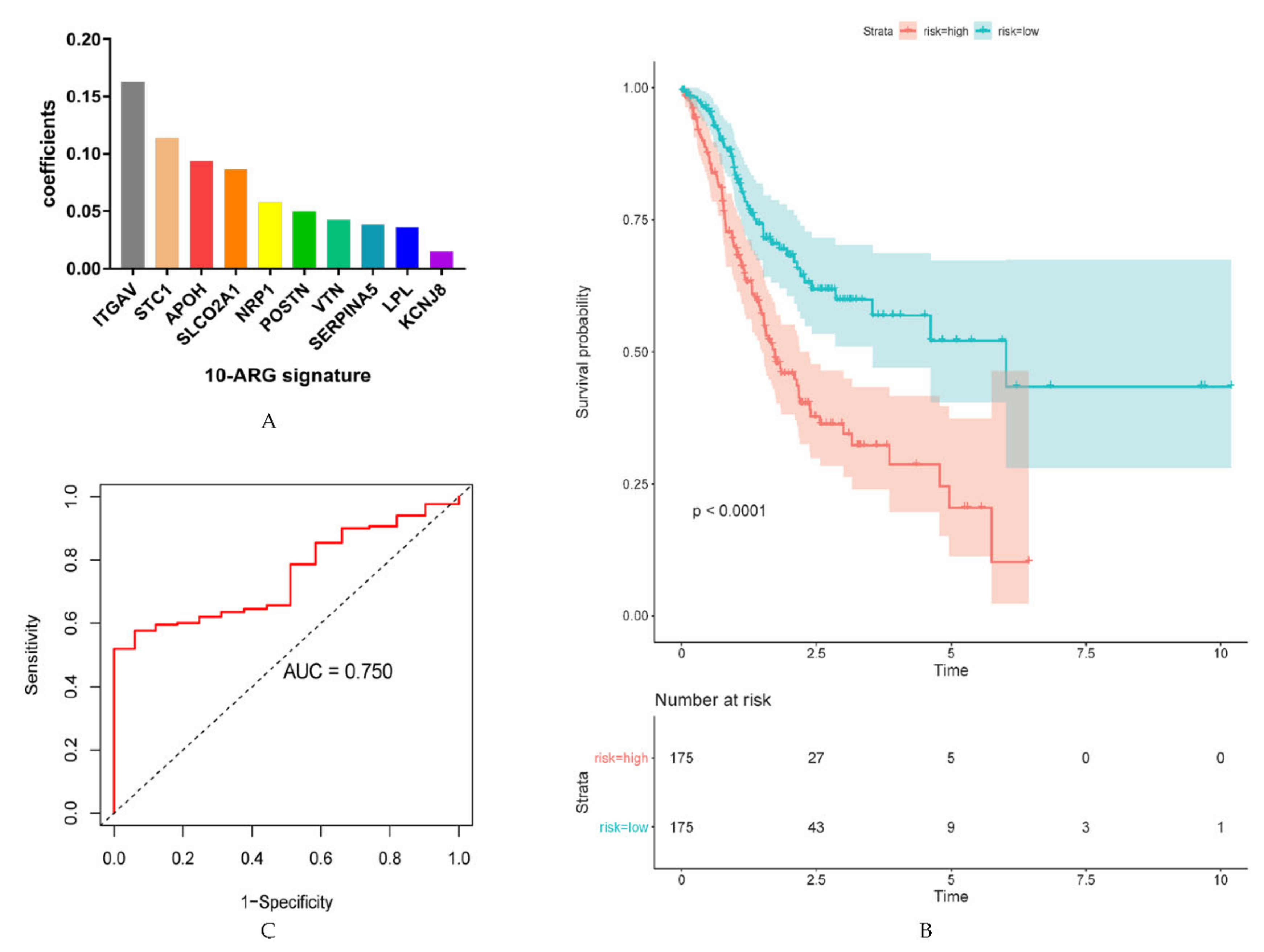
2.2. Identification of ARGs with Prognostic Value and Establishment of Prognostic Models
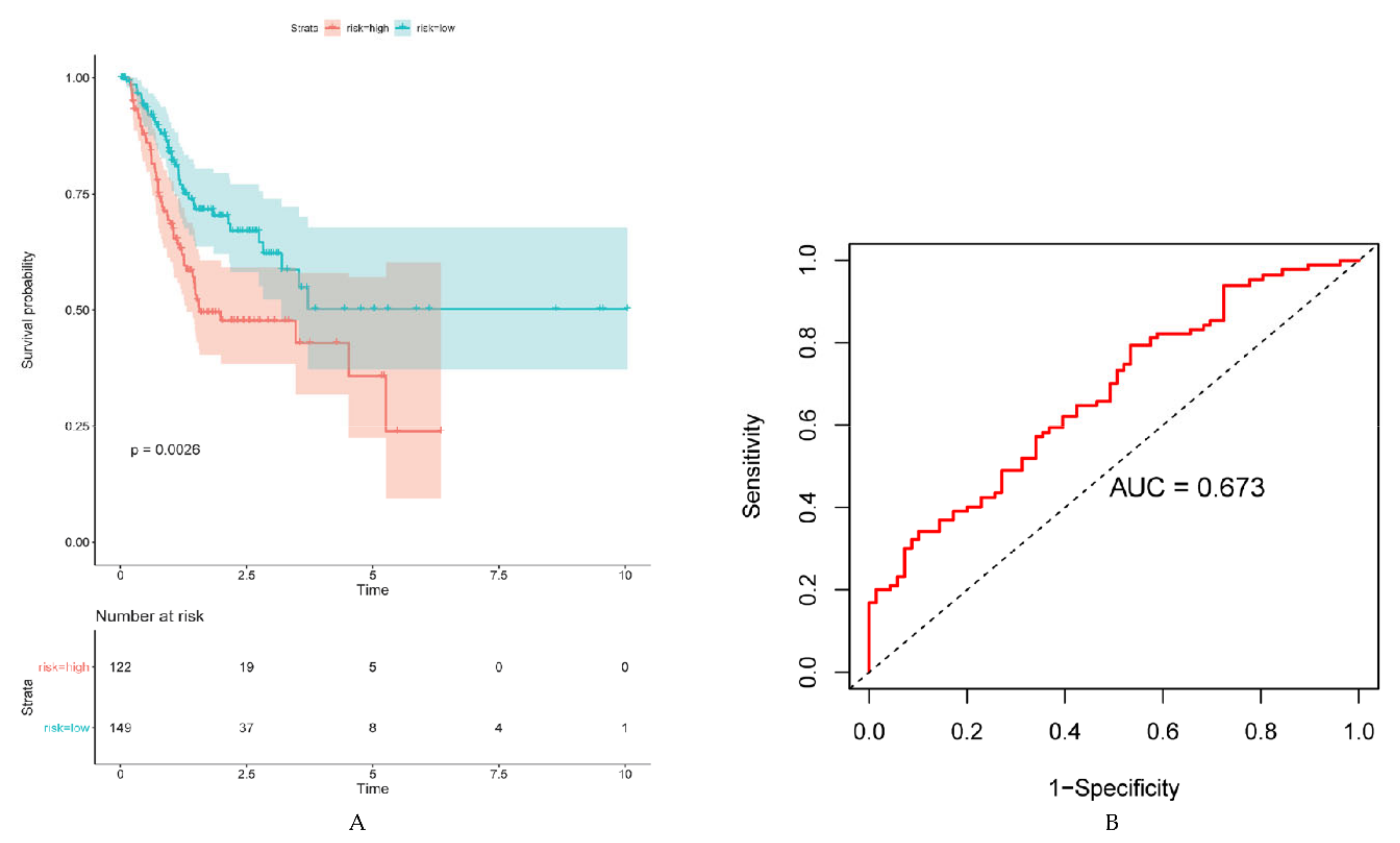
2.3. Validation of Prognostic ARG Signatures with External Dataset
2.4. ARG Signatures Independently Predict OS and DFS
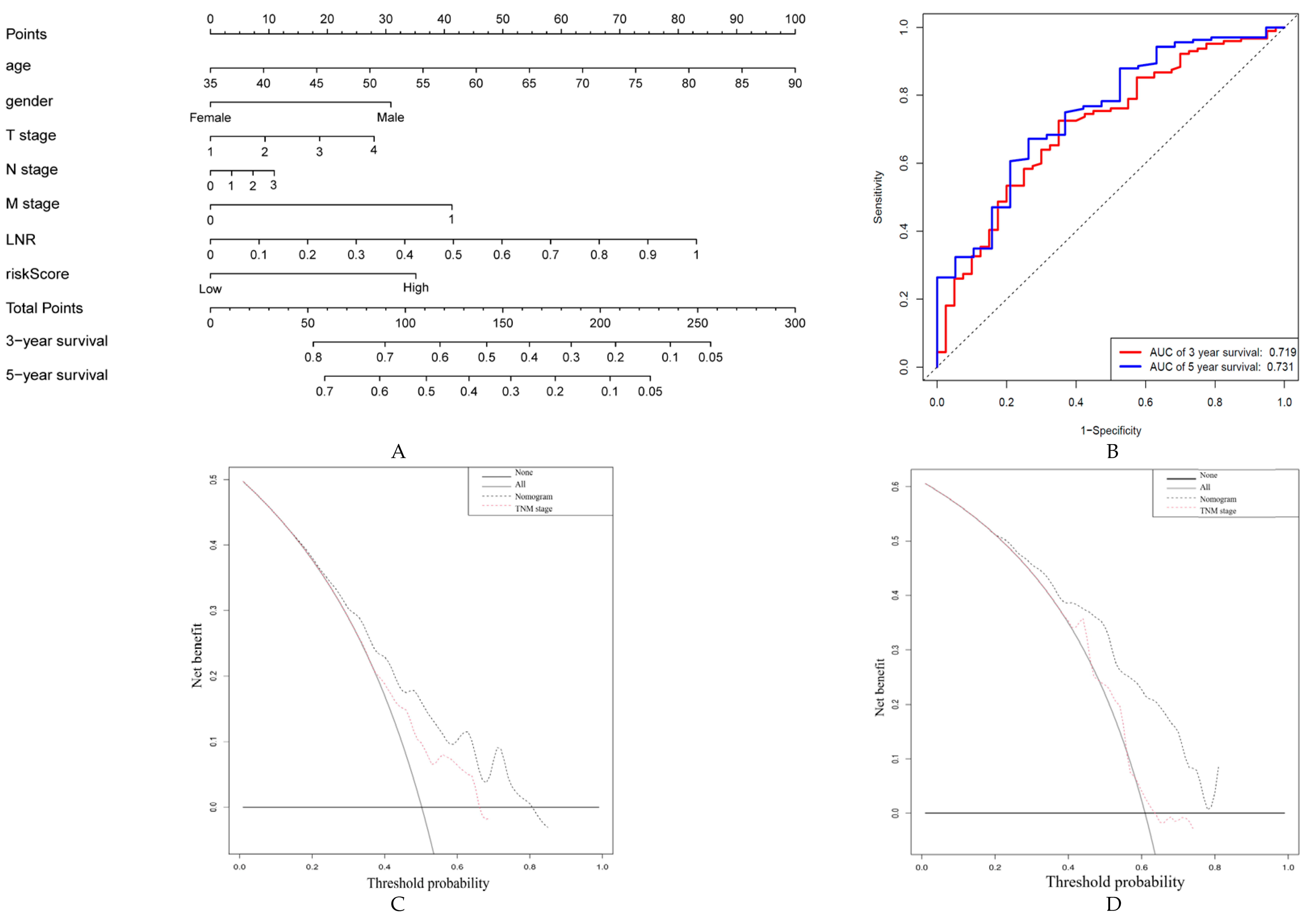
2.5. Construction and Validation of a Nomogram Based on ARG Signatures
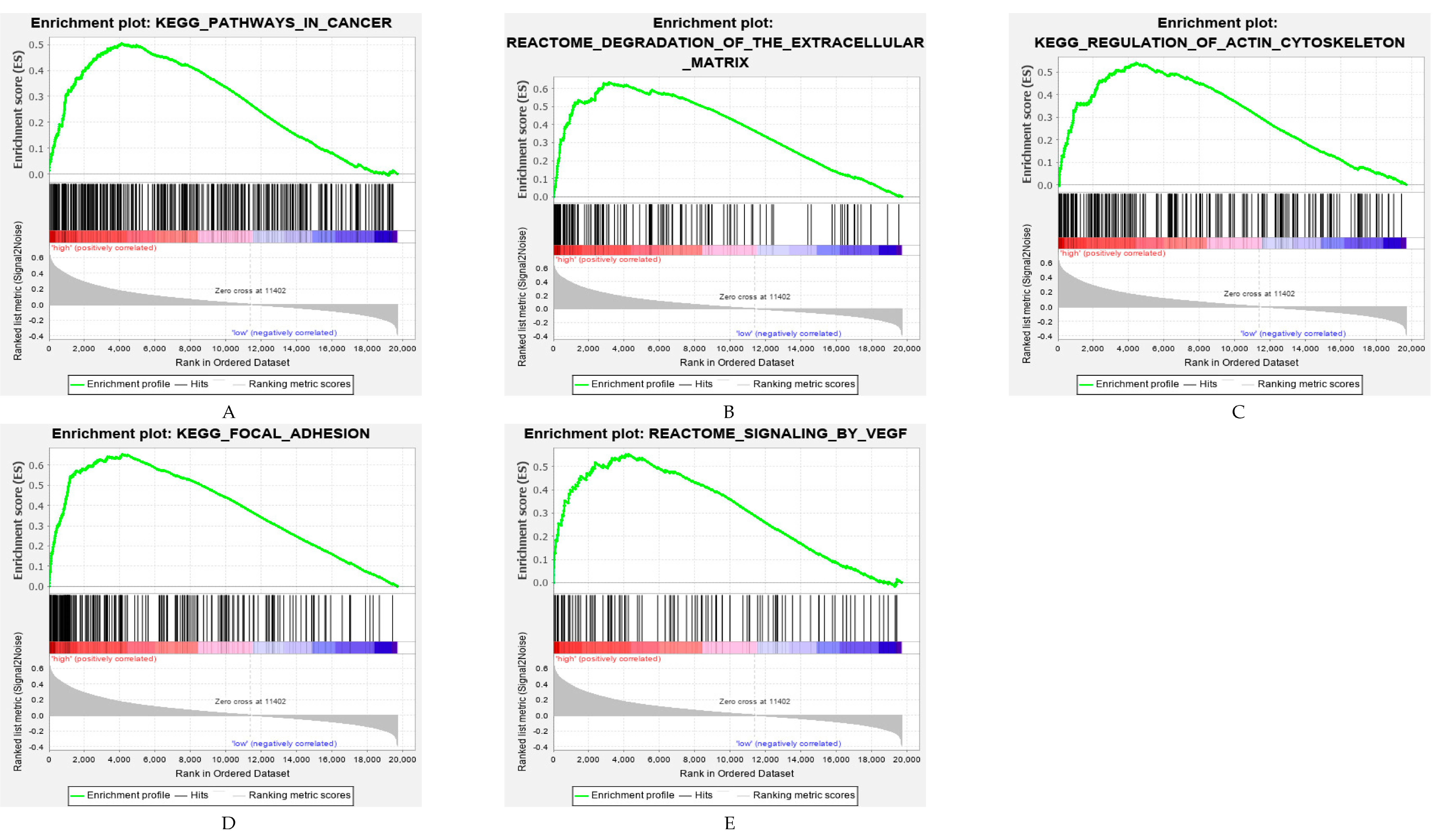
2.6. Functional Analysis of the ARG Signatures
3. Discussion
4. Materials and Methods
4.1. Gene Expression and Clinical Data Acquisition
4.2. Consensus Clustering Analysis
4.3. Development and Validation of Prognostic Signatures Based on ARGs
4.4. Construction and Evaluation of the Nomogram
4.5. Gene Set Enrichment Analysis (GSEA)
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eusebi, L.H.; Telese, A.; Marasco, G.; Bazzoli, F.; Zagari, R.M. Gastric cancer prevention strategies: A global perspective. J. Gastroenterol. Hepatol. 2020, 35, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Nikolaou, M.; Pavlopoulou, A.; Georgakilas, A.G.; Kyrodimos, E. The challenge of drug resistance in cancer treatment: A current overview. Clin. Exp. Metastasis 2018, 35, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Machlowska, J.; Baj, J.; Sitarz, M.; Maciejewski, R.; Sitarz, R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int. J. Mol. Sci. 2020, 21, 4012. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Sagaert, X.; Topal, B.; Haustermans, K.; Prenen, H. Gastric cancer. Lancet 2016, 388, 2654–2664. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Teleanu, R.I.; Chircov, C.; Grumezescu, A.M.; Teleanu, D.M. Tumor Angiogenesis and Anti-Angiogenic Strategies for Cancer Treatment. J. Clin. Med. 2019, 9, 84. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, H.L.; Tsai, M.M. Tumor progression-dependent angiogenesis in gastric cancer and its potential application. World J. Gastrointest. Oncol. 2019, 11, 686–704. [Google Scholar] [CrossRef]
- Ohtsu, A.; Shah, M.A.; Van Cutsem, E.; Rha, S.Y.; Sawaki, A.; Park, S.R.; Lim, H.Y.; Yamada, Y.; Wu, J.; Langer, B.; et al. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: A randomized, double-blind, placebo-controlled phase III study. J. Clin. Oncol. 2011, 29, 3968–3976. [Google Scholar] [CrossRef]
- Wilke, H.; Muro, K.; Van Cutsem, E.; Oh, S.C.; Bodoky, G.; Shimada, Y.; Hironaka, S.; Sugimoto, N.; Lipatov, O.; Kim, T.Y.; et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): A double-blind, randomised phase 3 trial. Lancet Oncol. 2014, 15, 1224–1235. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, X.; Jia, S.; Leng, Y.; Wang, Q.; Li, Z.; Dong, B.; Guo, T.; Cheng, X.; Du, H.; Hu, Y.; et al. An integrated classifier improves prognostic accuracy in non-metastatic gastric cancer. Oncoimmunology 2020, 9, 1792038. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, X.; Zheng, L.; Zhuang, C. Gene Signatures and Prognostic Values of m6A Regulators in Hepatocellular Carcinoma. Front. Genet. 2020, 11, 540186. [Google Scholar] [CrossRef] [PubMed]
- Blank, S.; Deck, C.; Dreikhausen, L.; Weichert, W.; Giese, N.; Falk, C.; Schmidt, T.; Ott, K. Angiogenic and growth factors in gastric cancer. J. Surg. Res. 2015, 194, 420–429. [Google Scholar] [CrossRef]
- Grunnet, M.; Mau-Sorensen, M.; Brunner, N. Tissue inhibitor of metalloproteinase 1 (TIMP-1) as a biomarker in gastric cancer: A review. Scand. J. Gastroenterol. 2013, 48, 899–905. [Google Scholar] [CrossRef]
- He, L.F.; Wang, T.T.; Gao, Q.Y.; Zhao, G.F.; Huang, Y.H.; Yu, L.K.; Hou, Y.Y. Stanniocalcin-1 promotes tumor angiogenesis through up-regulation of VEGF in gastric cancer cells. J. Biomed. Sci. 2011, 18, 39. [Google Scholar] [CrossRef] [Green Version]
- Zhong, H.; Li, X.; Zhang, J.; Wu, X. Overexpression of periostin is positively associated with gastric cancer metastasis through promoting tumor metastasis and invasion. J. Cell. Biochem. 2019, 120, 9927–9935. [Google Scholar] [CrossRef]
- Zhang, W. TCGA divides gastric cancer into four molecular subtypes: Implications for individualized therapeutics. Chin. J. Cancer 2014, 33, 469–470. [Google Scholar] [CrossRef]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhou, Q.; Yu, Z.; Wu, X.; Chen, X.; Li, J.; Li, C.; Yan, M.; Zhu, Z.; Liu, B.; et al. Cancer-associated fibroblast-derived Lumican promotes gastric cancer progression via the integrin β1-FAK signaling pathway. Int. J. Cancer 2017, 141, 998–1010. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Dai, Y.; Gong, Z.; Cheng, J.N.; Zhang, L.; Sun, C.; Zeng, X.; Jia, Q.; Zhu, B. Association between angiogenesis and cytotoxic signatures in the tumor microenvironment of gastric cancer. Onco Targets Ther. 2018, 11, 2725–2733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreuzzi, E.; Capuano, A.; Poletto, E.; Pivetta, E.; Fejza, A.; Favero, A.; Doliana, R.; Cannizzaro, R.; Spessotto, P.; Mongiat, M. Role of Extracellular Matrix in Gastrointestinal Cancer-Associated Angiogenesis. Int. J. Mol. Sci. 2020, 21, 3686. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Pumiglia, K.; LaFlamme, S.E. Laminin-511 and alpha6 integrins regulate the expression of CXCR4 to promote endothelial morphogenesis. J. Cell Sci. 2020, 133, jcs246595. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, H.; Jiang, Z.; Lin, Y.; Wang, X.; Xiang, J.; Peng, J. Integrin subunit alpha V promotes growth, migration, and invasion of gastric cancer cells. Pathol. Res. Pract. 2019, 215, 152531. [Google Scholar] [CrossRef] [PubMed]
- Cheuk, I.W.; Siu, M.T.; Ho, J.C.; Chen, J.; Shin, V.Y.; Kwong, A. ITGAV targeting as a therapeutic approach for treatment of metastatic breast cancer. Am. J. Cancer Res. 2020, 10, 211–223. [Google Scholar]
- Kang, C.L.; Qi, B.; Cai, Q.Q.; Fu, L.S.; Yang, Y.; Tang, C.; Zhu, P.; Chen, Q.W.; Pan, J.; Chen, M.H.; et al. LncRNA AY promotes hepatocellular carcinoma metastasis by stimulating ITGAV transcription. Theranostics 2019, 9, 4421–4436. [Google Scholar] [CrossRef]
- Pei, Y.; Zhang, Y.; Zheng, K.; Shang, G.; Wang, Y.; Wang, W.; Qiu, E.; Li, S.; Liu, F.; Zhang, X. Ilex hainanensis Merr targets ITGAV to suppress the proliferation and metastasis of osteosarcoma cells. Onco Targets Ther. 2019, 12, 4499–4507. [Google Scholar] [CrossRef] [Green Version]
- Kulbe, H.; Otto, R.; Darb-Esfahani, S.; Lammert, H.; Abobaker, S.; Welsch, G.; Chekerov, R.; Schäfer, R.; Dragun, D.; Hummel, M.; et al. Discovery and Validation of Novel Biomarkers for Detection of Epithelial Ovarian Cancer. Cells 2019, 8, 713. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Han, C.; Liao, X.; Yu, L.; Zhu, G.; Su, H.; Qin, W.; Lu, S.; Ye, X.; Peng, T. Genetic variants in the exon region of versican predict survival of patients with resected early-stage hepatitis B virus-associated hepatocellular carcinoma. Cancer Manag. Res. 2018, 10, 1027–1036. [Google Scholar] [CrossRef] [Green Version]
- Lian, L.; Li, X.L.; Xu, M.D.; Li, X.M.; Wu, M.Y.; Zhang, Y.; Tao, M.; Li, W.; Shen, X.M.; Zhou, C.; et al. VEGFR2 promotes tumorigenesis and metastasis in a pro-angiogenic-independent way in gastric cancer. BMC Cancer 2019, 19, 183. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Yang, G.; Feng, M.; Cao, Z.; Liu, Y.; Qiu, J.; You, L.; Zheng, L.; Zhang, T.; Zhao, Y. Expression, function and clinical application of stanniocalcin-1 in cancer. J. Cell. Mol. Med. 2020, 24, 7686–7696. [Google Scholar] [CrossRef]
- Li, L.; Jiang, X.; Zhang, Q.; Dong, X.; Gao, Y.; He, Y.; Qiao, H.; Xie, F.; Xie, X.; Sun, X. Neuropilin-1 is associated with clinicopathology of gastric cancer and contributes to cell proliferation and migration as multifunctional co-receptors. J. Exp. Clin. Cancer Res. 2016, 35, 16. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Qi, Z.; Zhou, M.; Yang, W.; Hu, R.; Li, G.; Ma, X.; Zhang, Z. Stanniocalcin1 promotes cell proliferation, chemoresistance and metastasis in hypoxic gastric cancer cells via Bcl2. Oncol. Rep. 2019, 41, 1998–2008. [Google Scholar] [CrossRef] [PubMed]
- Law, A.Y.; Wong, C.K. Stanniocalcin-1 and -2 promote angiogenic sprouting in HUVECs via VEGF/VEGFR2 and angiopoietin signaling pathways. Mol. Cell. Endocrinol. 2013, 374, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Mei, B.; Chen, J.; Yang, N.; Peng, Y. The regulatory mechanism and biological significance of the Snail-miR590-VEGFR-NRP1 axis in the angiogenesis, growth and metastasis of gastric cancer. Cell Death Dis. 2020, 11, 241. [Google Scholar] [CrossRef]
- Qiu, J.; Sun, M.; Wang, Y.; Chen, B. Identification and validation of an individualized autophagy-clinical prognostic index in gastric cancer patients. Cancer Cell Int. 2020, 20, 178. [Google Scholar] [CrossRef]
- Iasonos, A.; Schrag, D.; Raj, G.V.; Panageas, K.S. How to build and interpret a nomogram for cancer prognosis. J. Clin. Oncol. 2008, 26, 1364–1370. [Google Scholar] [CrossRef]
- Song, W.; Lv, C.G.; Miao, D.L.; Zhu, Z.G.; Wu, Q.; Wang, Y.G.; Chen, L. Development and validation of a nomogram for predicting survival in patients with gastrointestinal stromal tumours. Eur. J. Surg. Oncol. 2018, 44, 1657–1665. [Google Scholar] [CrossRef]
- Song, P.; Wu, L.; Jiang, B.; Liu, Z.; Cao, K.; Guan, W. Age-specific effects on the prognosis after surgery for gastric cancer: A SEER population-based analysis. Oncotarget 2016, 7, 48614–48624. [Google Scholar] [CrossRef]
- Kwon, K.J.; Shim, K.N.; Song, E.M.; Choi, J.Y.; Kim, S.E.; Jung, H.K.; Jung, S.A. Clinicopathological characteristics and prognosis of signet ring cell carcinoma of the stomach. Gastric Cancer 2014, 17, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Nakane, Y.; Iiyama, H.; Sato, M.; Kanbara, T.; Nakai, K.; Okumura, S.; Yamamichi, K.; Hioki, K. The superiority of ratio-based lymph node staging in gastric carcinoma. Ann. Surg. Oncol. 2002, 9, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Kano, K.; Yamada, T.; Yamamoto, K.; Komori, K.; Watanabe, H.; Hara, K.; Shimoda, Y.; Maezawa, Y.; Fujikawa, H.; Aoyama, T.; et al. Association Between Lymph Node Ratio and Survival in Patients with Pathological Stage II/III Gastric Cancer. Ann. Surg. Oncol. 2020, 27, 4235–4247. [Google Scholar] [CrossRef] [PubMed]
- Wittekind, C. The development of the TNM classification of gastric cancer. Pathol. Int. 2015, 65, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Development Core Team R (2012). R: A Language and Environment for Statistical Computing; Reference Index Version 2.15.1; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]


| Variable | Overall Survival in TCGA | |||
|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||
| Hazard Ratio (HR) (95%CI) | p-Value | HR (95%CI) | p-Value | |
| Age (≥60 vs. <60 years) | 1.63 (1.10−2.42) | 0.02 | 1.62 (1.09−2.41) | 0.02 |
| Gender (male vs. female) | 1.51 (1.03−2.21) | 0.03 | 1.43 (0.95−2.04) | 0.09 |
| Stage (III+IV vs. I+II) | 1.70 (1.18−2.43) | 0.004 | 1.64 (1.14−2.35) | 0.008 |
| Risk score (high vs. low) | 1.99 (1.39−2.83) | <0.001 | 1.84 (1.29−2.63) | <0.001 |
| (a) | ||||
| Variable | Disease-Free Survival in TCGA | |||
| Univariate Analysis | Multivariate Analysis | |||
| HR (95%CI) | p-Value | HR (95%CI) | p-Value | |
| Age (≥60 vs. <60 years) | 0.97 (0.64−1.47) | 0.89 | 1.10 (0.72−1.67) | 0.67 |
| Gender (male vs. female) | 1.82 (1.14−2.91) | 0.01 | 1.76 (1.10−2.81) | 0.02 |
| Stage (III+IV vs. I+II) | 1.44 (0.96−2.17) | 0.08 | 1.37 (0.91−2.07) | 0.13 |
| Risk score (high vs. low) | 1.98 (1.32−2.98) | <0.001 | 1.81 (1.21−2.72) | 0.004 |
| (b) | ||||
| Variable | Overall Survival in ACRG | |||
| Univariate Analysis | Multivariate Analysis | |||
| HR (95%CI) | p-Value | HR (95%CI) | p-Value | |
| Age (≥60 vs. <60 years) | 1.26 (0.89−1.77) | 0.19 | 1.58 (1.11−2.24) | 0.01 |
| Gender (male vs. female) | 0.92 (0.66−1.28) | 0.61 | 0.88 (0.63−1.23) | 0.45 |
| Stage (III+IV vs. I+II) | 3.41 (2.34−4.97) | <0.001 | 3.23 (2.21−4.72) | <0.001 |
| Risk score (high vs. low) | 1.83 (1.32−2.54) | <0.001 | 1.72 (1.23−2.41) | 0.002 |
| (c) | ||||
| Variable | Disease-Free Survival in ACRG | |||
| Univariate Analysis | Multivariate Analysis | |||
| HR (95%CI) | p-Value | HR (95%CI) | p-Value | |
| Age (≥60 vs. <60 years) | 1.09 (0.76−1.55) | 0.64 | 1.33 (0.84−2.12) | 0.23 |
| Gender (male vs. female) | 0.98 (0.68−1.42) | 0.93 | 0.87 (0.54−1.40) | 0.56 |
| Stage (III+IV vs. I+II) | 4.07 (2.62−6.33) | <0.001 | 3.76 (2.41−5.85) | <0.001 |
| Risk score (high vs. low) | 1.95 (1.35−2.80) | <0.001 | 1.68 (1.15−2.44) | 0.007 |
| (d) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, H.; Zhu, J.; Yu, H.; Bazhin, A.V.; Westphalen, C.B.; Renz, B.W.; Jacob, S.N.; Lampert, C.; Werner, J.; Angele, M.K.; et al. Angiogenesis-Related Gene Expression Signatures Predicting Prognosis in Gastric Cancer Patients. Cancers 2020, 12, 3685. https://doi.org/10.3390/cancers12123685
Ren H, Zhu J, Yu H, Bazhin AV, Westphalen CB, Renz BW, Jacob SN, Lampert C, Werner J, Angele MK, et al. Angiogenesis-Related Gene Expression Signatures Predicting Prognosis in Gastric Cancer Patients. Cancers. 2020; 12(12):3685. https://doi.org/10.3390/cancers12123685
Chicago/Turabian StyleRen, Haoyu, Jiang Zhu, Haochen Yu, Alexandr V. Bazhin, Christoph Benedikt Westphalen, Bernhard W. Renz, Sven N. Jacob, Christopher Lampert, Jens Werner, Martin K. Angele, and et al. 2020. "Angiogenesis-Related Gene Expression Signatures Predicting Prognosis in Gastric Cancer Patients" Cancers 12, no. 12: 3685. https://doi.org/10.3390/cancers12123685
APA StyleRen, H., Zhu, J., Yu, H., Bazhin, A. V., Westphalen, C. B., Renz, B. W., Jacob, S. N., Lampert, C., Werner, J., Angele, M. K., & Bösch, F. (2020). Angiogenesis-Related Gene Expression Signatures Predicting Prognosis in Gastric Cancer Patients. Cancers, 12(12), 3685. https://doi.org/10.3390/cancers12123685





