Investigating Serum and Tissue Expression Identified a Cytokine/Chemokine Signature as a Highly Effective Melanoma Marker
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
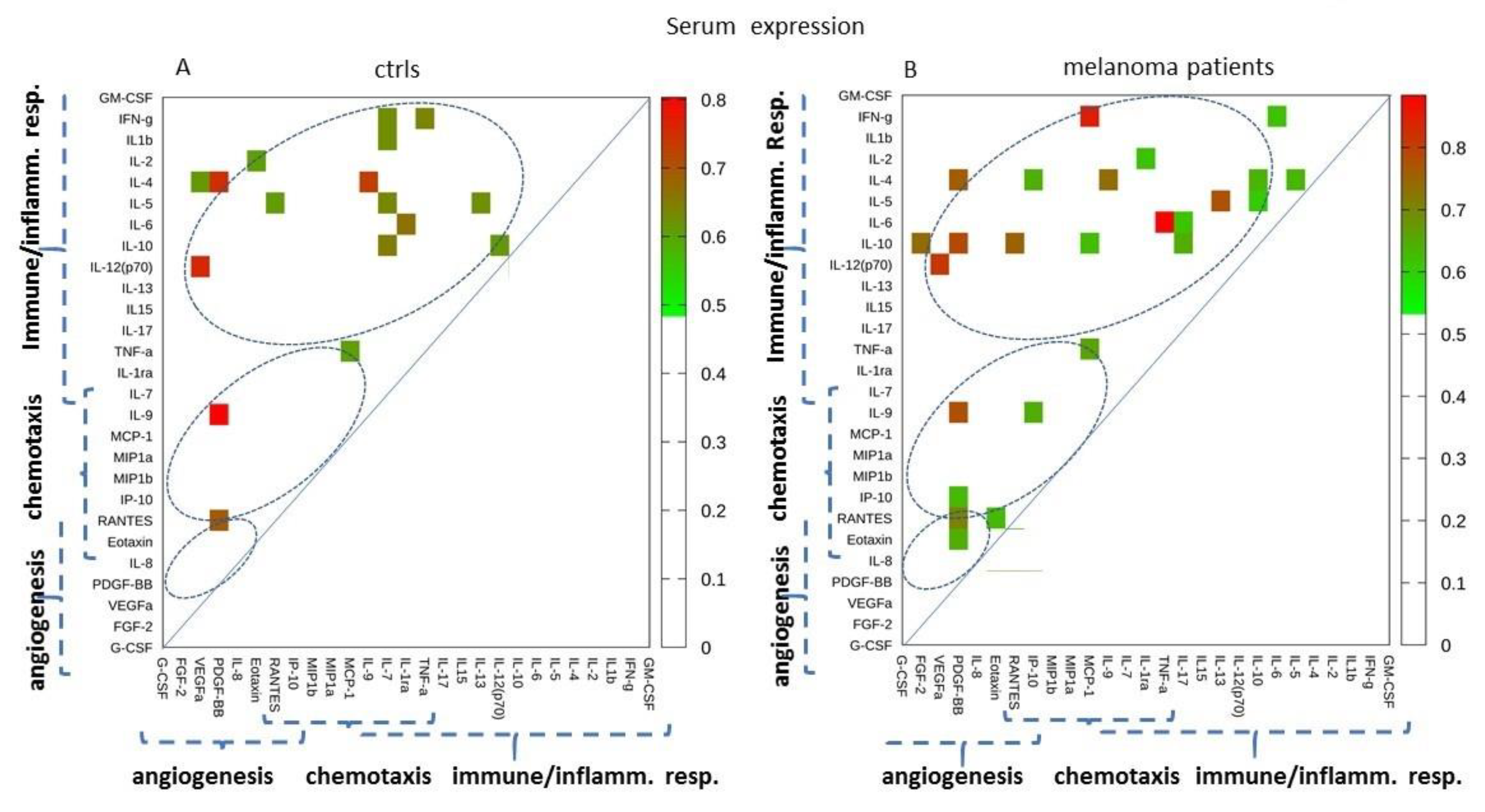
2.1. Serum Expression: Single-Molecule Analysis of the Cytokines/Chemokines in Melanoma Patients vs. Controls
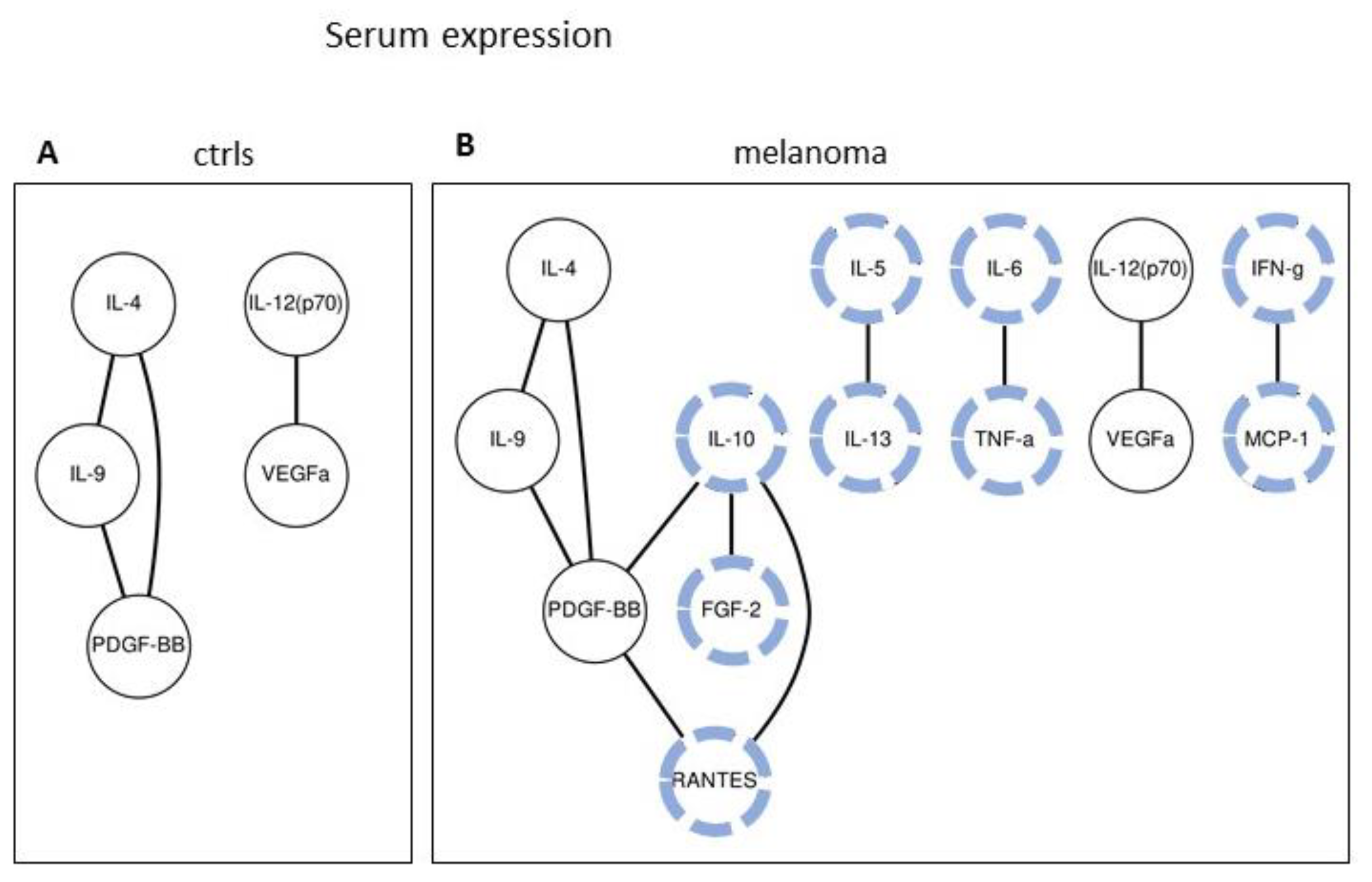
2.2. Serum Expression: Analysis of Paired Molecules by a Correlation Matrix
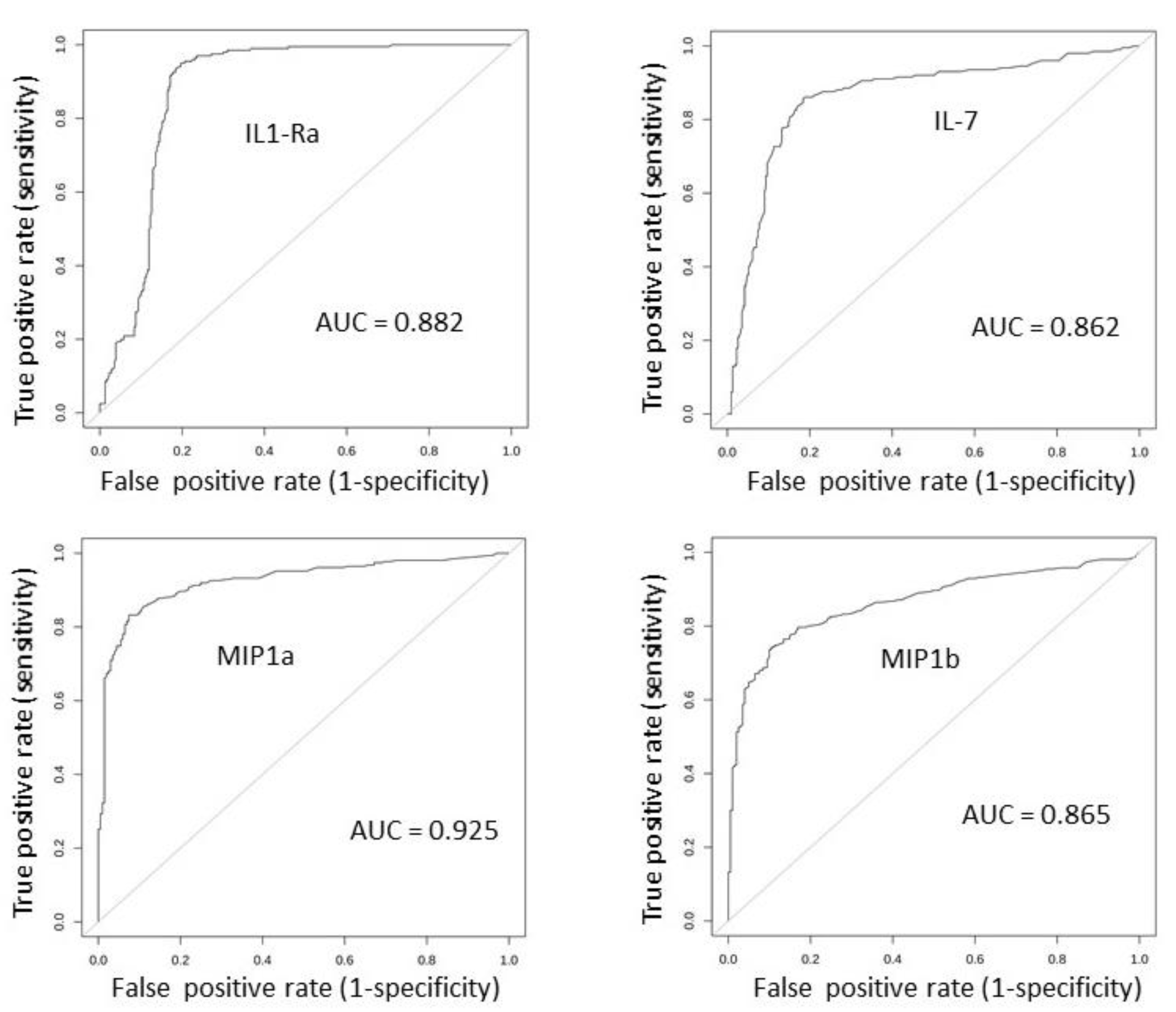
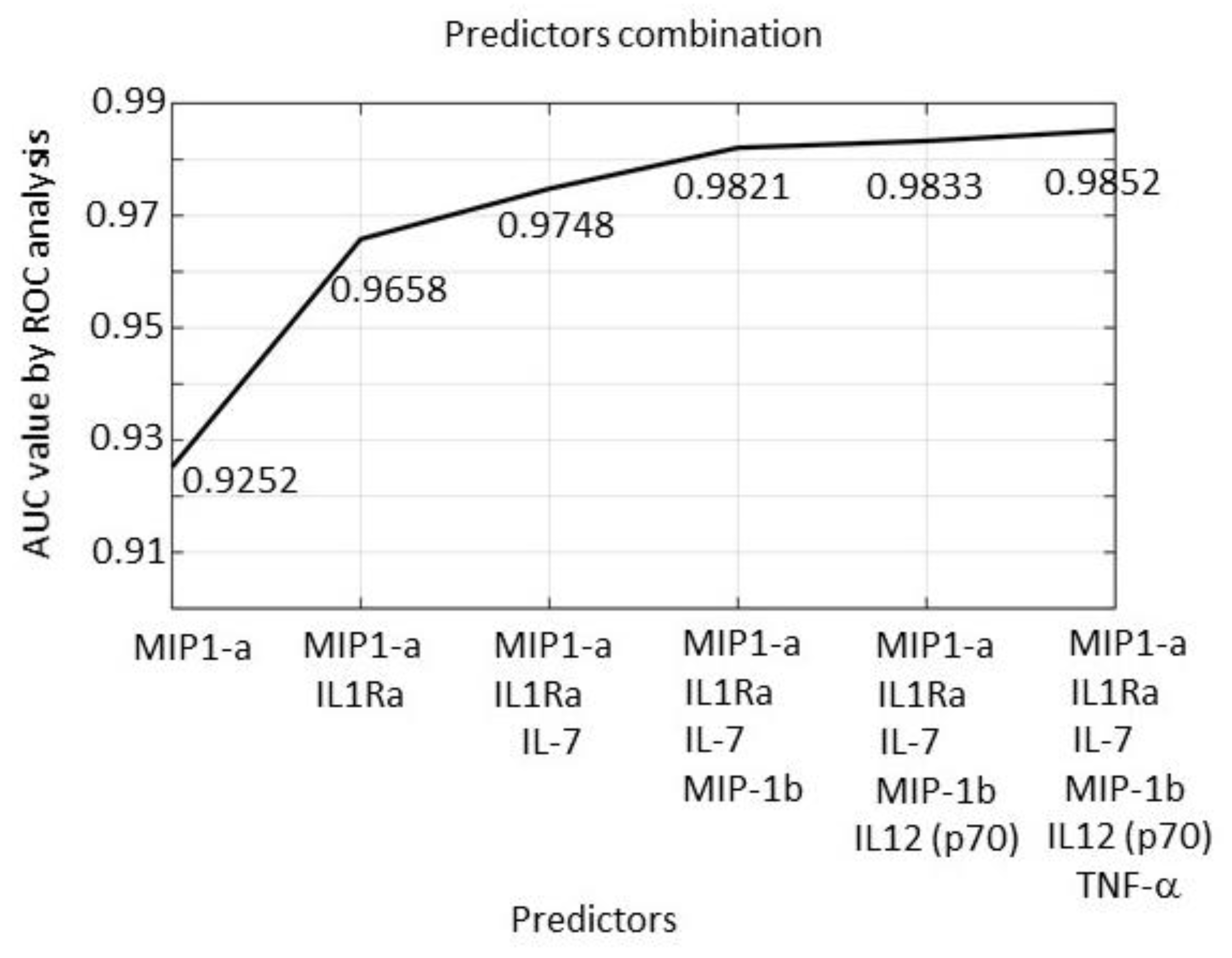
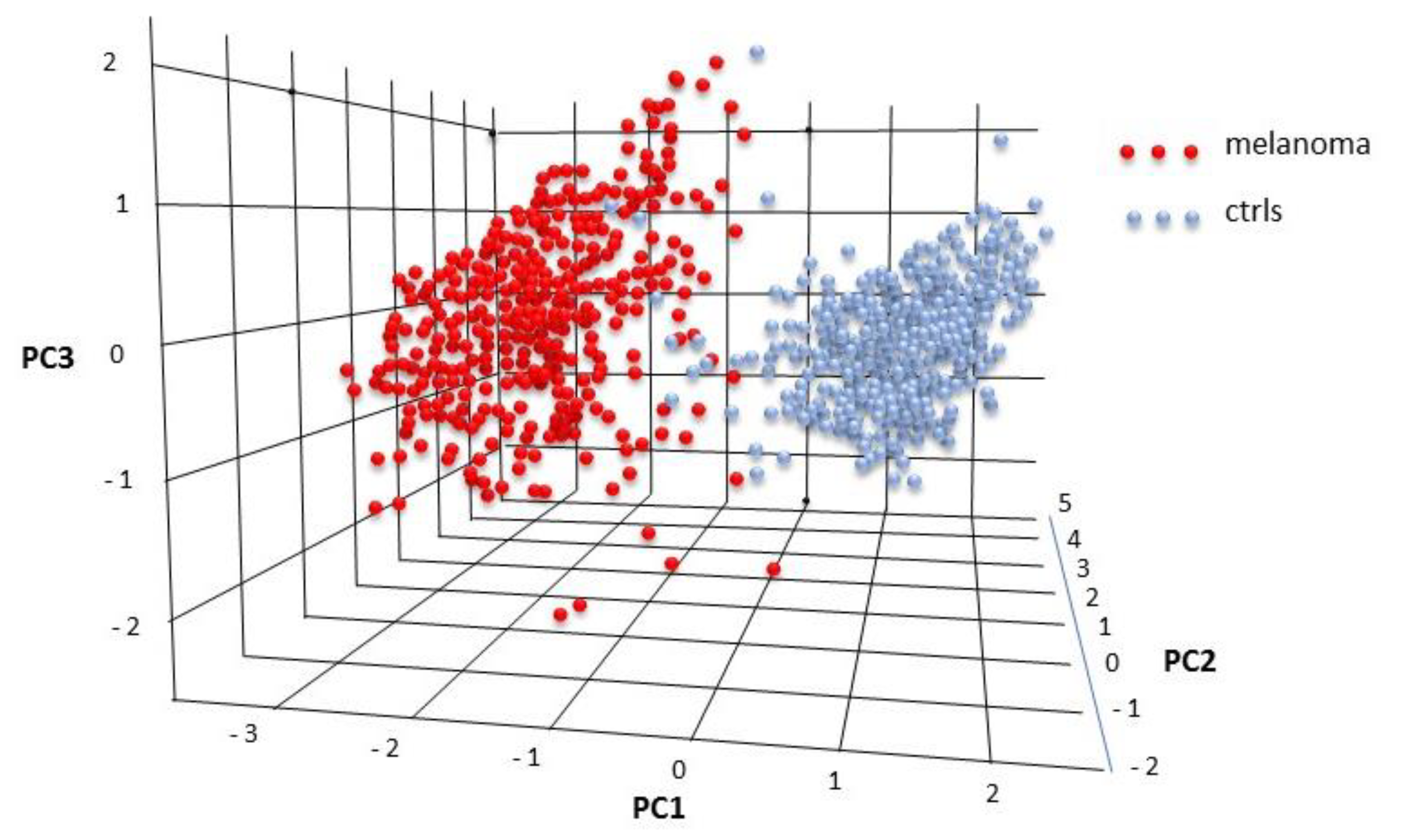
2.3. Serum Expression: Profile Analysis by SVM
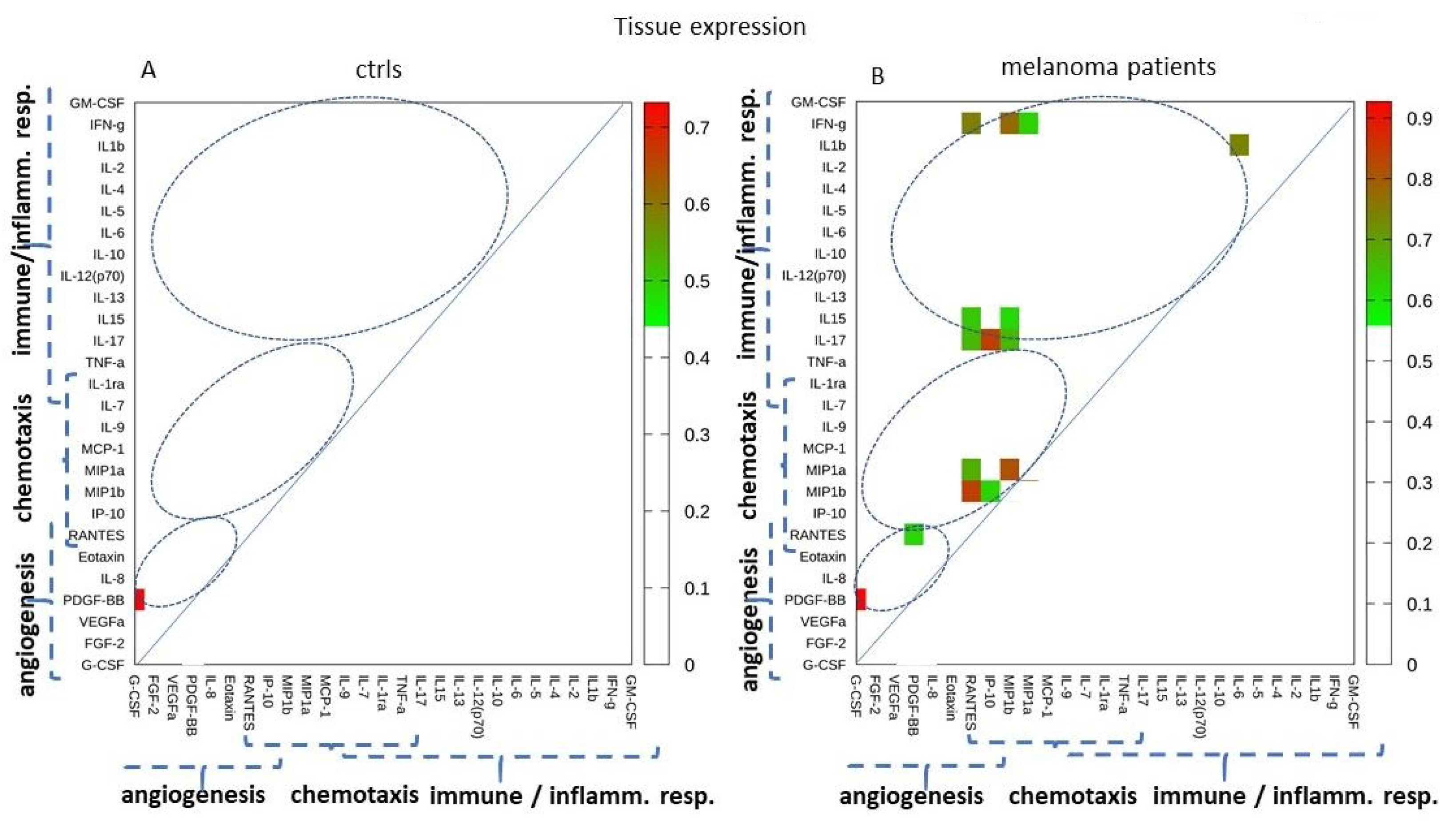
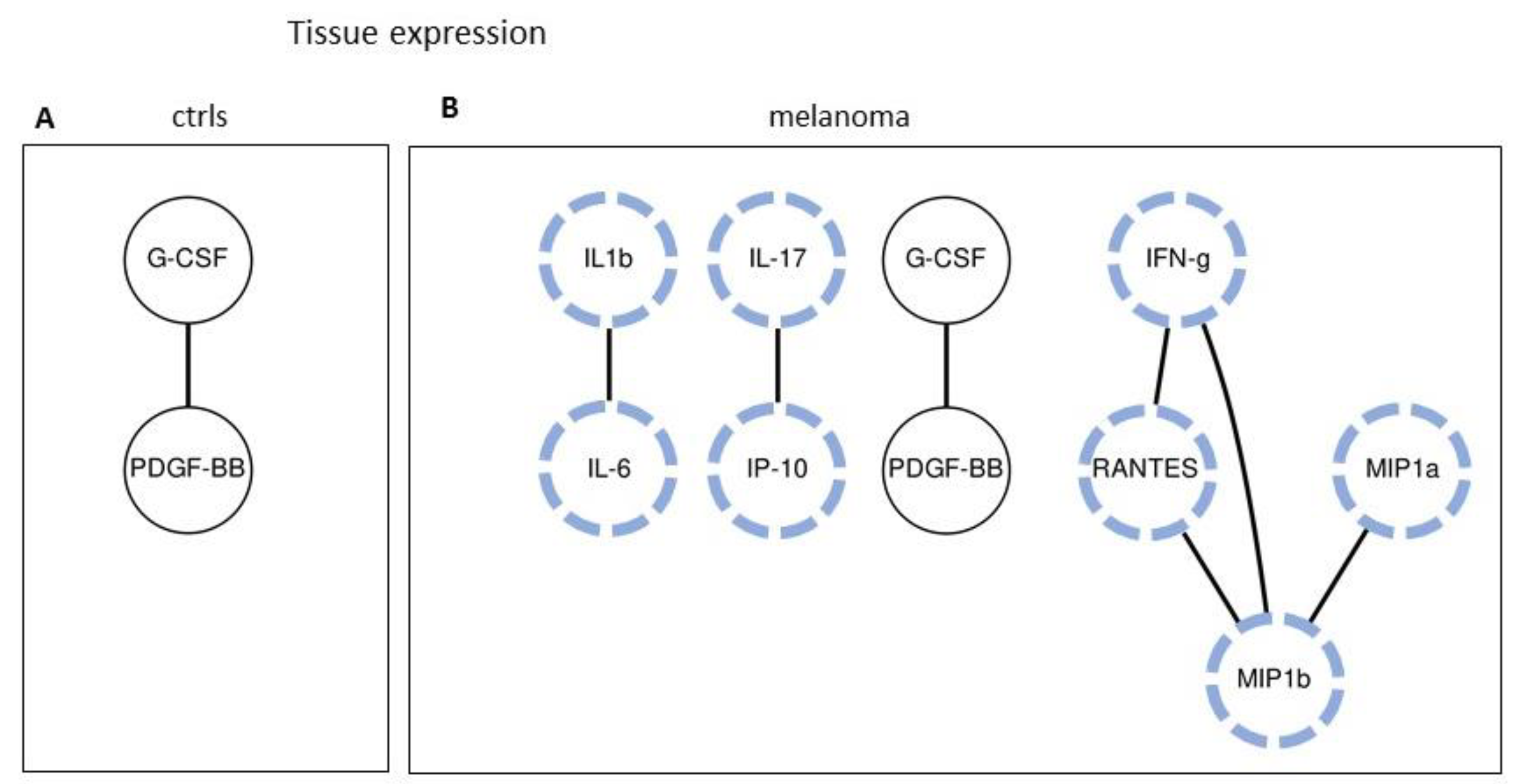
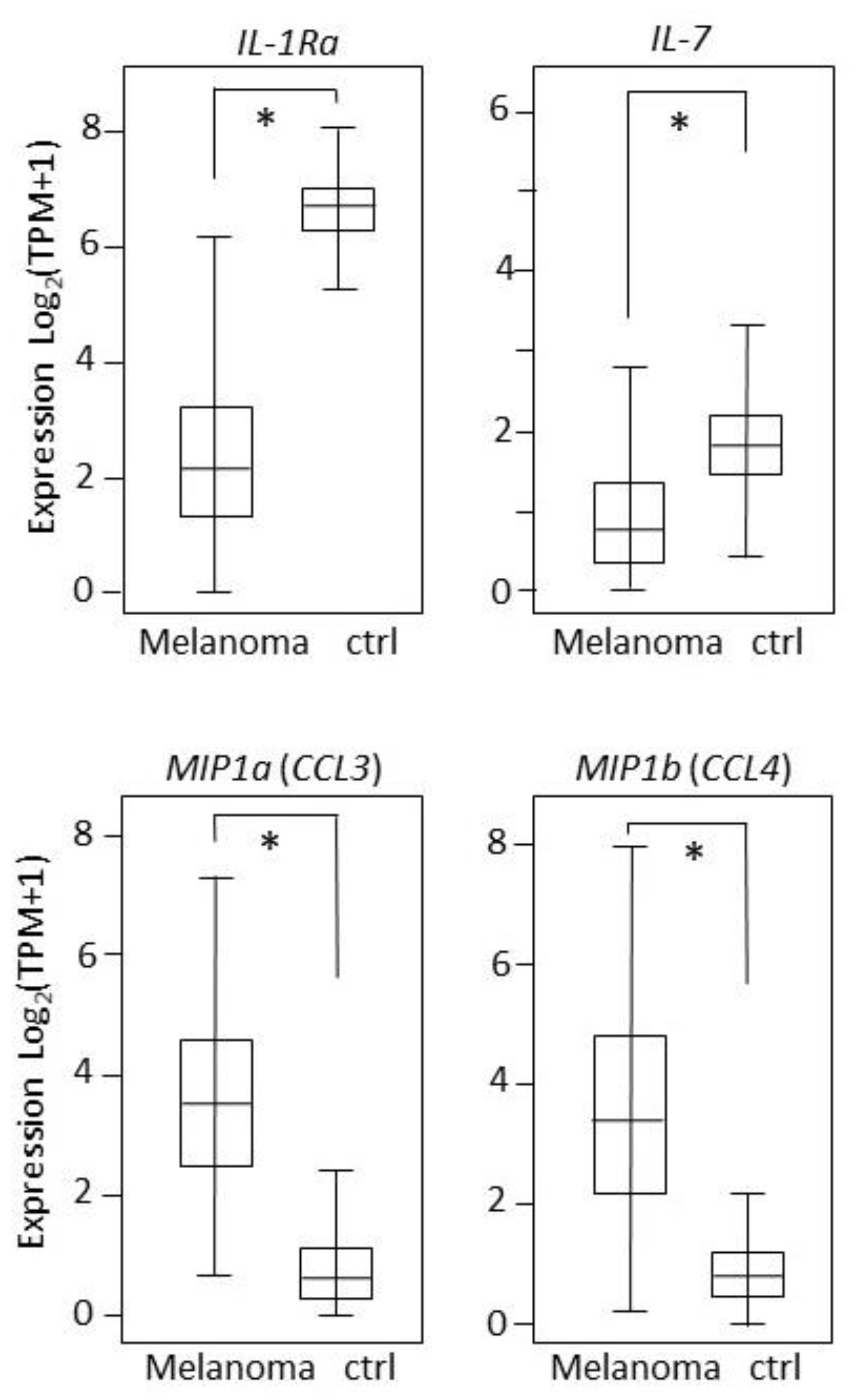
2.4. Tissues Expression: Single-Molecule Analysis of 27 Cytokines/Chemokines in Melanoma Patients and Controls
2.5. Tissue Expression: Analyzing Paired Molecules by a Matrix Correlation
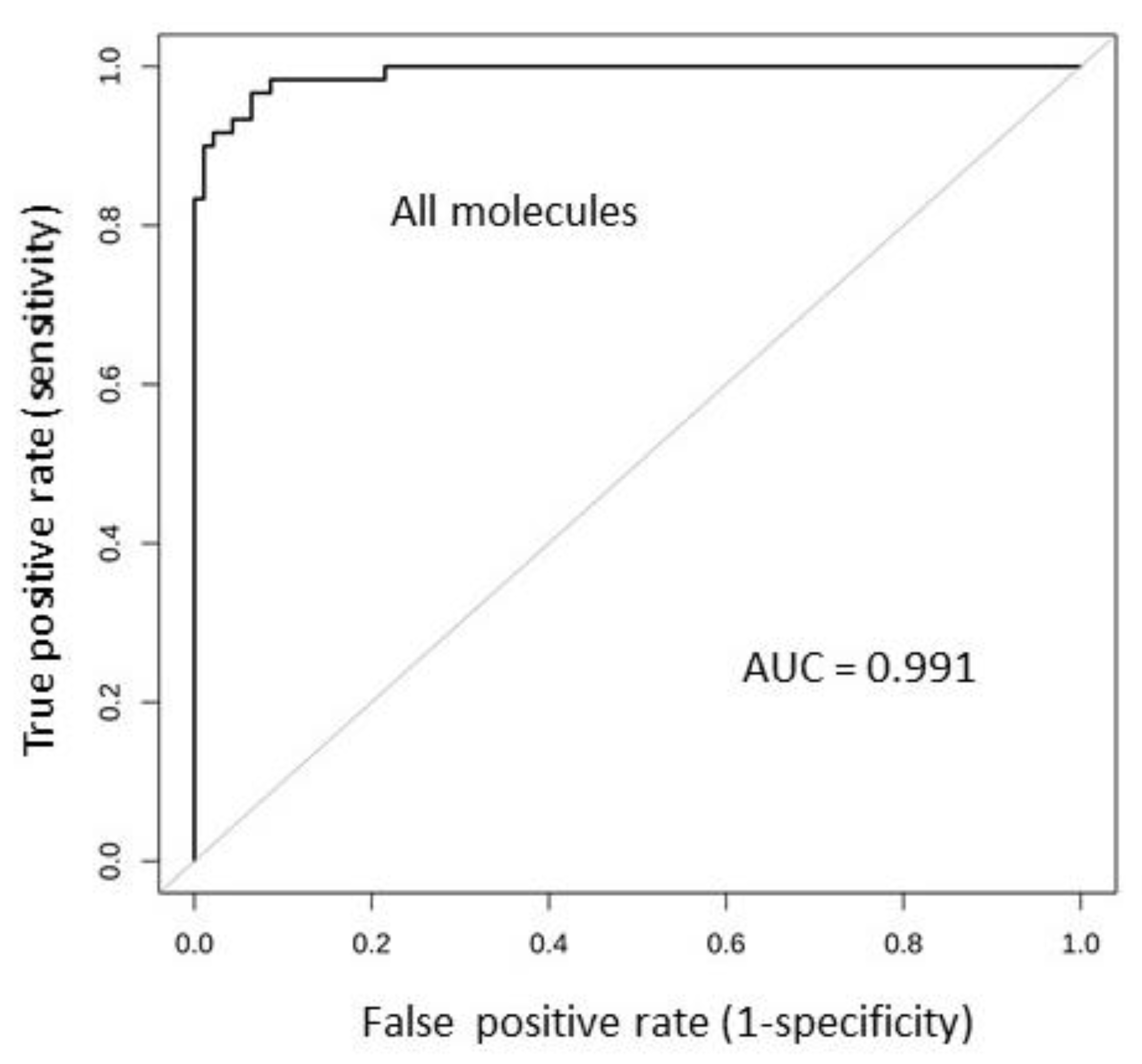
2.6. Tissue Expression: Profile Analysis by SVM
2.7. Results Validation
3. Discussion
4. Materials and Methods
4.1. Patients Selection and Recruitment
4.2. Serum Handling
4.3. Cytokines Quantification in Sera Samples
4.4. Serum Expression Data
4.5. Tissue Expression Data from GENT2 Database
4.6. Statistical Analyses
4.6.1. Single-Molecule Analysis
4.6.2. Paired-Molecule Analysis
4.6.3. Profile Analysis
4.7. Results Validation
4.7.1. Cross-Validation Procedure
4.7.2. Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Richmond, A.; Yang, J.; Su, Y. The good and the bad of chemokines/chemokine receptors in melanoma. Pigment. Cell Melanoma Res. 2009, 22, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, H.; Pourhanifeh, M.H.; Derakhshan, M.; Mahjoubin-Tehran, M.; Ghasemi, F.; Mousavi, S.; Rafiei, R.; Abbaszadeh-Goudarzi, K.; Mirzaei, H.R.; Mirzaei, H. CXCL-10: A new candidate for melanoma therapy? Cell Oncol. (Dordr) 2020, 43, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.S.; Cornelius, L.A. The Role of Chemokines in Melanoma Tumor Growth and Metastasis. J. Investig. Dermatol. 2002, 118, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Gerami, P.; Cook, R.W.; Wilkinson, J.; Russell, M.C.; Dhillon, N.; Amaria, R.N.; Gonzalez, R.; Lyle, S.; Johnson, C.E.; Oelschlager, K.M.; et al. Development of a Prognostic Genetic Signature to Predict the Metastatic Risk Associated with Cutaneous Melanoma. Clin. Cancer Res. 2015, 21, 175–183. [Google Scholar] [CrossRef]
- Qin, Y.; Verdegaal, E.M.; Siderius, M.; Bebelman, J.P.; Smit, M.J.; Leurs, R.; Willemze, R.; Tensen, C.P.; Osanto, S. Quantitative expression profiling of G-protein-coupled receptors (GPCRs) in metastatic melanoma: The constitutively active orphan GPCR GPR18 as novel drug target. Pigment. Cell Melanoma Res. 2010, 24, 207–218. [Google Scholar] [CrossRef]
- Harlin, H.; Meng, Y.; Peterson, A.C.; Zha, Y.; Tretiakova, M.; Slingluff, C.; McKee, M.; Gajewski, T.F. Chemokine Expression in Melanoma Metastases Associated with CD8+ T-Cell Recruitment. Cancer Res. 2009, 69, 3077–3085. [Google Scholar] [CrossRef]
- Kucera, R.; Topolcan, O.; Treskova, I.; Kinkorová, J.; Windrichova, J.; Fuchsova, R.; Svobodova, S.; Třeška, V.; Babuska, V.; Novák, J.; et al. Evaluation of IL-2, IL-6, IL-8 and IL-10 in Malignant Melanoma Diagnostics. Anticancer. Res. 2015, 35, 3537–3542. [Google Scholar]
- Tarhini, A.A.; Lin, Y.; Zahoor, H.; Shuai, Y.; Butterfield, L.H.; Ringquist, S.; Gogas, H.; Sander, C.; Lee, S.; Agarwala, S.S.; et al. Pro-Inflammatory Cytokines Predict Relapse-Free Survival after One Month of Interferon-α but Not Observation in Intermediate Risk Melanoma Patients. PLoS ONE 2015, 10, e0132745. [Google Scholar] [CrossRef]
- Tarhini, A.A.; Lin, Y.; Yeku, O.; LaFramboise, W.A.; Ashraf, M.; Sander, C.; Kirkwood, J.M.; Lee, S.J. A four-marker signature of TNF-RII, TGF-α, TIMP-1 and CRP is prognostic of worse survival in high-risk surgically resected melanoma. J. Transl. Med. 2014, 12, 19. [Google Scholar] [CrossRef]
- Lim, S.Y.; Lee, J.H.; Gide, T.N.; Menzies, A.M.; Guminski, A.; Carlino, M.S.; Breen, E.J.; Yang, J.Y.; Ghazanfar, S.; Kefford, R.F.; et al. Circulating Cytokines Predict Immune-Related Toxicity in Melanoma Patients Receiving Anti-PD-1–Based Immunotherapy. Clin. Cancer Res. 2019, 25, 1557–1563. [Google Scholar] [CrossRef]
- Samaniego, R.; Gutiérrez-González, A.; Gutiérrez-Seijo, A.; Sánchez-Gregorio, S.; García-Giménez, J.; Mercader, E.; Márquez-Rodas, I.; Avilés, J.A.; Relloso, M.; Sánchez-Mateos, P. CCL20 Expression by Tumor-Associated Macrophages Predicts Progression of Human Primary Cutaneous Melanoma. Cancer Immunol. Res. 2018, 6, 267–275. [Google Scholar] [CrossRef]
- Snow, H.; Hazell, S.; Francis, N.; Mohammed, K.; O’Neill, S.; Davies, E.; Mansfield, D.; Messiou, C.; Hujairi, N.; Nicol, D.; et al. Prostate-specific membrane antigen expression in melanoma metastases. J. Cutan. Pathol. 2020, 47, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Verdoliva, V.; Senatore, C.; Polci, M.L.; Rossi, S.; Cordella, M.; Carlucci, G.; Marchetti, P.; Antonini-Cappellini, G.; Facchiano, A.; D’Arcangelo, D.; et al. Differential Denaturation of Serum Proteome Reveals a Significant Amount of Hidden Information in Complex Mixtures of Proteins. PLoS ONE 2013, 8, e57104. [Google Scholar] [CrossRef] [PubMed]
- D’Arcangelo, D.; Scatozza, F.; Giampietri, C.; Marchetti, P.; Facchiano, F.; Facchiano, A. Ion Channel Expression in Human Melanoma Samples: In Silico Identification and Experimental Validation of Molecular Targets. Cancers 2019, 11, 446. [Google Scholar] [CrossRef] [PubMed]
- Biasiotta, A.; D’Arcangelo, D.; Passarelli, F.; Nicodemi, E.M.; Facchiano, A. Ion channels expression and function are strongly modified in solid tumors and vascular malformations. J. Transl. Med. 2016, 14, 285. [Google Scholar] [CrossRef] [PubMed]
- D’Arcangelo, D.; Giampietri, C.; Muscio, M.; Scatozza, F.; Facchiano, F.; Facchiano, A. WIPI1, BAG1, and PEX3 Autophagy-Related Genes Are Relevant Melanoma Markers. Oxidative Med. Cell. Longev. 2018, 2018, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Scatozza, F.; D’Arcangelo, D.; Giampietri, C.; Facchiano, F.; Facchiano, A. Melanogenesis and autophagy in melanoma. Melanoma Res. 2020, 30, 530–531. [Google Scholar] [CrossRef]
- Giampietri, C.; Tomaipitinca, L.; Scatozza, F.; Facchiano, A. Expression of genes related to lipid-handling may underlie the “obesity paradox” in melanoma: A public database-based approach. JMIR Cancer 2020, 6, e16974. [Google Scholar] [CrossRef]
- Vereecken, P.; Cornélis, F.; Van Baren, N.; Vandersleyen, V.; Baurain, J.-F. A Synopsis of Serum Biomarkers in Cutaneous Melanoma Patients. Dermatol. Res. Pr. 2012, 2012, 1–7. [Google Scholar] [CrossRef]
- Steppert, C.; Krugmann, J.; Sterlacci, W. Simultaneous endocrine expression and loss of melanoma markers in malignant melanoma metastases, a retrospective analysis. Pathol. Oncol. Res. 2019, 26, 1777–1779. [Google Scholar] [CrossRef]
- Weinstein, D.; Leininger, J.; Hamby, C.; Safai, B. Diagnostic and Prognostic Biomarkers in Melanoma. J. Clin. Aesthetic Dermatol. 2014, 7, 13–24. [Google Scholar]
- Welinder, C.; Pawłowski, K.; Sugihara, Y.; Yakovleva, M.; Jönsson, G.; Ingvar, C.; Lundgren, L.; Baldetorp, B.; Olsson, H.; Rezeli, M.; et al. A Protein Deep Sequencing Evaluation of Metastatic Melanoma Tissues. PLoS ONE 2015, 10, e0123661. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, D.; Aung, P.P.; Jour, G. The “OMICS” facet of melanoma: Heterogeneity of genomic, proteomic and metabolomic biomarkes. Semin. Cancer Biol. 2019, 59, 165–174. [Google Scholar] [CrossRef]
- Faraone, D.; Aguzzi, M.S.; Toietta, G.; Facchiano, A.M.; Facchiano, F.; Magenta, A.; Martelli, F.; Truffa, S.; Cesareo, E.; Ribatti, D.; et al. Platelet Derived Growth Factor- Receptor alpha strongly inhibits melanoma growth in vitro and in vivo. Neoplasia 2009, 11, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Aguzzi, M.S.; Faraone, D.; De Marchis, F.; Toietta, G.; Ribatti, D.; Parazzoli, A.; Colombo, P.; Capogrossi, M.C.; Facchiano, A. The FGF-2 Derived Peptide FREG Inhibits Melanoma Growth In Vitro And In Vivo”. Mol. Ther. 2011, 19, 266–273. [Google Scholar]
- D’Arcangelo, D.; Facchiano, F.; Nassa, G.; Stancato, A.; Antonini, A.; Rossi, S.; Senatore, C.; Cordella, M.; Tabolacci, C.; Salvati, A.; et al. PDGFR-alpha inhibits melanoma growth via CXCL10/IP-10: A multi-omics approach. Oncotarget 2016, 7, 77257–77275. [Google Scholar] [CrossRef]
- Rossi, S.; Cordella, M.; Tabolacci, C.; Nassa, G.; D’Arcangelo, D.; Senatore, C.; Pagnotto, P.; Magliozzi, R.; Salvati, A.; Weisz, A.; et al. TNF-alpha and metalloproteases as key players in melanoma cells aggressiveness. J. Exp. Clin. Cancer Res. 2018, 37, 1–17. [Google Scholar] [CrossRef]
- Behbahani, S.; Bs, S.M.; Ms, J.B.C.; Lambert, W.C.; Schwartz, R.A. Gender differences in cutaneous melanoma: Demographics, prognostic factors, and survival outcomes. Dermatol. Ther. 2020, 5, e14131. [Google Scholar] [CrossRef]
- U.S. Cancer Statistics Data Brief. Melanoma Incidence and Mortality, United States-2012–2016; U.S. Cancer Statistics Data Brief: Atlanta, GA, USA, 2019; Volume 9.
- Joosse, A.; De Vries, E.; Eckel, R.; Nijsten, T.; Eggermont, A.M.; Hölzel, D.; Coebergh, J.W.W.; Engel, J. Gender Differences in Melanoma Survival: Female Patients Have a Decreased Risk of Metastasis. J. Investig. Dermatol. 2011, 131, 719–726. [Google Scholar] [CrossRef]
- El Sharouni, M.-A.; Witkamp, A.J.; Sigurdsson, V.; Van Diest, P.J.; Louwman, M.W.J.; Kukutsch, N. Sex matters: Men with melanoma have a worse prognosis than women. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 2062–2067. [Google Scholar] [CrossRef]
- Enninga, E.A.L.; Moser, J.C.; Weaver, A.L.; Markovic, S.N.; Brewer, J.D.; Leontovich, A.A.; Hieken, T.J.; Shuster, L.; Kottschade, L.A.; Olariu, A.; et al. Survival of cutaneous melanoma based on sex, age, and stage in the United States, 1992–2011. Cancer Med. 2017, 6, 2203–2212. [Google Scholar] [CrossRef] [PubMed]
- Bellenghi, M.; Puglisi, R.; Pontecorvi, G.; De Feo, A.; Carè, A.; Mattia, G. Sex and Gender Disparities in Melanoma. Cancers 2020, 12, 1819. [Google Scholar] [CrossRef] [PubMed]
- Kučera, J.; Strnadová, K.; Dvořánková, B.; Lacina, L.; Krajsová, I.; Štork, J.; Kovářová, H.; Skalníková, H.K.; Vodička, P.; Motlík, J.; et al. Serum proteomic analysis of melanoma patients with immunohistochemical profiling of primary melanomas and cultured cells: Pilot study. Oncol. Rep. 2019, 42, 1793–1804. [Google Scholar] [CrossRef] [PubMed]
- Paganelli, A.; Garbarino, F.; Toto, P.; Di Martino, G.; D’Urbano, M.; Auriemma, M.; Di Giovanni, P.; Panarese, F.; Staniscia, T.; Amerio, P.; et al. Serological landscape of cytokines in cutaneous melanoma. Cancer Biomark. 2019, 26, 333–342. [Google Scholar] [CrossRef]
- Nagarkatti-Gude, N.; Bronkhorst, I.H.G.; Van Duinen, S.G.; Luyten, G.P.M.; Jager, M.J. Cytokines and Chemokines in the Vitreous Fluid of Eyes with Uveal Melanoma. Investig. Opthalmology Vis. Sci. 2012, 53, 6748–6755. [Google Scholar] [CrossRef]
- Reinert, C.P.; Gatidis, S.; Sekler, J.; Dittmann, H.; Pfannenberg, C.; la Fougere, C.; Nikolaou, K.; Forschner, A. Clinical and prognostic value of tumor volumetric parameters in melanoma patients undergoing 18 F-FDG-PET/CT: A comparison with serologic markers of tumor burden and inflammation. Cancer Imaging 2020, 20, 44. [Google Scholar] [CrossRef]
- Nomiyama, H.; Osada, N.; Yoshie, O. Systematic classification of vertebrate chemokines based on conserved synteny and evolutionary history. Genes Cells 2013, 18, 1–16. [Google Scholar] [CrossRef]
- De Marchis, F.; Ribatti, D.; Giampietri, C.; Lentini, A.; Faraone, D.; Scoccianti, M.; Capogrossi, M.C.; Facchiano, A. Platelet-Derived Growth Factor Inhibits Basic Fibroblast Growth Factor Angiogenic Properties in vitro and in vivo, via its alpha receptor. Blood 2002, 99, 2045–2053. [Google Scholar] [CrossRef]
- Faraone, D.; Aguzzi, M.S.; Ragone, G.; Russo, K.; Capogrossi, M.C.; Facchiano, A. Heterodimerization of FGF-receptor 1 and PDGF-receptor-α: A novel mechanism underlying the inhibitory effect of PDGF-BB on FGF-2 in human cells. Blood 2006, 107, 1896–1902. [Google Scholar] [CrossRef][Green Version]
- Zhang, J.M.; An, J. Cytokines, Inflammation and Pain. Int. Anesth. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nat. Cell Biol. 2015, 523, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, C.; Du, J.-X.; Zhao, J.; Shi, M.-T.; Jin, M.-W.; Liu, H. Adipocytes promote tumor progression and induce PD-L1 expression via TNF-α/IL-6 signaling. Cancer Cell Int. 2020, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Seeja, R.D.; Suresh, A. Deep Learning Based Skin Lesion Segmentation and Classification of Melanoma Using Support Vector Machine (SVM). Asian Pac. J. Cancer Prev. 2019, 20, 1555–1561. [Google Scholar] [CrossRef]
- Tiwari, K.A.; Raišutis, R.; Liutkus, J.; Valiukevičienė, S. Diagnostics of Melanocytic Skin Tumours by a Combination of Ultrasonic, Dermatoscopic and Spectrophotometric Image Parameters. Diagnostics 2020, 10, 632. [Google Scholar] [CrossRef]
- Shofty, B.; Artzi, M.; Shtrozberg, S.; Fanizzi, C.; DiMeco, F.; Haim, O.; Hason, S.P.; Ram, Z.; Ben Bashat, D.; Grossman, R. Virtual biopsy using MRI radiomics for prediction of BRAF status in melanoma brain metastasis. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef]
- Szyc, Ł.; Hillen, U.; Scharlach, C.; Kauer, F.; Garbe, C. Diagnostic Performance of a Support Vector Machine for Dermatofluoroscopic Melanoma Recognition: The Results of the Retrospective Clinical Study on 214 Pigmented Skin Lesions. Diagnostics 2019, 9, 103. [Google Scholar] [CrossRef]
- Mancuso, F.; Lage, S.; Rasero, J.; Díaz-Ramón, J.L.; Apraiz, A.; Pérez-Yarza, G.; Ezkurra, P.A.; Penas, C.; Sánchez-Diez, A.; García-Vazquez, M.D.; et al. Serum markers improve current prediction of metastasis development in early-stage melanoma patients: A machine learning-based study. Mol. Oncol. 2020, 14, 1705–1718. [Google Scholar] [CrossRef]
- Yurkovetsky, Z.R.; Kirkwood, J.M.; Edington, H.D.; Marrangoni, A.M.; Velikokhatnaya, L.; Winans, M.T.; Gorelik, E.; Lokshin, A.E. Multiplex Analysis of Serum Cytokines in Melanoma Patients Treated with Interferon-2b. Clin. Cancer Res. 2007, 13, 2422–2428. [Google Scholar] [CrossRef]
- Byrum, S.D. Quantitative Proteomics Identifies Activation of Hallmark Pathways of Cancer in Patient Melanoma. J. Proteom. Bioinform. 2013, 6, 043–050. [Google Scholar] [CrossRef]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Allen, F.; Bobanga, I.D.; Rauhe, P.; Barkauskas, D.S.; Teich, N.; Tong, C.; Myers, J.; Huang, A.Y. CCL3 augments tumor rejection and enhances CD8+ T cell infiltration through NK and CD103+ dendritic cell recruitment via IFNγ. OncoImmunology 2018, 7, e1393598. [Google Scholar] [CrossRef] [PubMed]
- R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing. R Core Team. 2020. Available online: http://www.r-project.org (accessed on 4 December 2020).
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A Training Algorithm for Optimal Margin Classifiers; ACM: New York, NY, USA, 1992; pp. 144–152. [Google Scholar]
- Cortes, C.; Vapnik, V.N. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Little, R.; Rubin, D. Statistical Analysis with Missing Data, 2nd ed.; Wiley-Interscience: Hoboken, NJ, USA, 2002. [Google Scholar]
- Hsu, C.W.; Chang, C.C.; Lin, C.J. A Practical Guide to Support Vector Classification; Bioinformatics 1; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Luor, D.-C. A comparative assessment of data standardization on support vector machine for classification problems. Intell. Data Anal. 2015, 19, 529–546. [Google Scholar] [CrossRef]

| Patient Type | Number | Mean Age | Mean Thickness (mm) | Thickness Distribution | |
|---|---|---|---|---|---|
| <1 mm * Number | ≥1 mm * Number | ||||
| Female controls | 72 | 41.3 | 0.00 | 0 | 0 |
| Male controls | 64 | 45.3 | 0.00 | 0 | 0 |
| Female melanoma | 48 | 54.5 | 1.60 | 23 | 22 |
| Male melanoma | 48 | 58.0 | 1.08 | 31 | 14 |
| Total | 232 | ||||
| Cytokines | Controls Median (n = 136) | Melanoma Median (n = 96) | p-Value Mann–Whitney Controls vs. Melanoma | AUC ± S.E. by ROC Analysis |
|---|---|---|---|---|
| IL-1b | 0.53 | 0.65 | 0.04 | 0.61 ± 0.05 |
| IL-1Ra | 26.77 | 17.83 | 0.14 | 0.56 ± 0.04 |
| IL-2 | 3.45 | 2.14 | 0.14 | 0.66 ± 0.10 |
| IL-4 | 2.95 | 2.88 | 0.47 | 0.53 ± 0.04 |
| IL-5 | 2.77 | 2.34 | 0.22 | 0.60 ± 0.08 |
| IL-6 | 5.37 | 3.17 | 0.04 | 0.70 ± 0.09 |
| IL-7 | 2.24 | 2.24 | 0.72 | 0.52 ± 0.05 |
| IL-8 | 6.63 | 6.4 | 0.55 | 0.52 ± 0.04 |
| IL-9 | 45.58 | 42.05 | 0.29 | 0.54 ± 0.04 |
| IL-10 | 7.08 | 4.56 | 0.10 | 0.63 ± 0.08 |
| IL-12(p70) | 15.74 | 16.07 | 1.00 | 0.50 ± 0.05 |
| IL-13 | 2.43 | 2.99 | 0.83 | 0.52 ± 0.09 |
| IL-15 | 52.22 | 30.11 | 1.00 | 0.50 ± 0.27 |
| IL-17 | 16.72 | 13.1 | 0.37 | 0.54 ± 0.04 |
| Eotaxin | 95.28 | 106.28 | 0.38 | 0.53 ± 0.04 |
| FGF-2 | 32.68 | 30.01 | 0.26 | 0.55 ± 0.04 |
| G-CSF | 4.73 | 5.41 | 0.33 | 0.55 ± 0.05 |
| GM-CSF | 10.21 | 10.63 | 0.67 | 0.53 ± 0.06 |
| IFN-γ | 19.1 | 23.2 | 0.48 | 0.53 ± 0.04 |
| IP-10 (CXCL10) | 438.69 | 501.41 | 0.04 | 0.58 ± 0.04 |
| MCP-1(MCAF) | 18.59 | 12.49 | 0.24 | 0.58 ± 0.07 |
| MIP-1a (CCL3) | 1.78 | 1.74 | 0.59 | 0.52 ± 0.04 |
| MIP-1b (CCL4) | 54.36 | 56.26 | 0.43 | 0.53 ± 0.04 |
| PDGF-BB | 1603.74 | 1033.41 | 0.01 | 0.61 ± 0.05 |
| RANTES (CCL5) | 11,353.34 | 8735.27 | 0.01 | 0.57 ± 0.06 |
| TNF-α | 16.4 | 18.06 | 0.26 | 0.52 ± 0.06 |
| VEGF | 59.75 | 57.98 | 0.88 | 0.54 ± 0.06 |
| Cytokines | Melanoma Breslow Thickness <1 mm | Melanoma Breslow Thickness ≥1 mm | Mann-Whitney <1 mm vs. ≥1 mm | ||
|---|---|---|---|---|---|
| Count | Median | Count | Median | p Value | |
| IL-1b | 26 | 0.65 | 16 | 0.72 | 0.66 |
| IL-1Ra | 44 | 18.24 | 27 | 17.83 | 0.59 |
| IL-2 | 5 | 1.9 | 7 | 2.38 | 0.25 |
| IL-4 | 54 | 2.85 | 35 | 3.06 | 0.81 |
| IL-5 | 15 | 2.34 | 9 | 1.7 | 0.86 |
| IL-6 | 7 | 0.83 | 8 | 3.34 | 0.18 |
| IL-7 | 31 | 2.24 | 15 | 2.9 | 0.34 |
| IL-8 | 47 | 7.8 | 30 | 5.78 | 0.01 |
| IL-9 | 52 | 42.55 | 35 | 41.91 | 0.64 |
| IL-10 | 11 | 3.84 | 10 | 5.12 | 0.92 |
| IL-12(p70) | 40 | 20.48 | 20 | 14.64 | 0.51 |
| IL-13 | 11 | 2.43 | 8 | 3.2 | 0.60 |
| IL-15 | 2 | 13.02 | 1 | 42.31 | 0.67 |
| IL-17 | 38 | 13.15 | 30 | 11.79 | 0.94 |
| Eotaxin | 54 | 108.94 | 35 | 93.24 | 0.71 |
| FGF-2 | 49 | 29.73 | 34 | 31.46 | 0.77 |
| G-CSF | 27 | 5.45 | 10 | 4.12 | 0.30 |
| GM-CSF | 17 | 14.06 | 25 | 9.78 | 0.22 |
| IFN-γ | 51 | 23.49 | 31 | 22.13 | 0.51 |
| IP-10 (CXCL10) | 53 | 491.81 | 35 | 574.3 | 0.88 |
| MCP-1(MCAF) | 18 | 10.56 | 11 | 24.83 | 0.02 |
| MIP-1a (CCL3) | 54 | 1.82 | 35 | 1.71 | 0.56 |
| MIP-1b (CCL4) | 53 | 55.51 | 35 | 53.52 | 0.73 |
| PDGF-BB | 53 | 1048.16 | 35 | 1080.98 | 0.47 |
| RANTES (CCL5) | 53 | 10,341.43 | 35 | 7534.18 | 0.03 |
| TNF-α | 43 | 16.65 | 19 | 22.78 | 0.06 |
| VEGF | 51 | 63.58 | 35 | 52.26 | 0.45 |
| Cytokines | No. of Pairs | Spearman R Correlation of Serum Expression with Breslow Thickness | p-Value (2-Tails) |
|---|---|---|---|
| IL-1b | 42 | 0.04 | 0.80 |
| IL-1Ra | 71 | 0.04 | 0.75 |
| IL-2 | 12 | 0.01 | 0.97 |
| IL-4 | 89 | −0.02 | 0.88 |
| IL-5 | 24 | 0.01 | 0.96 |
| IL-6 | 15 | 0.24 | 0.40 |
| IL-7 | 46 | 0.28 | 0.06 |
| IL-8 | 77 | −0.23 | 0.05 |
| IL-9 | 87 | −0.09 | 0.42 |
| IL-10 | 21 | −0.14 | 0.55 |
| IL-12(p70) | 60 | −0.02 | 0.86 |
| IL-13 | 19 | −0.09 | 0.72 |
| IL-15 | 3 | 1.00 | 0.33 |
| IL-17 | 68 | 0.05 | 0.67 |
| Eotaxin | 89 | −0.01 | 0.96 |
| FGF-2 | 83 | 0.03 | 0.81 |
| G-CSF | 37 | 0.04 | 0.83 |
| GM-CSF | 42 | −0.40 | 0.01 |
| IFN-γ | 82 | 0.07 | 0.53 |
| IP-10 (CXCL10) | 88 | 0.04 | 0.72 |
| MCP-1(MCAF) | 29 | 0.39 | 0.04 |
| MIP-1a (CCL3) | 89 | −0.09 | 0.40 |
| MIP-1b (CCL4) | 88 | −0.02 | 0.86 |
| PDGF-BB | 88 | −0.10 | 0.35 |
| RANTES (CCL5) | 88 | −0.20 | 0.06 |
| TNF-α | 62 | 0.31 | 0.01 |
| VEGF | 86 | −0.02 | 0.87 |
| Cytokines | All Controls (Male + Female) | All Melanoma (Male + Female) | ||||
|---|---|---|---|---|---|---|
| No. of Pairs | Spearman R Correlation of Serum Expression with Age | p Value (2 Tails) | No. of Pairs | Spearman R Correlation of Serum Expression with Age | p-Value (2 Tails) | |
| IL-1b | 85 | 0.14 | 0.21 | 44 | −0.13 | 0.40 |
| IL-1Ra | 121 | 0.15 | 0.10 | 75 | −0.08 | 0.50 |
| IL-2 | 19 | −0.21 | 0.38 | 13 | 0.23 | 0.46 |
| IL-4 | 135 | 0.08 | 0.36 | 95 | −0.05 | 0.66 |
| IL-5 | 30 | 0.34 | 0.07 | 25 | −0.20 | 0.33 |
| IL-6 | 21 | 0.00 | 0.99 | 16 | −0.01 | 0.96 |
| IL-7 | 81 | 0.35 | 0.001 | 48 | −0.09 | 0.53 |
| IL-8 | 128 | 0.12 | 0.17 | 83 | −0.20 | 0.07 |
| IL-9 | 135 | 0.12 | 0.17 | 93 | 0.05 | 0.61 |
| IL-10 | 39 | 0.05 | 0.77 | 22 | −0.09 | 0.69 |
| IL-12(p70) | 110 | 0.30 | 0.002 | 62 | −0.08 | 0.55 |
| IL-13 | 27 | 0.39 | 0.04 | 19 | 0.21 | 0.38 |
| IL-15 | 2 | - | - | 4 | - | - |
| IL-17 | 108 | 0.12 | 0.23 | 73 | −0.01 | 0.93 |
| Eotaxin | 132 | 0.13 | 0.13 | 95 | −0.01 | 0.97 |
| FGF-2 | 129 | 0.02 | 0.85 | 88 | 0.01 | 0.94 |
| G-CSF | 92 | −0.03 | 0.76 | 40 | −0.38 | 0.02 |
| GM-CSF | 48 | −0.05 | 0.72 | 46 | −0.10 | 0.53 |
| IFN-γ | 136 | 0.09 | 0.31 | 86 | −0.12 | 0.26 |
| IP-10 (CXCL10) | 136 | 0.22 | 0.01 | 94 | 0.20 | 0.05 |
| MCP-1(MCAF) | 47 | 0.11 | 0.45 | 30 | −0.17 | 0.38 |
| MIP-1a (CCL3) | 133 | 0.23 | 0.01 | 95 | −0.16 | 0.13 |
| MIP-1b (CCL4) | 136 | 0.30 | 0.0005 | 94 | −0.15 | 0.15 |
| PDGF-BB | 135 | 0.02 | 0.86 | 94 | −0.11 | 0.29 |
| RANTES (CCL5) | 136 | −0.02 | 0.84 | 94 | −0.08 | 0.42 |
| TNF-α | 120 | 0.17 | 0.06 | 65 | −0.14 | 0.28 |
| VEGF | 135 | 0.17 | 0.05 | 92 | 0.09 | 0.41 |
| Cytokines | Male Melanoma | Female Melanoma | Mann–Whitney |
|---|---|---|---|
| Median Value | Median Value | p-Value | |
| IL-1b | 0.63 | 0.785 | 0.07 |
| IL-1Ra | 15.17 | 24.125 | 0.25 |
| IL-2 | 1.9 | 3.09 | 0.14 |
| IL-4 | 3.05 | 2.75 | 0.06 |
| IL-5 | 2.34 | 2.02 | 0.36 |
| IL-6 | 3.34 | 3.0 | 0.73 |
| IL-7 | 2.24 | 2.31 | 0.49 |
| IL-8 | 6.57 | 6.32 | 0.64 |
| IL-9 | 44.35 | 38.84 | 0.14 |
| IL-10 | 3.84 | 6.31 | 0.13 |
| IL-12(p70) | 16.94 | 14.57 | 0.98 |
| IL-13 | 2.19 | 3.23 | 0.60 |
| IL-15 | - | 30.11 | - |
| IL-17 | 13.1 | 12.96 | 0.67 |
| Eotaxin | 135.15 | 91.8 | 0.002 |
| FGF-2 | 32.55 | 29.73 | 0.25 |
| G-CSF | 5.09 | 5.45 | 0.60 |
| GM-CSF | 11.71 | 9.97 | 0.47 |
| IFN-γ | 20.83 | 27.33 | 0.26 |
| IP-10 (CXCL10) | 567.19 | 468.93 | 0.13 |
| MCP-1(MCAF) | 10.995 | 25.6 | 0.05 |
| MIP-1a (CCL3) | 1.86 | 1.7 | 0.49 |
| MIP-1b (CCL4) | 56.89 | 51.09 | 0.30 |
| PDGF-BB | 1145.64 | 900.76 | 0.06 |
| RANTES (CCL5) | 9536.67 | 7879.5 | 0.19 |
| TNF-α | 17.355 | 20.11 | 0.34 |
| VEGF | 65.345 | 50.515 | 0.49 |
| Missing Values | Num. Melanoma | Num. Controls | Training Set Size | Testing Set Size | Predictors: Sex or Age | AUC (ROC) | Accuracy | No Info Rate | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| Removed * | 72 | 124 | 138 | 58 | Sex, Age | 0.674 | 0.621 | 0.64 | 0.66 |
| Sex | 0.658 | 0.638 | 0.64 | 0.56 | |||||
| Age | 0.761 | 0.724 | 0.64 | 0.11 | |||||
| None | 0.615 | 0.586 | 0.64 | 0.83 | |||||
| Set to 0 ** | 96 | 136 | 164 | 68 | Sex, Age | 0.621 | 0.588 | 0.59 | 0.55 |
| Sex | 0.510 | 0.588 | 0.59 | 0.55 | |||||
| Age | 0.704 | 0.662 | 0.59 | 0.13 | |||||
| None | 0.619 | 0.588 | 0.59 | 0.55 |
| Cytokines | Median | p-Value (Mann–Whitney) | ||||||
|---|---|---|---|---|---|---|---|---|
| Ctrls (201) | Melanoma | Ctrls vs. all | (with Bonferroni Correction) | |||||
| All (310) | Prim. (83) | Metast. (227) | Ctrls vs. Prim. | Ctrls vs. Metast. | Prim. vs. Metast. | |||
| IL-1b | 6.66 | 7.04 | 6.73 | 7.15 | <0.0001 | 0.15 | <0.0001 | 0.71 |
| IL-1Ra | 9.38 | 7.02 | 7.03 | 7.01 | <0.0001 | <0.0001 | <0.0001 | 1.31 |
| IL-2 | 2.58 | 2.81 | 2.58 | 3.00 | 0.59 | 0.96 | 1.00 | 1.00 |
| IL-4 | 3.46 | 3.46 | 3.17 | 3.58 | 0.96 | 0.45 | 1.00 | 0.15 |
| IL-5 | 2.81 | 3.00 | 3.00 | 3.00 | 0.48 | 1.00 | 1.00 | 1.00 |
| IL-6 | 5.61 | 6.39 | 5.98 | 6.64 | <0.0001 | 0.06 | <0.0001 | 0.06 |
| IL-7 | 7.22 | 5.49 | 5.17 | 5.73 | <0.0001 | <0.0001 | <0.0001 | 0.01 |
| IL-8 | 3.32 | 3.32 | 3.17 | 3.32 | 0.99 | 1.00 | 1.00 | 1.00 |
| IL-9 | 2.58 | 2.81 | 2.58 | 2.81 | 0.76 | 1.00 | 1.00 | 0.48 |
| IL-10 | 3.70 | 4.88 | 4.75 | 5.04 | <0.0001 | <0.0001 | <0.0001 | 0.66 |
| IL-12(p70) | 4.25 | 2.32 | 2.81 | 2.00 | <0.0001 | <0.0001 | <0.0001 | 0.12 |
| IL-13 | 5.64 | 5.52 | 5.55 | 5.49 | 0.96 | 1.00 | 1.00 | 1.00 |
| IL-15 | 6.95 | 6.83 | 6.30 | 6.97 | 0.44 | 0.001 | 1.00 | 0.01 |
| IL-17 | 5.67 | 6.83 | 6.07 | 7.37 | <0.0001 | 0.08 | <0.0001 | 0.05 |
| Eotaxin | 4.75 | 5.29 | 4.95 | 5.39 | <0.0001 | 0.21 | <0.0001 | 0.06 |
| FGF-2 | 7.11 | 7.03 | 6.25 | 7.24 | 0.51 | <0.0001 | 0.53 | <0.0001 |
| G-CSF | 11.83 | 11.55 | 12.25 | 11.37 | 0.95 | 0.001 | 0.30 | 0.006 |
| GM-CSF | 4.81 | 4.64 | 4.52 | 4.64 | 0.83 | 1.00 | 1.00 | 0.99 |
| IFN-γ | 4.70 | 5.58 | 4.91 | 5.83 | <0.0001 | 0.03 | <0.0001 | 0.01 |
| IP-10 (CXCL10) | 2.81 | 3.86 | 3.17 | 4.25 | <0.0001 | 0.36 | <0.0001 | 0.01 |
| MCP-1(MCAF) | 3.32 | 3.46 | 3.00 | 3.58 | 0.03 | 1.00 | 0.03 | 0.01 |
| MIP-1a (CCL3) | 5.13 | 8.11 | 7.76 | 8.24 | <0.0001 | <0.0001 | <0.0001 | 0.07 |
| MIP-1b (CCL4) | 5.29 | 7.90 | 7.35 | 8.13 | <0.0001 | <0.0001 | <0.0001 | 0.12 |
| PDGF-BB | 13.21 | 13.01 | 13.21 | 12.83 | 0.02 | 0.60 | 0.001 | 0.003 |
| RANTES (CCL5) | 6.86 | 8.16 | 7.81 | 8.22 | <0.0001 | <0.0001 | <0.0001 | 1.00 |
| TNF-α | 6.13 | 7.54 | 7.27 | 7.57 | <0.0001 | <0.0001 | <0.0001 | 0.003 |
| VEGF | 8.95 | 8.97 | 8.67 | 9.03 | 0.37 | 0.001 | 1.00 | 0.002 |
| Cytokines | AUC ± S.E. of ROC Analysis | |||
|---|---|---|---|---|
| Ctrls vs. all Melanoma | Ctrls vs. Primary | Ctrls vs. Metastatic | Primary vs. Metastatic | |
| IL-1b | 0.62 ± 0.03 | 0.57 ± 0.04 | 0.63 ± 0.03 | 0.54 ± 0.04 |
| IL-1Ra | 0.88 ± 0.02 | 0.88 ± 0.03 | 0.88 ± 0.02 | 0.53 ± 0.04 |
| IL-2 | 0.51 ± 0.03 | 0.54 ± 0.04 | 0.51 ± 0.03 | 0.53 ± 0.04 |
| IL-4 | 0.50 ± 0.03 | 0.55 ± 0.04 | 0.52 ± 0.03 | 0.57 ± 0.04 |
| IL-5 | 0.52 ± 0.03 | 0.51 ± 0.04 | 0.52 ± 0.03 | 0.51 ± 0.04 |
| IL-6 | 0.64 ± 0.03 | 0.59 ± 0.04 | 0.66 ± 0.03 | 0.59 ± 0.04 |
| IL-7 | 0.86 ± 0.02 | 0.91 ± 0.03 | 0.85 ± 0.02 | 0.61 ± 0.04 |
| IL-8 | 0.50 ± 0.03 | 0.52 ± 0.04 | 0.51 ± 0.03 | 0.52 ± 0.04 |
| IL-9 | 0.51 ± 0.03 | 0.53 ± 0.04 | 0.52 ± 0.03 | 0.55 ± 0.04 |
| IL-10 | 0.68 ± 0.03 | 0.65 ± 0.03 | 0.69 ± 0.03 | 0.55 ± 0.04 |
| IL-12(p70) | 0.78 ± 0.02 | 0.77 ± 0.03 | 0.79 ± 0.02 | 0.58 ± 0.04 |
| IL-13 | 0.50 ± 0.03 | 0.50 ± 0.04 | 0.50 ± 0.03 | 0.50 ± 0.04 |
| IL-15 | 0.52 ± 0.03 | 0.64 ± 0.01 | 0.52 ± 0.03 | 0.61 ± 0.04 |
| IL-17 | 0.61 ± 0.03 | 0.54 ± 0.04 | 0.63 ± 0.03 | 0.59 ± 0.04 |
| Eotaxin | 0.63 ± 0.03 | 0.57 ± 0.04 | 0.65 ± 0.03 | 0.59 ± 0.04 |
| FGF-2 | 0.52 ± 0.03 | 0.67 ± 0.04 | 0.54 ± 0.03 | 0.66 ± 0.04 |
| G-CSF | 0.50 ± 0.03 | 0.63 ± 0.04 | 0.55 ± 0.03 | 0.61 ± 0.04 |
| GM-CSF | 0.51 ± 0.03 | 0.52 ± 0.04 | 0.52 ± 0.03 | 0.54 ± 0.04 |
| IFN-γ | 0.69 ± 0.03 | 0.60 ± 0.04 | 0.72 ± 0.03 | 0.61 ± 0.04 |
| IP-10 (CXCL10) | 0.64 ± 0.03 | 0.56 ± 0.04 | 0.67 ± 0.03 | 0.61 ± 0.04 |
| MCP-1(MCAF) | 0.56 ± 0.03 | 0.54 ± 0.04 | 0.59 ± 0.03 | 0.62 ± 0.04 |
| MIP-1a (CCL3) | 0.93 ± 0.01 | 0.91 ± 0.02 | 0.93 ± 0.02 | 0.58 ± 0.04 |
| MIP-1b (CCL4) | 0.87 ± 0.02 | 0.87 ± 0.02 | 0.86 ± 0.02 | 0.58 ± 0.04 |
| PDGF-BB | 0.56 ± 0.03 | 0.55 ± 0.04 | 0.60 ± 0.03 | 0.62 ± 0.04 |
| RANTES (CCL5) | 0.73 ± 0.03 | 0.73 ± 0.03 | 0.72 ± 0.02 | 0.53 ± 0.04 |
| TNF-α | 0.77 ± 0.03 | 0.68 ± 0.03 | 0.80 ± 0.02 | 0.62 ± 0.04 |
| VEGF | 0.52 ± 0.03 | 0.63 ± 0.04 | 0.52 ± 0.03 | 0.63 ± 0.04 |
| Num. Melanoma | Num. Controls | Training Set Size | Testing Set Size | AUC (ROC) | Accuracy | No Info Rate | p-Value |
|---|---|---|---|---|---|---|---|
| 310 | 201 | 358 | 153 | 0.99 | 0.95 | 0.61 | <0.00001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cesati, M.; Scatozza, F.; D’Arcangelo, D.; Antonini-Cappellini, G.C.; Rossi, S.; Tabolacci, C.; Nudo, M.; Palese, E.; Lembo, L.; Di Lella, G.; et al. Investigating Serum and Tissue Expression Identified a Cytokine/Chemokine Signature as a Highly Effective Melanoma Marker. Cancers 2020, 12, 3680. https://doi.org/10.3390/cancers12123680
Cesati M, Scatozza F, D’Arcangelo D, Antonini-Cappellini GC, Rossi S, Tabolacci C, Nudo M, Palese E, Lembo L, Di Lella G, et al. Investigating Serum and Tissue Expression Identified a Cytokine/Chemokine Signature as a Highly Effective Melanoma Marker. Cancers. 2020; 12(12):3680. https://doi.org/10.3390/cancers12123680
Chicago/Turabian StyleCesati, Marco, Francesca Scatozza, Daniela D’Arcangelo, Gian Carlo Antonini-Cappellini, Stefania Rossi, Claudio Tabolacci, Maurizio Nudo, Enzo Palese, Luigi Lembo, Giovanni Di Lella, and et al. 2020. "Investigating Serum and Tissue Expression Identified a Cytokine/Chemokine Signature as a Highly Effective Melanoma Marker" Cancers 12, no. 12: 3680. https://doi.org/10.3390/cancers12123680
APA StyleCesati, M., Scatozza, F., D’Arcangelo, D., Antonini-Cappellini, G. C., Rossi, S., Tabolacci, C., Nudo, M., Palese, E., Lembo, L., Di Lella, G., Facchiano, F., & Facchiano, A. (2020). Investigating Serum and Tissue Expression Identified a Cytokine/Chemokine Signature as a Highly Effective Melanoma Marker. Cancers, 12(12), 3680. https://doi.org/10.3390/cancers12123680






