Genomic Fabric Remodeling in Metastatic Clear Cell Renal Cell Carcinoma (ccRCC): A New Paradigm and Proposal for a Personalized Gene Therapy Approach
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Independent Characteristics of the Quantified Genes in Each Profiled Region
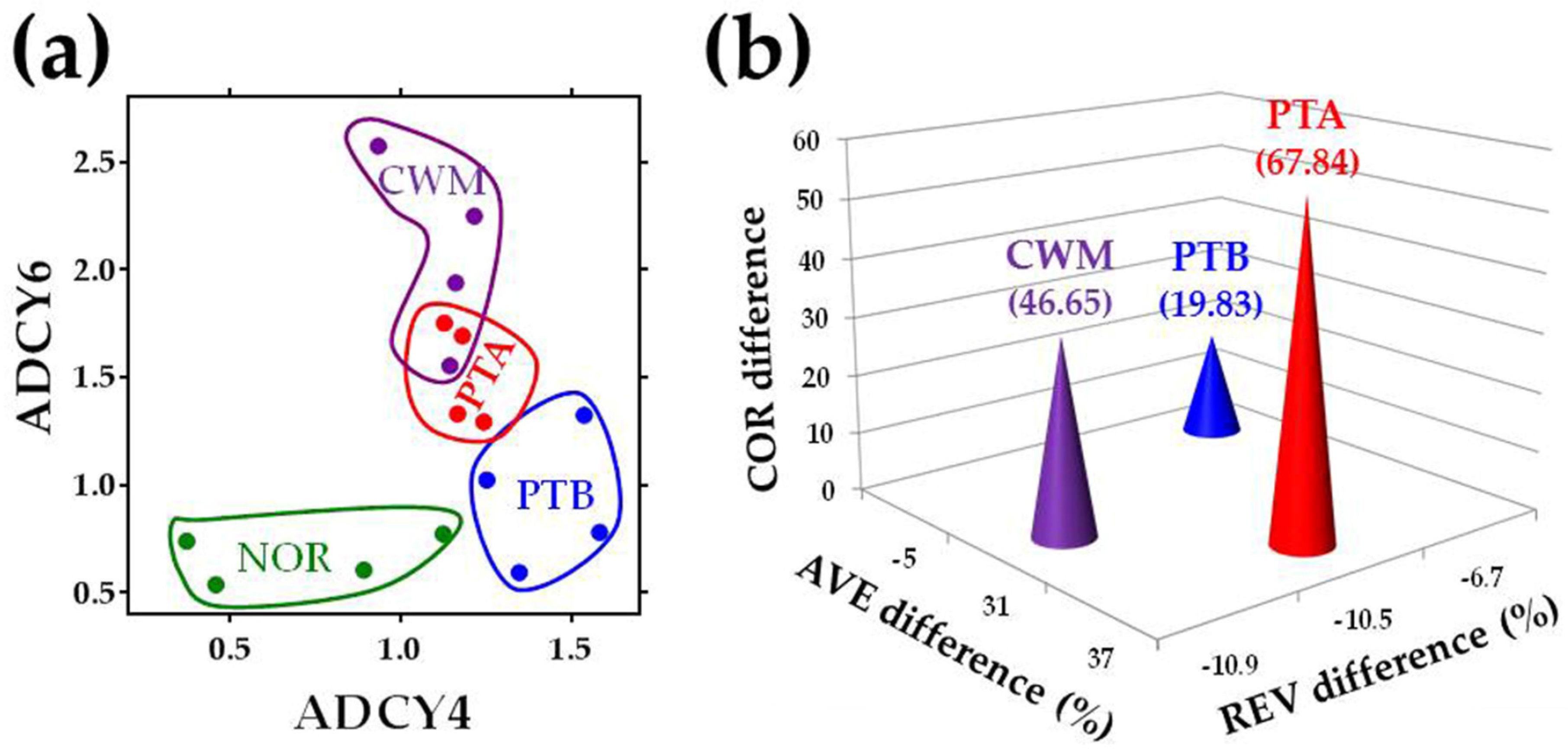
2.2. Transcriptomic Differences between Cancer Nodules
2.3. Regulation of the ccRCC Functional Pathway
2.4. Regulation of the Apoptosis Pathway
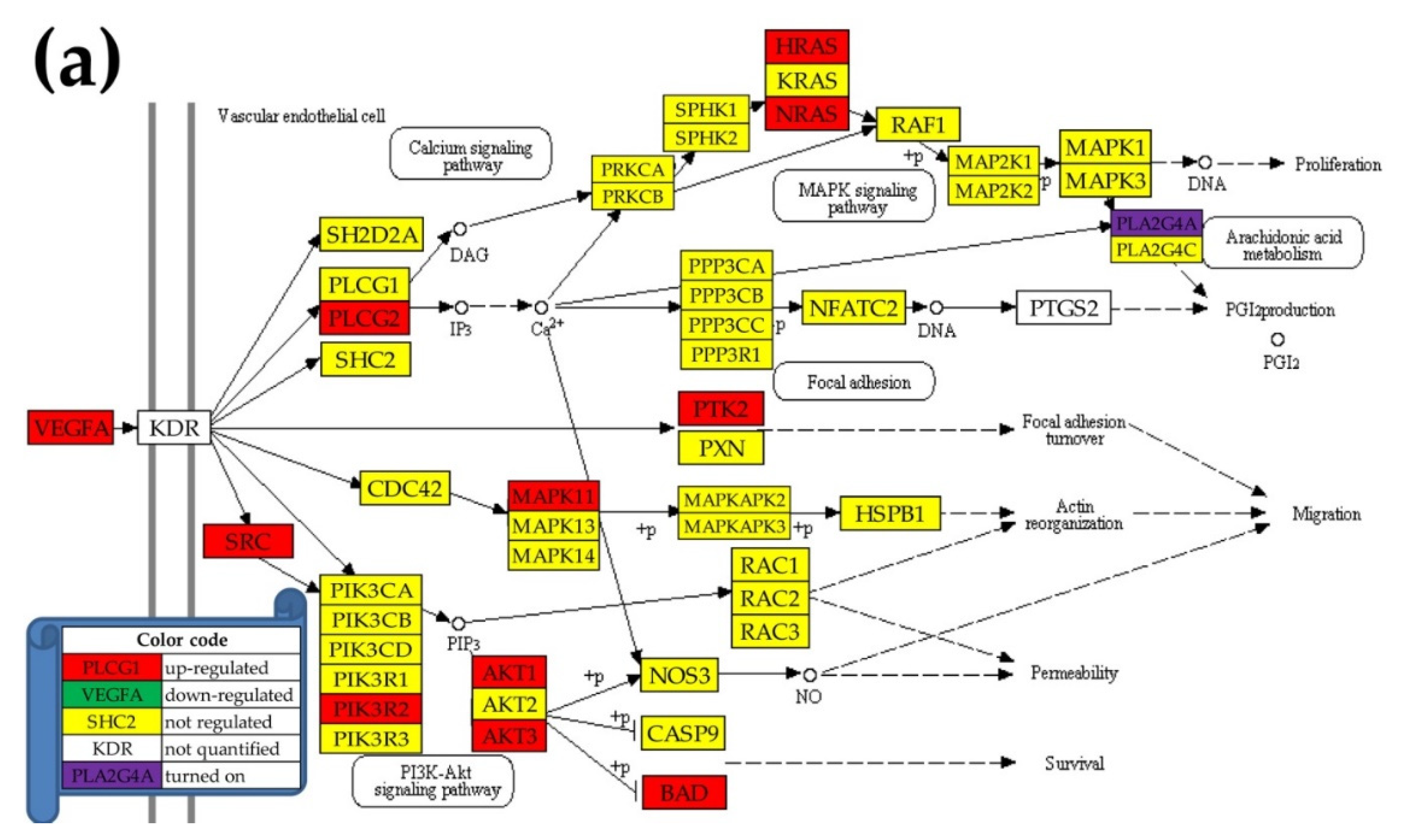
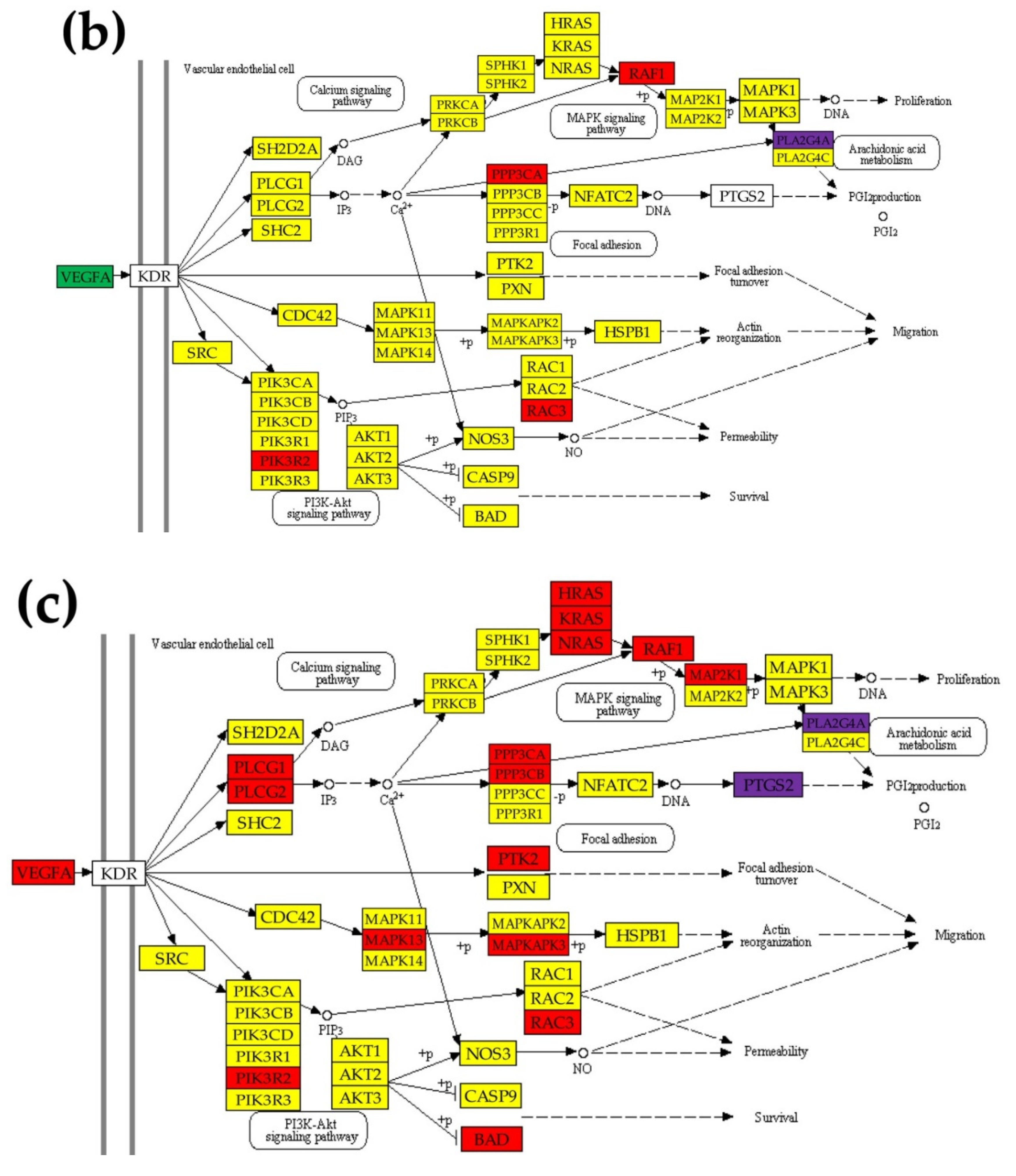
2.5. Regulation of the VEGF Signaling Pathway
2.6. Alteration of the Homeostatic Mechanisms That Control the Transcript Abundances
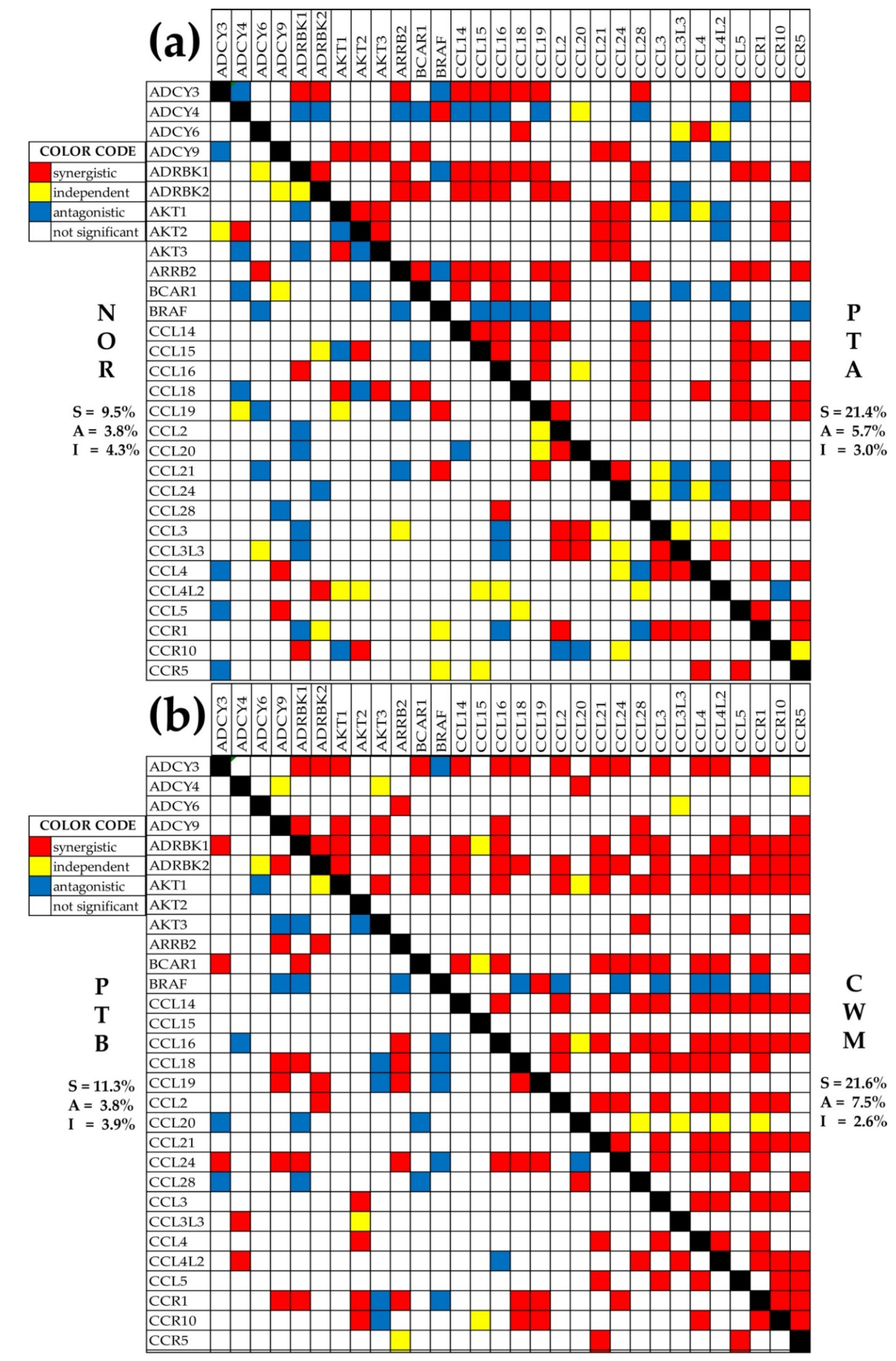
2.7. Changes in Gene Networking
2.8. Changes of the Transcriptomic Landscapes and Genomic Fabrics Interplay
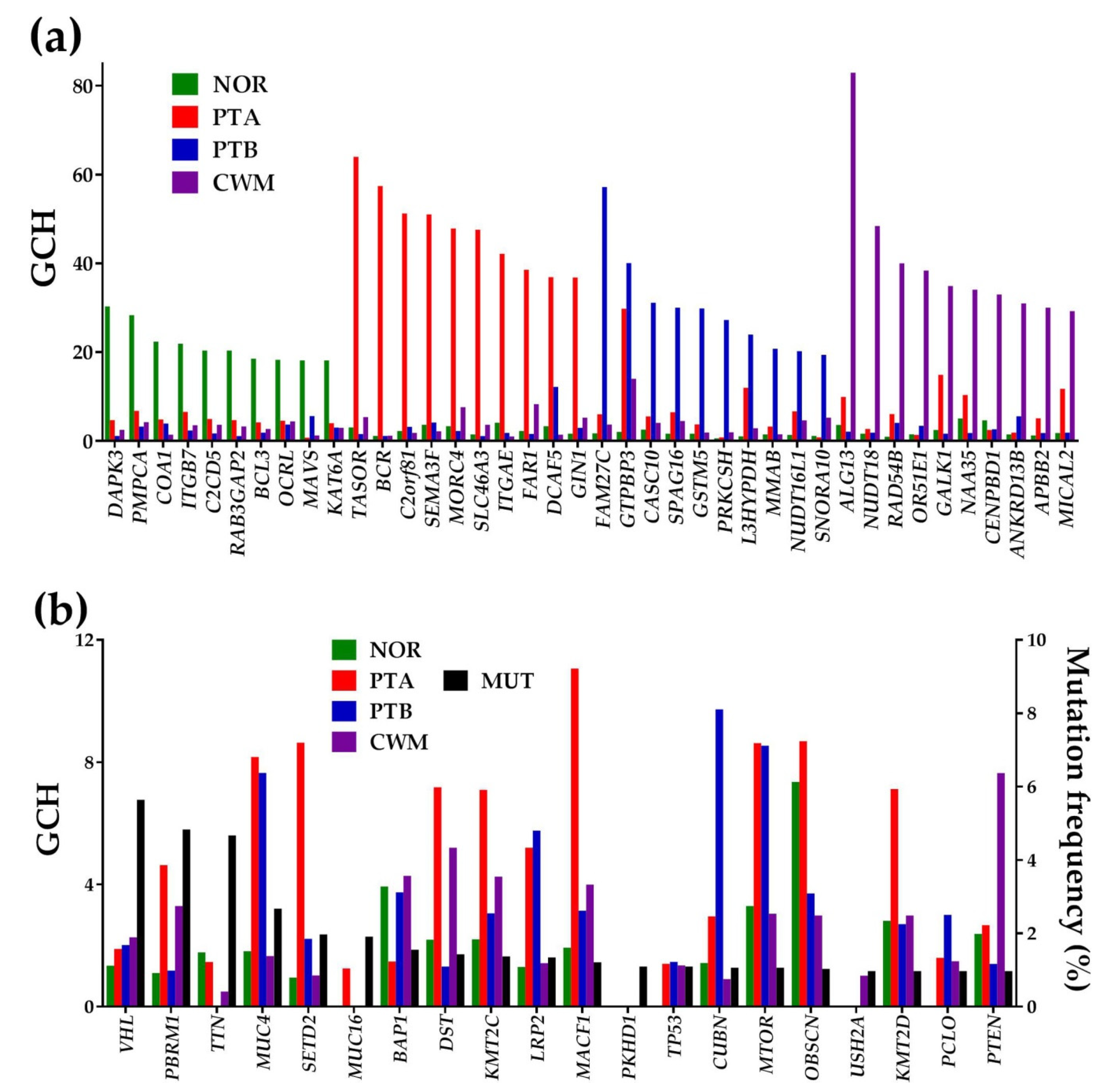
2.9. Gene Hierarchies and Gene Master Regulators
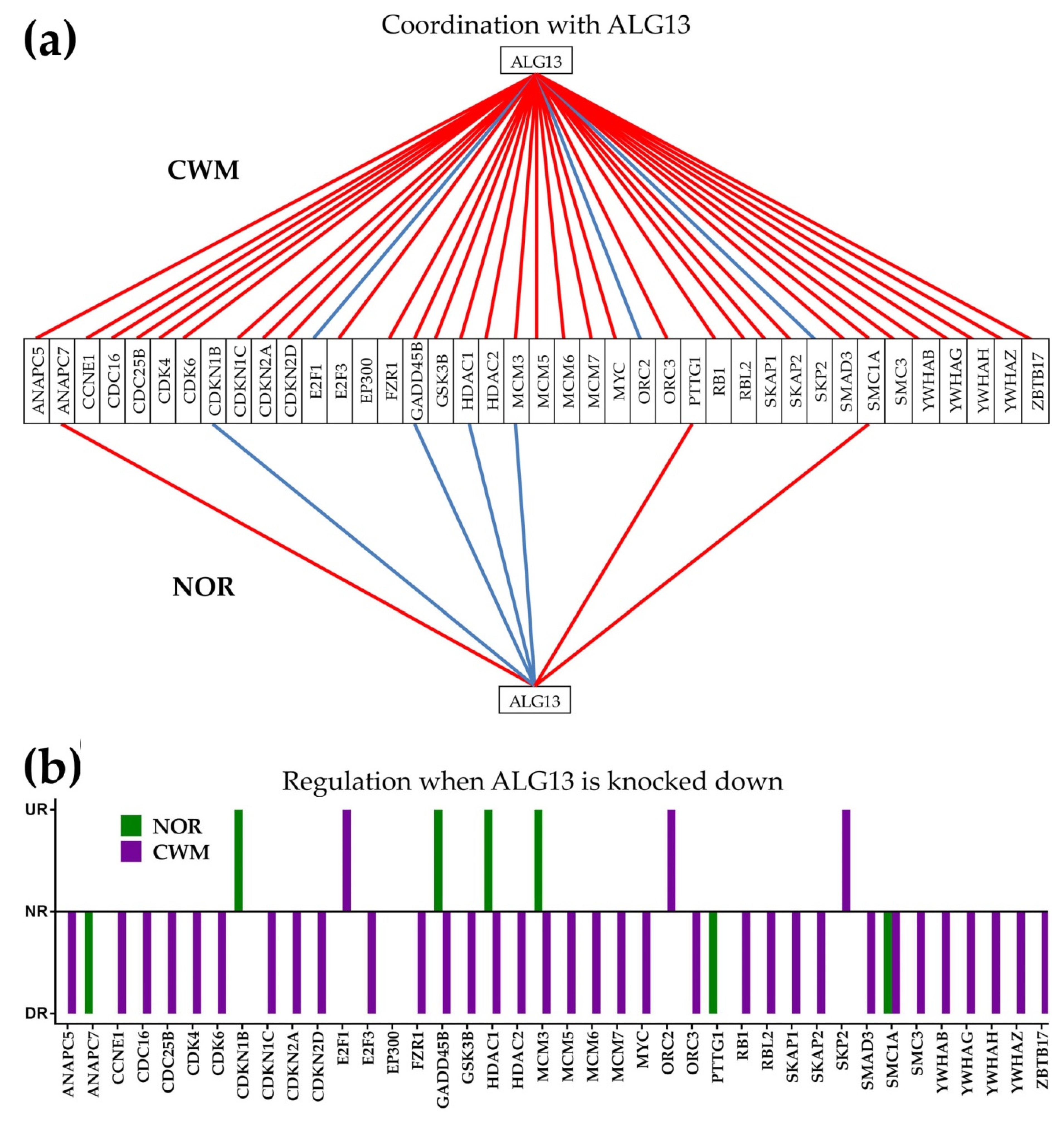
2.10. GMR at Work
3. Discussion
4. Materials and Methods
4.1. Gene Expression Data
4.2. Average Expression Level (AVE)
4.3. Relative Expression Variability
4.4. Expression Coordination
4.5. Expression Regulation
4.6. Transcriptomic Separation
4.7. Pair-Wise Relevance
4.8. Gene Commanding Height (GCH) and Identification of Gene Master Regulator (GMR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Key Statistics about Kidney Cancer. Available online: https://www.cancer.org/cancer/kidney-cancer.html (accessed on 8 February 2020).
- McLaughlin, J.K.; Lipworth, L. Epidemiologic aspects of renal cell cancer. Semin. Oncol. 2000, 27, 115–123. [Google Scholar] [PubMed]
- TNM Kidney Cancer Staging Made Simple. Available online: https://kidneycancerinfo.weebly.com/staging-and-grading.html (accessed on 15 September 2020).
- Rioux-Leclercq, N. Le Grade Nucleaire de Fuhrman, Facteur Pronostique du Cancer du Rein Depuis 25 ans [The Fuhrman Grading System for Kidney Cancer Prognosis]. Prog. Urol. 2006, 16, 5–8. [Google Scholar] [PubMed]
- Laskar, R.S.; Muller, D.C.; Li, P.; Machiela, M.J.; Ye, Y.; Gaborieau, V.; Foll, M.; Hofmann, J.N.; Colli, L.; Sampson, J.N.; et al. Sex specific associations in genome wide association analysis of renal cell carcinoma. Eur. J. Hum. Genet. 2019, 27, 1589–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, J.; Syed, W. Renal Cancer. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK558975/ (accessed on 20 August 2020).
- Purdue, M.P.; Song, L.; Scélo, G.; Houlston, R.S.; Wu, X.; Sakoda, L.C.; Thai, K.; Graff, R.E.; Rothman, N.; Brennan, P.; et al. Pathway Analysis of Renal Cell Carcinoma Genome-Wide Association Studies Identifies Novel Associations. Cancer Epidemiol. Biomark. Prev. 2020. [Google Scholar] [CrossRef]
- Cui, H.; Xu, L.; Li, Z.; Hou, K.Z.; Che, X.F.; Liu, B.F.; Liu, Y.P.; Qu, X.J. Integrated bioinformatics analysis for the identification of potential key genes affecting the pathogenesis of clear cell renal cell carcinoma. Oncol. Lett. 2020, 20, 1573–1584. [Google Scholar] [CrossRef]
- Tornberg, S.V.; Nisen, H.; Järvinen, P.; Järvinen, R.; Kilpeläinen, T.P.; Taari, K.; Stenman, U.-H.; Visapää, H. Serum tumour associated trypsin inhibitor, as a biomarker for survival in renal cell carcinoma. Scand. J. Urol. 2020, 4, 1–7. [Google Scholar] [CrossRef]
- Harmonized Cancer Datasets Genomic Data Commons Data Portal. Available online: https://portal.gdc.cancer.gov (accessed on 11 February 2020).
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 3698. [Google Scholar] [CrossRef]
- Linehan, W.M.; Schmidt, L.S.; Crooks, D.R.; Wei, D.; Srinivasan, R.; Lang, M.; Ricketts, C.J. The Metabolic Basis of Kidney Cancer. Cancer Discov. 2019, 9, 1006–1021. [Google Scholar] [CrossRef] [Green Version]
- Linehan, W.M.; Walther, M.M.; Zbar, B. The Genetic Basis of Cancer of the Kidney. J. Urol. 2003, 170, 2163–2172. [Google Scholar] [CrossRef]
- Mery, B.; Jones, S.; Vallard, A.; Rowinski, E.; Guillot, A.; Magné, N. Cancer du rein métastatique: Recommandations et perspectives en 1re ligne. Bull. Cancer 2018, 105, S235–S241. [Google Scholar] [CrossRef]
- Spirina, L.V.; Yurmazov, Z.A.; Gorbunov, A.K.; Usynin, E.A.; Lushnikova, N.A.; Kovaleva, I.V. Molecular Protein and Expression Profile in the Primary Tumors of Clear Cell Renal Carcinoma and Metastases. Cells 2020, 9, 1680. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Li, L.; Ding, P.; Li, L.; Ge, X.; Zheng, L.; Wang, X.; Wang, J.; Zhang, W.; Wang, N.; et al. Complement Inhibitor CRIg/FH Ameliorates Renal Ischemia Reperfusion Injury via Activation of PI3K/AKT Signaling. J. Immunol. 2018, 201, 3717–3730. [Google Scholar] [CrossRef] [PubMed]
- Al Kafri, N.; Hafizi, S. Tumour-Secreted Protein S (ProS1) Activates a Tyro3-Erk Signalling Axis and Protects Cancer Cells from Apoptosis. Cancers 2019, 11, 1843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasinska, A.; Jaszczynski, J.; Adamczyk, A.; Janecka-Widła, A.; Wilk, W.; Cichocka, A.; Stelmach, A. Biomarkers of epithelial-mesenchymal transition in localized, surgically treated clear-cell renal cell carcinoma. Folia Histochem. Cytobiol. 2018, 56, 195–206. [Google Scholar] [CrossRef]
- Alcaraz, E.; Vilardell, J.; Borgo, C.; Sarró, E.; Plana, M.; Marin, O.; Pinna, L.A.; Bayascas, J.; Meseguer, A.; Salvi, M.; et al. Effects of CK2β subunit down-regulation on Akt signalling in HK-2 renal cells. PLoS ONE 2020, 15, e0227340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.Y.; Lin, P.Y.; Weiss, R.H. Targeting the PI3K–Akt pathway in kidney cancer. Expert Rev. Anticancer Ther. 2007, 7, 863–870. [Google Scholar] [CrossRef]
- Damayanti, N.P.; Budka, J.A.; Khella, H.W.Z.; Ferris, M.W.; Ku, S.Y.; Kauffman, E.; Wood, A.C.; Ahmed, K.; Chintala, V.N.; Adelaiye-Ogala, R.; et al. Therapeutic Targeting of TFE3/IRS-1/PI3K/mTOR Axis in Translocation Renal Cell Carcinoma. Clin. Cancer Res. 2018, 24, 5977–5989. [Google Scholar] [CrossRef] [Green Version]
- Gargalionis, A.N.; Sarlani, E.; Stofas, A.; Malakou, L.S.; Adamopoulos, C.; Bamias, A.; Boutati, E.; Constantinides, C.A.; Stravodimos, K.G.; Piperi, C.; et al. Polycystin-1 induces activation of the PI3K/AKT/mTOR pathway and promotes angiogenesis in renal cell carcinoma. Cancer Lett. 2020, 489, 135–143. [Google Scholar] [CrossRef]
- Iacobas, S.; Iacobas, D.A.; Spray, D.C.; Scemes, E. The connexin43 transcriptome during brain development: Importance of genetic background. Brain Res. 2012, 1487, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Thomas, N.M.; Jasmin, J.F.; Lisanti, M.P.; Iacobas, D.A. Sex differences in expression and subcellular localization of heart rhythm determinant proteins. Biochem. Biophys. Res. Commun. 2011, 406, 117–122. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Fan, C.; Iacobas, S.; Spray, D.C.; Haddad, G.G. Transcriptomic changes in developing kidney exposed to chronic hypoxia. Biochem. Biophys. Res. Commun. 2006, 349, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Desruisseaux, M.; Iacobas, D.A.; Iacobas, S.; Mukherjee, S.; Weiss, L.M.; Tanowitz, H.B.; Spray, D.C. Alterations in the Brain Transcriptome in Plasmodium Berghei ANKA Infected Mice. J. Neuroparasitol. 2010, 1, 74–81. [Google Scholar] [CrossRef] [Green Version]
- Iacobas, D.A.; Iacobas, S.; Nebieridze, N.; Velíšek, L.; Velíšková, J. Estrogen Protects Neurotransmission Transcriptome during Status Epilepticus. Front. Neurosci. 2018, 12, 332. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Iacobas, D.A.; Zhou, D.; Chen, Q.; Gavrialov, O.; Haddad, G.G. Gene expression and phenotypic characterization of mouse heart after chronic constant and intermittent hypoxia. Physiol. Genom. 2005, 22, 292–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thi, M.M.; Iacobas, D.A.; Iacobas, S.; Spray, D.C. Fluid Shear Stress Regulates Vascular Endothelial Growth Factor Gene in Osteoblasts. Ann. N. Y. Acad. Sci. 2007, 1117, 73–81. [Google Scholar] [CrossRef]
- Kobets, T.; Iatropoulos, M.J.; Duan, J.D.; Brunnemann, K.D.; Iacobas, D.A.; Iacobas, S.; Vock, E.; Deschl, U.; Williams, G.M. Effects of Nitrosamines on the Expression of Genes Involved in Xenobiotic Metabolism in the Chicken Egg Alternative Genotoxicity Model. Toxicol. Sci. 2018, 166, 82–96. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Chachua, T.; Iacobas, S.; Benson, M.J.; Borges, K.; Velísková, J.; Velísek, L. ACTH and PMX53 recover the normal synaptic transcriptome in a rat model of infantile spasms. Sci. Rep. 2018, 8, 5722. [Google Scholar] [CrossRef]
- Lee, P.R.; Cohen, J.E.; Iacobas, D.A.; Iacobas, S.; Fields, R.D. Gene networks activated by pattern-specific generation of action potentials in dorsal root ganglia neurons. Sci. Rep. 2017, 7, 43765. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, D.K.; Xi, Y.; Kapur, P.; Madhuranthakam, A.J.; Lewis, M.A.; Udayakumar, D.; Rasmussen, R.; Yuan, Q.; Bagrodia, A.; Margulis, V.; et al. Magnetic Resonance Imaging Radiomics Analyses for Prediction of High-Grade Histology and Necrosis in Clear Cell Renal Cell Carcinoma: Preliminary Experience. Clin. Genitourin. Cancer 2020, 1558–7673. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Iacobas, S. Towards a Personalized Cancer Gene Therapy: A Case of Clear Cell Renal Cell Carcinoma. Cancer Oncol. Res. 2017, 5, 45–52. [Google Scholar] [CrossRef]
- Noon, A.P.; Vlatković, N.; Polański, R.; Maguire, M.; Shawki, H.; Parsons, K.; Boyd, M.T. p53 and MDM2 in renal cell carcinoma: Biomarkers for disease progression and future therapeutic targets? Cancer 2010, 116, 780–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harlander, S.; Schönenberger, D.; Toussaint, N.C.; Prummer, M.; Catalano, A.; Brandt, L.; Moch, H.; Wild, P.J.; Frew, I.J. Combined mutation in Vhl, Trp53 and Rb1 causes clear cell renal cell carcinoma in mice. Nat. Med. 2017, 23, 869–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacobas, D.A. The Genomic Fabric Perspective on the Transcriptome Between Universal Quantifiers and Personalized Genomic Medicine. Biol. Theory 2016, 11, 123–137. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Lee, P.R.; Cohen, J.E.; Fields, R.D. Coordinated Activity of Transcriptional Networks Responding to the Pattern of Action Potential Firing in Neurons. Genes 2019, 10, 754. [Google Scholar] [CrossRef] [Green Version]
- Iacobas, D.A.; Tuli, N.; Iacobas, S.; Rasamny, J.K.; Moscatello, A.; Geliebter, J.; Tiwari, R.M. Gene master regulators of papillary and anaplastic thyroid cancer phenotypes. Oncotarget 2018, 9, 2410–2424. [Google Scholar] [CrossRef] [Green Version]
- Iacobas, S.; Ede, N.; Iacobas, D.A. The Gene Master Regulators (GMR) Approach Provides Legitimate Targets for Personalized, Time-Sensitive Cancer Gene Therapy. Genes 2019, 10, 560. [Google Scholar] [CrossRef] [Green Version]
- Remodeling of Major Genomic Fabrics and Their Interplay in Metastatic Clear Cell Renal Cell Carcinoma. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72304 (accessed on 6 February 2020).
- Chemokine Signaling Pathway. Available online: https://www.kegg.jp/kegg-bin/show_pathway?hsa04062maphsa04062 (accessed on 7 March 2020).
- Hussain, M.; Adah, D.; Tariq, M.; Lu, Y.; Zhang, J.; Liu, J. CXCL13/CXCR5 signaling axis in cancer. Life Sci. 2019, 227, 175–186. [Google Scholar] [CrossRef]
- Kadomoto, S.; Izumi, K.; Mizokami, A. The CCL20-CCR6 Axis in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 5186. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Cheng, E.H. Exploiting the circuit breaker cancer evolution model in human clear cell renal cell carcinoma. Cell Stress 2020, 4, 191–198. [Google Scholar] [CrossRef]
- Renal Cell Carcinoma Pathway. Available online: https://www.kegg.jp/kegg-bin/show_pathway?hsa05211 (accessed on 7 March 2020).
- Prickett, D.; Watson, M. Use of GenMAPP and MAPPFinder to analyse pathways involved in chickens infected with the protozoan parasite Eimeria. BMC Proc. 2009, 3, S7. [Google Scholar] [CrossRef] [Green Version]
- Apoptosis Pathway. Available online: https://www.kegg.jp/kegg-bin/show_pathway?hsa04210 (accessed on 6 March 2020).
- VEGF Signaling Pathway. Available online: https://www.kegg.jp/kegg-bin/show_pathway?hsa04370 (accessed on 7 March 2020).
- Rini, B.I.; Battle, D.; Figlin, R.A.; George, D.J.; Hammers, H.; Hutson, T.; Jonasch, E.; Joseph, R.W.; McDermott, D.F.; Motzer, R.J.; et al. The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of advanced renal cell carcinoma (RCC). J. Immunother. Cancer 2019, 7, 354. [Google Scholar] [CrossRef] [PubMed]
- Melincovici, C.S.; Boşca, A.B.; Şuşman, S.; Mărginean, M.; Mihu, C.; Istrate, M.; Moldovan, I.M.; Roman, A.L.; Mihu, C.M. Vascular endothelial growth factor (VEGF)—Key factor in normal and pathological angiogenesis. Rom. J. Morphol. Embryol. 2018, 59, 455–467. [Google Scholar] [PubMed]
- Yarla, N.S.; Bishayee, A.; Vadlakonda, L.; Chintala, R.; Duddukuri, G.R.; Reddanna, P.; Dowluru, K.S. Phospholipase A2 Isoforms as Novel Targets for Prevention and Treatment of Inflammatory and Oncologic Diseases. Curr. Drug Targets 2016, 17, 1940–1962. [Google Scholar] [CrossRef]
- Li, S.; Jiang, M.; Wang, L.; Yu, S. Combined chemotherapy with cyclooxygenase-2 (COX-2) inhibitors in treating human cancers: Recent advancement. Biomed. Pharmacother. 2020, 129, 110389. [Google Scholar] [CrossRef]
- Iacobas, D.A. Biomarkers, Master Regulators and Genomic Fabric Remodeling in a Case of Papillary Thyroid Carcinoma. Genes 2020, 11, 1030. [Google Scholar] [CrossRef]
- Iacobas, D.A. Powerful quantifiers for cancer transcriptomics. World J. Clin. Oncol. 2020, 11, 679–704. [Google Scholar] [CrossRef] [PubMed]
- Iacobas, S.; Thomas, N.M.; Iacobas, D.A. Plasticity of the myelination genomic fabric. Mol. Genet. Genom. 2012, 287, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Oxidative Phosphorylation Pathway. Available online: https://www.kegg.jp/kegg-bin/show_pathway?hsa00190 (accessed on 20 August 2020).
- Iacobas, D.A.; Iacobas, S.; Stout, R.; Spray, D.C. Cellular Environment Remodels the Genomic Fabrics of Functional Pathways in Astrocytes. Genes 2020, 11, 520. [Google Scholar] [CrossRef]
- Bladder Cancer Pathway. Available online: https://www.kegg.jp/kegg-bin/show_pathway?hsa05219+1613 (accessed on 17 September 2020).
- N-Glycan Biosynthesis. Available online: https://www.kegg.jp/kegg-bin/show_pathway?hsa00510+79868 (accessed on 17 September 2020).
- Basal Transcription Factors. Available online: https://www.kegg.jp/kegg-bin/show_pathway?hsa03022 (accessed on 20 September 2020).
- RNA Polymerase Pathway. Available online: https://www.kegg.jp/kegg-bin/show_pathway?hsa03020 (accessed on 20 September 2020).
- Cell Cycle Pathway. Available online: https://www.kegg.jp/kegg-bin/show_pathway?hsa04110 (accessed on 20 September 2020).
- NCBI PubMed Microarray Studies on Renal Cancers. Available online: https://pubmed.ncbi.nlm.nih.gov/?term=renal+cancer%2C+microarray&sort=date (accessed on 26 September 2020).
- Cui, Y.; Miao, C.; Hou, C.; Wang, Z.; Liu, B. Apolipoprotein C1 (APOC1): A Novel Diagnostic and Prognostic Biomarker for Clear Cell Renal Cell Carcinoma. Front. Oncol. 2020, 10, 1436. [Google Scholar] [CrossRef]
- Ma, C.G.; Xu, W.H.; Xu, Y.; Wang, J.; Liu, W.R.; Cao, D.L.; Wang, H.K.; Shi, G.H.; Zhu, Y.P.; Qu, Y.Y.; et al. Identification and validation of novel metastasis-related signatures of clear cell renal cell carcinoma using gene expression databases. Am. J. Transl. Res. 2020, 12, 4108–4126. [Google Scholar]
- Allen, A.; Gau, D.; Francoeur, P.; Sturm, J.; Wang, Y.; Martin, R.; Maranchie, J.; Duensing, A.; Kaczorowski, A.; Duensing, S.; et al. Actin-binding protein profilin1 promotes aggressiveness of clear-cell renal cell carcinoma cells. J. Biol. Chem. 2020, 295, 15636–15649. [Google Scholar] [CrossRef] [PubMed]
- Teng, H.; Mao, F.; Liang, J.; Xue, M.; Wei, W.; Li, X.; Zhang, K.; Feng, D.; Liu, B.; Sun, Z. Transcriptomic signature associated with carcinogenesis and aggressiveness of papillary thyroid carcinoma. Theranostics 2018, 8, 4345–4358. [Google Scholar] [CrossRef] [PubMed]
- Iacobas, S.; Iacobas, D.A. Astrocyte proximity modulates the myelination gene fabric of oligodendrocytes. Neuron Glia Biol. 2010, 6, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Shanmugasundaram, K.; Block, K. Renal Carcinogenesis, Tumor Heterogeneity, and Reactive Oxygen Species: Tactics Evolved. Antioxid. Redox Signal. 2016, 25, 685–701. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Udayakumar, D.; Cai, L.; Hu, Z.; Kapur, P.; Kho, E.-Y.; Pavía-Jiménez, A.; Fulkerson, M.; De Leon, A.D.; Yuan, Q.; et al. Addressing metabolic heterogeneity in clear cell renal cell carcinoma with quantitative Dixon MRI. JCI Insight 2017, 2, e94278. [Google Scholar] [CrossRef]
- Stanta, G.; Bonin, S. Overview on Clinical Relevance of Intra-Tumor Heterogeneity. Front. Med. 2018, 5, 85. [Google Scholar] [CrossRef] [Green Version]
- Dzobo, K.; Senthebane, D.A.; Thomford, N.E.; Rowe, A.; Dandara, C.; Parker, M.I. Not Everyone Fits the Mold: Intratumor and Intertumor Heterogeneity and Innovative Cancer Drug Design and Development. OMICS J. Integr. Biol. 2018, 22, 17–34. [Google Scholar] [CrossRef]
- Taverna, G.; Di Francesco, S.; Borroni, E.M.; Yiu, D.; Toniato, E.; Milanesi, S.; Chiriva-Internati, M.; Bresalier, R.S.; Zanoni, M.; Vota, P.; et al. The kidney, COVID-19, and the chemokine network: An intriguing trio. Int. Urol. Nephrol. 2020, 27, 1–8. [Google Scholar] [CrossRef]
- Rosen, L.S. Clinical Experience with Angiogenesis Signaling Inhibitors: Focus on Vascular Endothelial Growth Factor (VEGF) Blockers. Cancer Control 2002, 9, 36–44. [Google Scholar] [CrossRef]
- Douse, C.H.; Tchasovnikarova, I.A.; Timms, R.T.; Protasio, A.V.; Seczynska, M.; Prigozhin, D.M.; Albecka, A.; Wagstaff, J.; Williamson, J.C.; Freund, S.M.V.; et al. TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control. Nat. Commun. 2020, 11, 4940. [Google Scholar] [CrossRef]
- Gresakova, V.; Novosadova, V.; Prochazkova, M.; Bhargava, S.; Jenickova, I.; Prochazka, J.; Sedlacek, R. Fam208a orchestrates interaction protein network essential for early embryonic development and cell division. Exp. Cell Res. 2019, 382, 111437. [Google Scholar] [CrossRef] [PubMed]
- Esposito, T.; De Stefano, G.; Reccia, M.G.; Di Lorenzo, I.; Napolitano, F.; Scalabrì, F.; Lombardi, A.; Gianfrancesco, F.; Saleem, M.A.; Griffiths, L.R. Dysregulation of the Expression of Asparagine-Linked Glycosylation 13 Short Isoform 2 Affects Nephrin Function by Altering Its N-Linked Glycosylation. Nephron 2017, 136, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Iacobas, D.A.; Iacobas, S.; Werner, P.; Scemes, E.; Spray, D.C. Alteration of transcriptomic networks in adoptive-transfer experimental autoimmune encephalomyelitis. Front. Integr. Neurosci. 2007, 1, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spray, D.C.; Iacobas, D.A. Organizational Principles of the Connexin-Related Brain Transcriptome. J. Membr. Biol. 2007, 218, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, S.; De Luca, G.; Grassi, L.; Federici, G.; Alfonsi, R.; Signore, M.; Addario, A.; De Salvo, L.; Francescangeli, F.; Sanchez, M.; et al. Renal cancer: New models and approach for personalizing therapy. J. Exp. Clin. Cancer Res. 2018, 37, 217. [Google Scholar] [CrossRef]
- Chen, S.L.; Hu, F.; Wang, D.W.; Qin, Z.Y.; Liang, Y.; Dai, Y.J. Prognosis and regulation of an adenylyl cyclase network in acute myeloid leukemia. Aging 2020, 12, 11864–11877. [Google Scholar] [CrossRef] [PubMed]
- Barth, D.A.; Slaby, O.; Klec, C.; Juracek, J.; Drula, R.; Calin, G.A.; Pichler, M. Current Concepts of Non-Coding RNAs in the Pathogenesis of Non-Clear Cell Renal Cell Carcinoma. Cancers 2019, 11, 1580. [Google Scholar] [CrossRef] [Green Version]
- Jilaveanu, L.B.; Shuch, B.; Zito, C.R.; Parisi, F.; Barr, M.; Kluger, Y.; Chen, L.; Kluger, H.M. PD-L1 Expression in Clear Cell Renal Cell Carcinoma: An Analysis of Nephrectomy and Sites of Metastases. J. Cancer 2014, 5, 166–172. [Google Scholar] [CrossRef] [Green Version]
- Corrò, C.; Moch, H. Biomarker discovery for renal cancer stem cells. J. Pathol. Clin. Res. 2018, 4, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Paz-Yaacov, N.; Bazak, L.; Buchumenski, I.; Porath, H.T.; Danan-Gotthold, M.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep. 2015, 13, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Zhou, X.; Xu, P.; Li, Y.; Shi, H.; Chen, D.; Li, R.; Shi, H. Deficiency of apoptosis-stimulating protein two of p53 ameliorates acute kidney injury induced by ischemia reperfusion in mice through upregulation of autophagy. J. Cell. Mol. Med. 2019, 23, 2457–2467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamenova, I.; Mukherjee, P.; Conic, S.; Mueller, F.; El-Saafin, F.; Bardot, P.; Garnier, J.-M.; Dembele, D.; Capponi, S.; Timmers, H.T.M.; et al. Co-translational assembly of mammalian nuclear multisubunit complexes. Nat. Commun. 2019, 10, 1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grierson, P.M.; Lillard, K.; Behbehani, G.K.; Combs, K.A.; Bhattacharyya, S.; Acharya, S.; Groden, J. BLM helicase facilitates RNA polymerase I-mediated ribosomal RNA transcription. Hum. Mol. Genet. 2012, 21, 1172–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Liu, Y.; Li, Y.; Wang, H.; Stewart, S.; Van Der Jeught, K.; Agarwal, P.; Zhang, Y.; Liu, S.; Zhao, G.; et al. Precise targeting of POLR2A as a therapeutic strategy for human triple negative breast cancer. Nat. Nanotechnol. 2019, 14, 388–397. [Google Scholar] [CrossRef]
- Finlay-Schultz, J.; Gillen, A.E.; Brechbuhl, H.M.; Ivie, J.J.; Matthews, S.B.; Jacobsen, B.M.; Bentley, D.L.; Kabos, P.; Sartorius, C.A. Breast Cancer Suppression by Progesterone Receptors Is Mediated by Their Modulation of Estrogen Receptors and RNA Polymerase III. Cancer Res. 2017, 77, 4934–4946. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Hou, G.; Fang, Z.; Liu, J.; Lv, X.D. Distinct expression and prognostic value of OTU domain-containing proteins in non-small-cell lung cancer. Oncol. Lett. 2019, 18, 5417–5427. [Google Scholar] [CrossRef] [Green Version]
- Cronin, N.B.; Yang, J.; Zhang, Z.; Kulkarni, K.; Chang, L.; Yamano, H.; Barford, D. Atomic-Resolution Structures of the APC/C Subunits Apc4 and the Apc5 N-Terminal Domain. J. Mol. Biol. 2015, 427, 3300–3315. [Google Scholar] [CrossRef] [Green Version]
- Agilent-026652 Whole Human Genome Microarray 4x44K v2. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL13497 (accessed on 12 February 2020).
- Statistical Significance of the Pearson Correlation Coefficient. Available online: https://www.youtube.com/watch?v=Kc3M5x7125A (accessed on 12 November 2020).
- Choueiri, T.K.; Motzer, R.J. Systemic Therapy for Metastatic Renal-Cell Carcinoma. N. Engl. J. Med. 2017, 376, 354–366. [Google Scholar] [CrossRef]
- Zbar, B.; Klausner, R.; Linehan, W.M. Studying Cancer Families to Identify Kidney Cancer Genes. Annu. Rev. Med. 2003, 54, 217–233. [Google Scholar] [CrossRef]
- Di Nunno, V.; Mollica, V.; Brunelli, M.; Gatto, L.; Schiavina, R.; Fiorentino, M.; Santoni, M.; Montironi, R.; Caliò, A.; Eccher, A.; et al. A Meta-Analysis Evaluating Clinical Outcomes of Patients with Renal Cell Carcinoma Harboring Chromosome 9P Loss. Mol. Diagn. Ther. 2019, 23, 569–577. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacobas, D.A.; Mgbemena, V.E.; Iacobas, S.; Menezes, K.M.; Wang, H.; Saganti, P.B. Genomic Fabric Remodeling in Metastatic Clear Cell Renal Cell Carcinoma (ccRCC): A New Paradigm and Proposal for a Personalized Gene Therapy Approach. Cancers 2020, 12, 3678. https://doi.org/10.3390/cancers12123678
Iacobas DA, Mgbemena VE, Iacobas S, Menezes KM, Wang H, Saganti PB. Genomic Fabric Remodeling in Metastatic Clear Cell Renal Cell Carcinoma (ccRCC): A New Paradigm and Proposal for a Personalized Gene Therapy Approach. Cancers. 2020; 12(12):3678. https://doi.org/10.3390/cancers12123678
Chicago/Turabian StyleIacobas, Dumitru A., Victoria E. Mgbemena, Sanda Iacobas, Kareena M. Menezes, Huichen Wang, and Premkumar B. Saganti. 2020. "Genomic Fabric Remodeling in Metastatic Clear Cell Renal Cell Carcinoma (ccRCC): A New Paradigm and Proposal for a Personalized Gene Therapy Approach" Cancers 12, no. 12: 3678. https://doi.org/10.3390/cancers12123678
APA StyleIacobas, D. A., Mgbemena, V. E., Iacobas, S., Menezes, K. M., Wang, H., & Saganti, P. B. (2020). Genomic Fabric Remodeling in Metastatic Clear Cell Renal Cell Carcinoma (ccRCC): A New Paradigm and Proposal for a Personalized Gene Therapy Approach. Cancers, 12(12), 3678. https://doi.org/10.3390/cancers12123678






