Exploring the Role of Osteosarcoma-Derived Extracellular Vesicles in Pre-Metastatic Niche Formation and Metastasis in the 143-B Xenograft Mouse Osteosarcoma Model
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
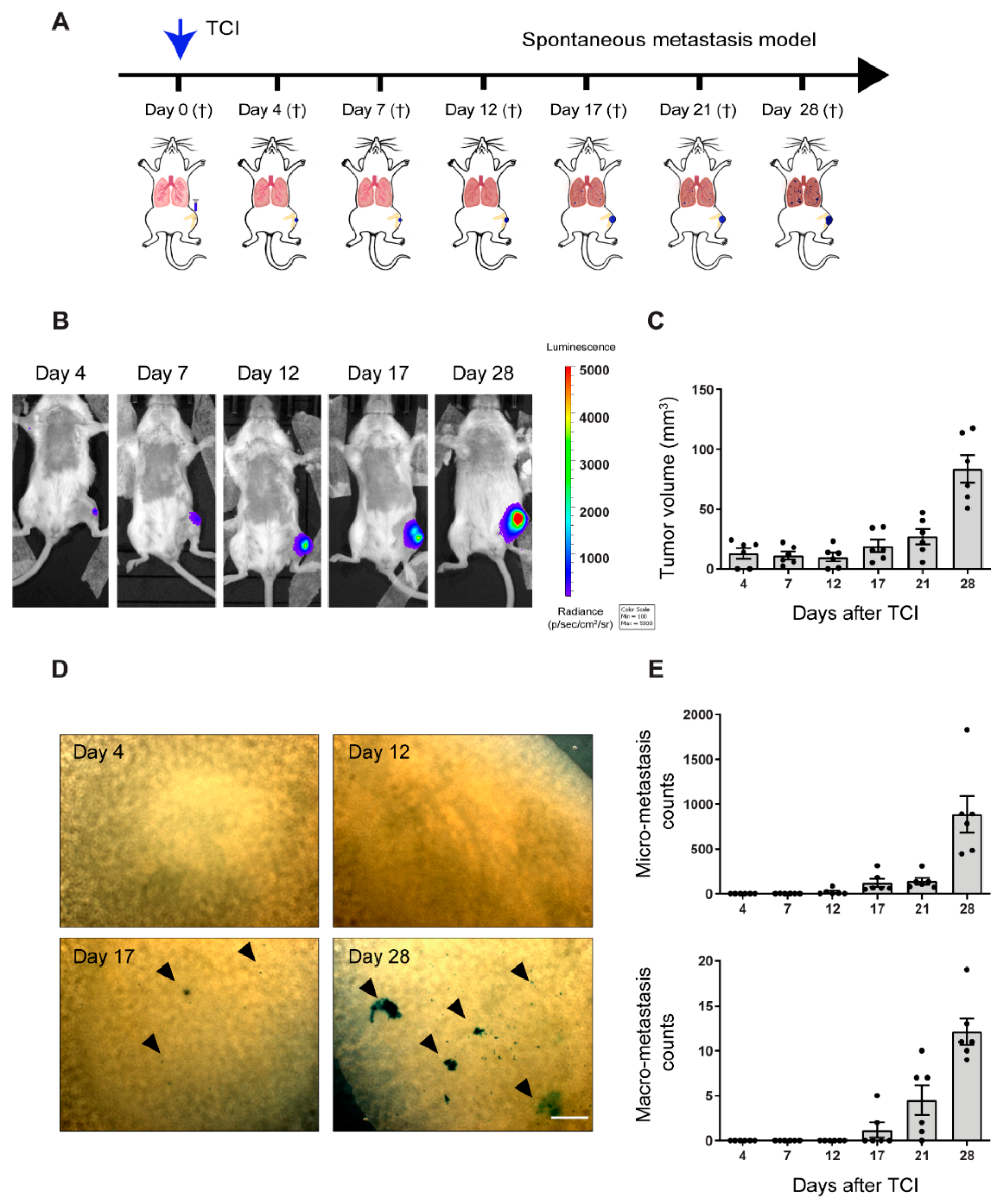
2.1. Pre-Metastatic Phase Determination in the Orthotopic 143-B Osteosarcoma Model in SCID Mice
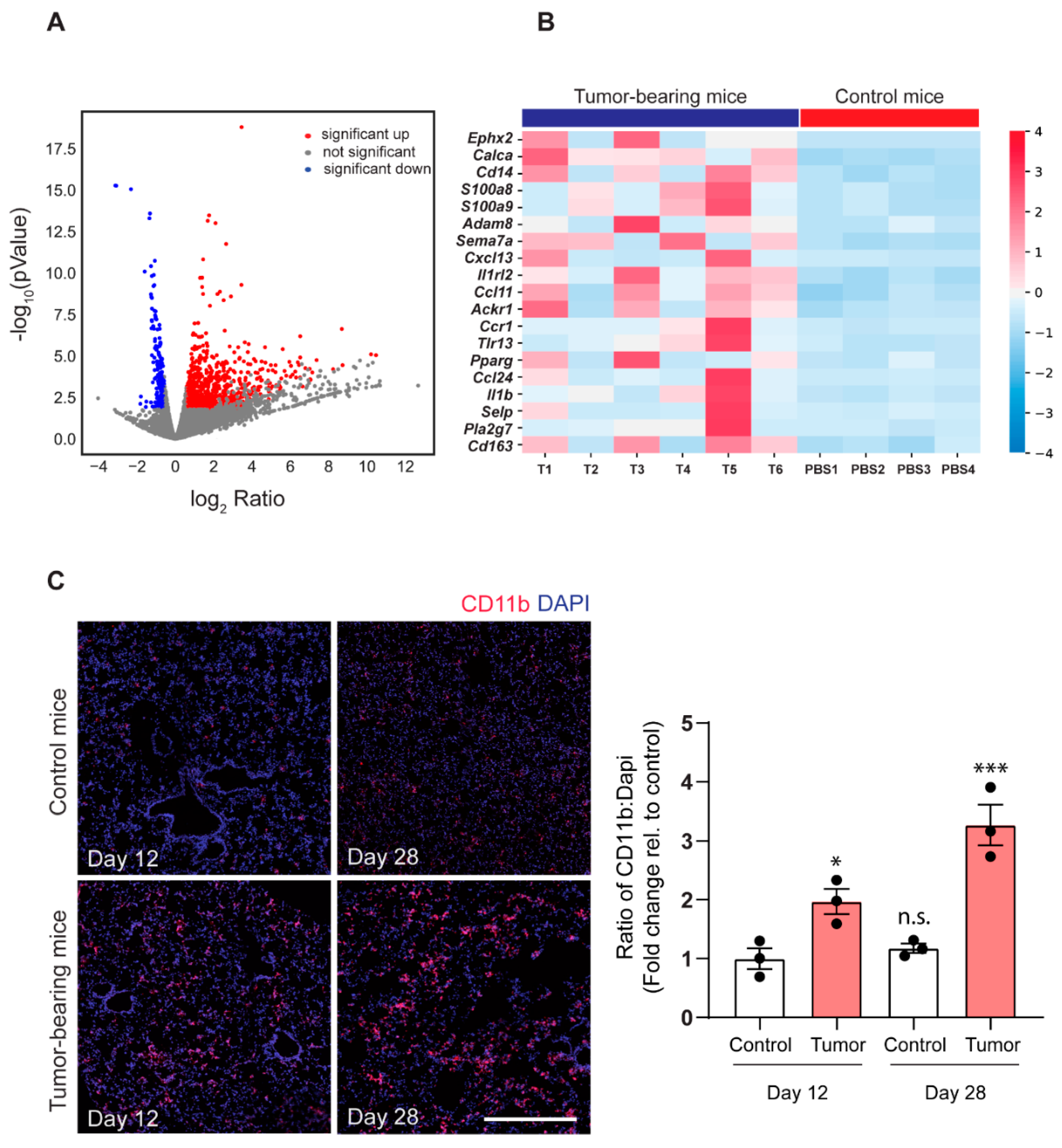
2.2. 143-B Osteosarcoma Primary Tumors Drive Immune Cell Infiltration into the Pre-Metastatic Lungs
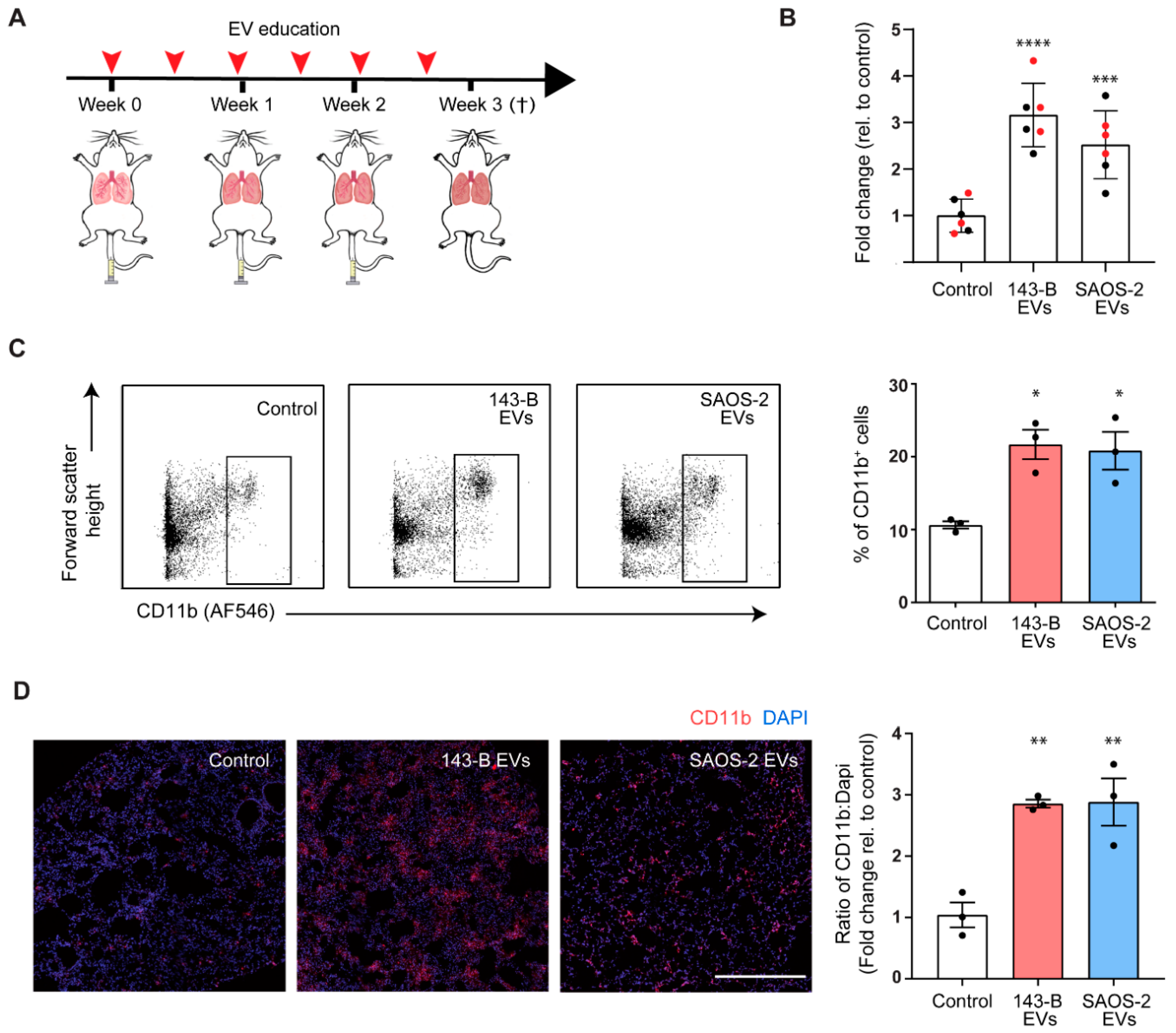
2.3. Osteosarcoma Cell-Derived EV Education Drives Inflammatory Myeloid Cell Infiltration into Lungs of Naïve Mice
2.4. Osteosarcoma Cell-Derived EV Education Does Not Alter the Primary Tumor Growth and Metastasis of Osteosarcoma Cells in a Spontaneous Metastatic 143-B Mouse Model
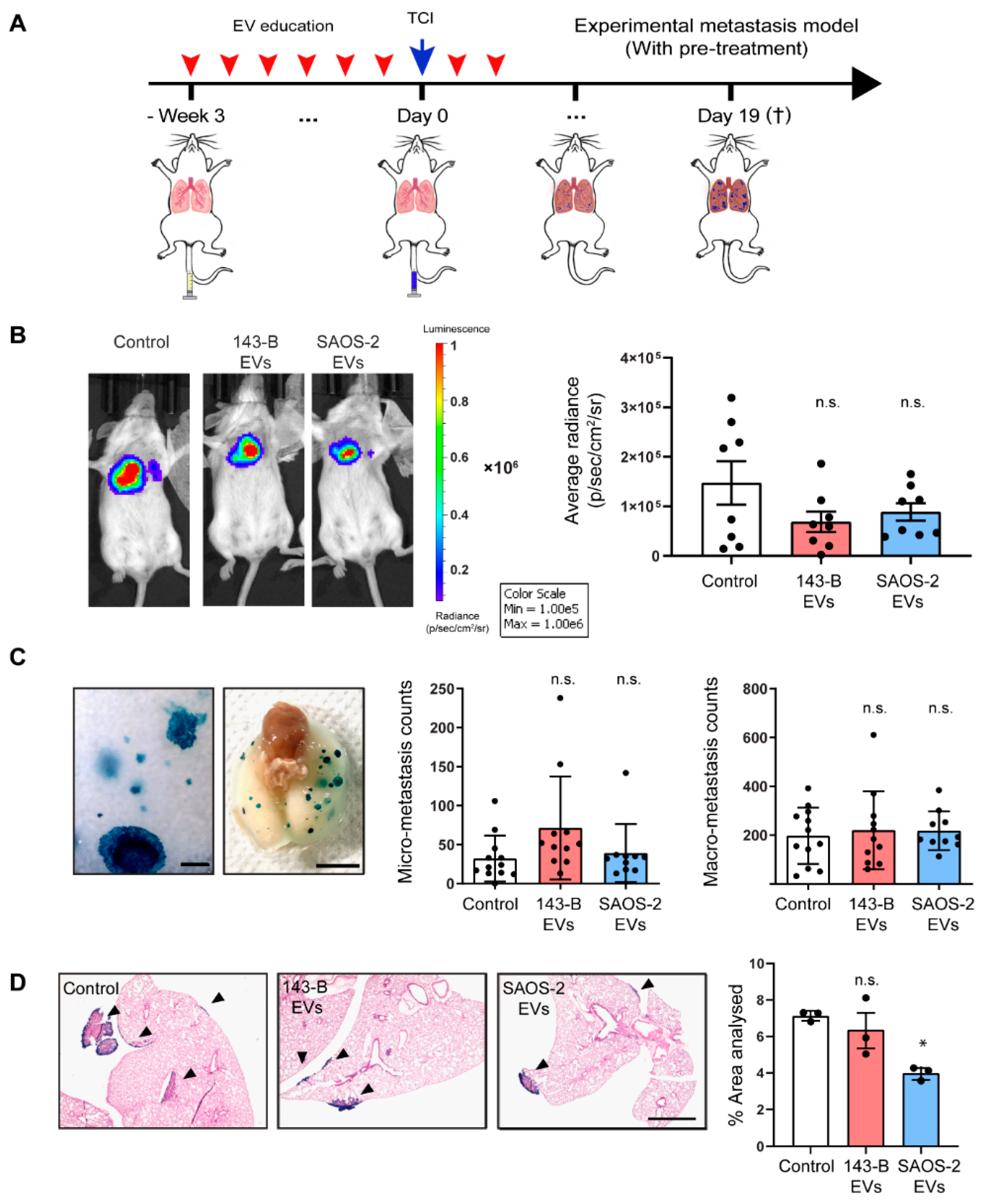
2.5. Osteosarcoma Cell-Derived EV Education Does Not Alter the Experimental Metastatic Potential of the 143-B Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. EV Isolation, Fibercell Hollow-Fiber Bioreactor Culture and Bioreactor EV Purification
4.3. Animal Care and Orthotopic Induction of 143-B Osteosarcoma Tumors in SCID Mice
4.4. Ex Vivo Visualization of LacZ-Transduced Metastatic Tumor Cells in the Lung
4.5. RNA Sequencing, Data Analysis and qRT-PCR
4.6. EV Education and Orthotopic/Experimental Metastasis Induction of Osteosarcoma Tumors in SCID Mice
4.7. Flow Cytometry Analysis of Lung Tissues
4.8. Immunoflurescent and Histological Tissue Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| EVs | Extracellular vesicles |
| MDSCs | Myeloid-derived suppressor cells |
| PMN | Pre-metastatic niche |
| SCID | Severe-combined immunodeficient mice |
| SEM | Standard error of the mean |
| TCI | Tumor cell injection |
| ECM | Extracellular matrix |
| GO | Gene ontology |
References
- Picci, P. Osteosarcoma (Osteogenic sarcoma). Orphanet J. Rare Dis. 2007, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Chambers, A.F.; Groom, A.C.; Macdonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Fidler, I.J. The pathogenesis of cancer metastasis: The ’seed and soil’ hypothesis revisited. Nat. Rev. Cancer 2003, 3, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Langley, R.R.; Fidler, I.J. Tumor Cell-Organ Microenvironment Interactions in the Pathogenesis of Cancer Metastasis. Endocr. Rev. 2007, 28, 297–321. [Google Scholar] [CrossRef]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef]
- Psaila, B.; Lyden, D. The metastatic niche: Adapting the foreign soil. Nat. Rev. Cancer 2009, 9, 285–293. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell 2016, 30, 668–681. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, Y.; Han, Y.; Zhang, Q.; Jiang, Z.; Zhang, X.; Huang, B.; Xu, X.; Zheng, J.; Cao, X. Tumor Exosomal RNAs Promote Lung Pre-metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell 2016, 30, 243–256. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.M.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-Secreted miR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Chicón-Bosch, M.; Tirado, O.M. Exosomes in Bone Sarcomas: Key Players in Metastasis. Cells 2020, 9, 241. [Google Scholar] [CrossRef]
- Jerez, S.; Araya, H.; Thaler, R.; Charlesworth, M.C.; López-Solís, R.; Kalergis, A.M.; Céspedes, P.F.; Dudakovic, A.; Stein, G.S.; Van Wijnen, A.J.; et al. Proteomic Analysis of Exosomes and Exosome-Free Conditioned Media From Human Osteosarcoma Cell Lines Reveals Secretion of Proteins Related to Tumor Progression. J. Cell. Biochem. 2017, 118, 351–360. [Google Scholar] [CrossRef]
- Garimella, R.; Washington, L.; Isaacson, J.; Vallejo, J.; Spence, M.; Tawfik, O.; Rowe, P.; Brotto, M.; Perez, R. Extracellular Membrane Vesicles Derived from 143B Osteosarcoma Cells Contain Pro-Osteoclastogenic Cargo: A Novel Communication Mechanism in Osteosarcoma Bone Microenvironment. Transl. Oncol. 2014, 7, 331–340. [Google Scholar] [CrossRef]
- Raimondi, L.; De Luca, A.; Gallo, A.; Costa, V.; Russelli, G.; Cuscino, N.; Manno, M.; Raccosta, S.; Carina, V.; Bellavia, D.; et al. Osteosarcoma cell-derived exosomes affect tumor microenvironment by specific packaging of microRNAs. Carcinogenesis 2020, 41, 666–677. [Google Scholar] [CrossRef]
- Gong, L.; Bao, Q.; Hu, C.; Wang, J.; Zhou, Q.; Wei, L.; Tong, L.; Zhang, W.; Shen, Y. Exosomal miR-675 from metastatic osteosarcoma promotes cell migration and invasion by targeting CALN1. Biochem. Biophys. Res. Commun. 2018, 500, 170–176. [Google Scholar] [CrossRef]
- Mazumdar, A.; Urdinez, J.; Boro, A.; Migliavacca, J.; Arlt, M.J.; Muff, R.; Fuchs, B.; Snedeker, J.G.; Gvozdenovic, A. Osteosarcoma-Derived Extracellular Vesicles Induce Lung Fibroblast Reprogramming. Int. J. Mol. Sci. 2020, 21, 5451. [Google Scholar] [CrossRef]
- Baglio, S.R.; Lagerweij, T.; Pérez-Lanzón, M.; Ho, X.D.; Léveillé, N.; Melo, S.A.; Cleton-Jansen, A.-M.; Jordanova, E.S.; Roncuzzi, L.; Greco, M.; et al. Blocking Tumor-Educated MSC Paracrine Activity Halts Osteosarcoma Progression. Clin. Cancer Res. 2017, 23, 3721–3733. [Google Scholar] [CrossRef]
- Macklin, R.; Wang, H.; Loo, D.; Martin, S.; Cumming, A.; Cai, N.; Lane, R.; Ponce, N.S.; Topkas, E.; Inder, K.; et al. Extracellular vesicles secreted by highly metastatic clonal variants of osteosarcoma preferentially localize to the lungs and induce metastatic behaviour in poorly metastatic clones. Oncotarget 2016, 7, 43570–43587. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, P.; Arlt, M.J.E.; Campanile, C.; Husmann, K.; Gvozdenovic, A.; Apuzzo, T.; Thelen, M.; Born, W.; Fuchs, B. CXCR4 antibody treatment suppresses metastatic spread to the lung of intratibial human osteosarcoma xenografts in mice. Clin. Exp. Metastasis 2014, 31, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Rankin, E.B.; Giaccia, A.J. Hypoxic control of metastasis. Science 2016, 352, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Kusmartsev, S.; Eruslanov, E.; Kübler, H.; Tseng, T.; Sakai, Y.; Su, Z.; Kaliberov, S.; Heiser, A.; Rosser, C.; Dahm, P.; et al. Oxidative Stress Regulates Expression of VEGFR1 in Myeloid Cells: Link to Tumor-Induced Immune Suppression in Renal Cell Carcinoma. J. Immunol. 2008, 181, 346–353. [Google Scholar] [CrossRef]
- Sceneay, J.; Chow, M.T.; Chen, A.; Halse, H.M.; Wong, C.S.; Andrews, D.M.; Sloan, E.K.; Parker, B.S.; Bowtell, D.D.; Smyth, M.J. Primary tumor hypoxia recruits CD11b+/Ly6Cmed/Ly6G+ immune suppressor cells and compromises NK cell cytotoxicity in the premetastatic niche. Cancer Res. 2012, 72, 3906–3911. [Google Scholar] [CrossRef]
- Yang, L.; Edwards, C.M.; Mundy, G.R. Gr-1+CD11b+ myeloid-derived suppressor cells: Formidable partners in tumor metastasis. J. Bone Miner. Res. 2010, 25, 1701–1706. [Google Scholar] [CrossRef]
- Yan, H.H.; Pickup, M.; Pang, Y.; Gorska, A.E.; Li, Z.; Chytil, A.; Geng, Y.; Gray, J.W.; Moses, H.L.; Yang, L. Gr-1+CD11b+ Myeloid Cells Tip the Balance of Immune Protection to Tumor Promotion in the Premetastatic Lung. Cancer Res. 2010, 70, 6139–6149. [Google Scholar] [CrossRef]
- Plebanek, M.P.; Angeloni, N.L.; Vinokour, E.; Dalia, M.-M.; Henkin, A.; Martinez-Marin, D.; Filleur, S.; Bhowmick, R.; Henkin, J.; Miller, S.D.; et al. Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Hiratsuka, S.; Watanabe, A.; Aburatani, H.; Maru, Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat. Cell Biol. 2006, 8, 1369–1375. [Google Scholar] [CrossRef]
- Liu, Y.; Xiang, X.; Zhuang, X.; Zhang, S.; Liu, C.; Cheng, Z.; Michalek, S.; Grizzle, W.; Zhang, H.-G. Contribution of MyD88 to the Tumor Exosome-Mediated Induction of Myeloid Derived Suppressor Cells. Am. J. Pathol. 2010, 176, 2490–2499. [Google Scholar] [CrossRef]
- Ortiz, A.; Gui, J.; Zahedi, F.; Yu, P.; Cho, C.; Bhattacharya, S.; Carbone, C.J.; Yu, Q.; Katlinski, K.V.; Katlinskaya, Y.V.; et al. An Interferon-Driven Oxysterol-Based Defense against Tumor-Derived Extracellular Vesicles. Cancer Cell 2019, 35, 33–45.e6. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, R.N.; Riba, R.D.; Zacharoulis, S.; Bramley, A.H.; Vincent, L.; Costa, C.; Macdonald, D.D.; Jin, D.K.; Shido, K.; Kerns, S.A.; et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 2005, 438, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zou, J.; Li, S.; Topper, M.J.; Tao, Y.; Zhang, H.; Jiao, X.; Xie, W.; Kong, X.; Vaz, M.; et al. Epigenetic therapy inhibits metastases by disrupting premetastatic niches. Nature 2020, 579, 284–290. [Google Scholar] [CrossRef]
- Granot, Z.; Henke, E.; Comen, E.A.; King, T.A.; Norton, L.; Benezra, R. Tumor Entrained Neutrophils Inhibit Seeding in the Premetastatic Lung. Cancer Cell 2011, 20, 300–314. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Liu, Y.; Lee, H.; Herrmann, A.; Zhang, W.; Zhang, C.; Shen, S.; Priceman, S.J.; Kujawski, M.; Pal, S.K.; et al. S1PR1-STAT3 Signaling Is Crucial for Myeloid Cell Colonization at Future Metastatic Sites. Cancer Cell 2012, 21, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Ostrand-Rosenberg, S.; Sinha, P. Myeloid-Derived Suppressor Cells: Linking Inflammation and Cancer. J. Immunol. 2009, 182, 4499–4506. [Google Scholar] [CrossRef]
- Jiang, K.; Li, J.; Zhang, J.; Wang, L.; Zhang, Q.; Ge, J.; Guo, Y.; Wang, B.; Huang, Y.; Yang, T. SDF-1/CXCR4 axis facilitates myeloid-derived suppressor cells accumulation in osteosarcoma microenvironment and blunts the response to anti-PD-1 therapy. Int. Immunopharmacol. 2019, 75, 105818. [Google Scholar] [CrossRef]
- Kansara, M.; Thomson, K.; Pang, P.; Dutour, A.; Mirabello, L.; Acher, F.; Pin, J.-P.; Demicco, E.G.; Yan, J.; Teng, M.; et al. Infiltrating Myeloid Cells Drive Osteosarcoma Progression via GRM4 Regulation of IL23. Cancer Discov. 2019, 9, 1511–1519. [Google Scholar] [CrossRef]
- Charan, M.; Dravid, P.; Cam, M.; Setty, B.; Roberts, R.D.; Houghton, P.J.; Cam, H. Tumor secreted ANGPTL2 facilitates recruitment of neutrophils to the lung to promote lung pre-metastatic niche formation and targeting ANGPTL2 signaling affects metastatic disease. Oncotarget 2020, 11, 510–522. [Google Scholar] [CrossRef]
- Uehara, T.; Eikawa, S.; Nishida, M.; Kunisada, Y.; Yoshida, A.; Fujiwara, T.; Kunisada, T.; Ozaki, T.; Udono, H. Metformin induces CD11b+-cell-mediated growth inhibition of an osteosarcoma: Implications for metabolic reprogramming of myeloid cells and anti-tumor effects. Int. Immunol. 2019, 31, 187–198. [Google Scholar] [CrossRef]
- Bajpai, J.; Sharma, M.; Sreenivas, V.; Kumar, R.; Gamnagatti, S.; Alam Khan, S.; Rastogi, S.; Malhotra, A.; Bakhshi, S. VEGF expression as a prognostic marker in osteosarcoma. Pediatric Blood Cancer 2009, 53, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- LaMora, A.; Talbot, J.; Bougras, G.; Amiaud, J.; LeDuc, M.; Chesneau, J.; Taurelle, J.; Stresing, V.; Le Deley, M.C.; Heymann, M.F.; et al. Overexpression of Smad7 Blocks Primary Tumor Growth and Lung Metastasis Development in Osteosarcoma. Clin. Cancer Res. 2014, 20, 5097–5112. [Google Scholar] [CrossRef] [PubMed]
- Peng, N.; Gao, S.; Guo, X.; Wang, G.; Cheng, C.; Li, M.; Liu, K. Silencing of VEGF inhibits human osteosarcoma angiogenesis and promotes cell apoptosis via VEGF/PI3K/AKT signaling pathway. Am. J. Transl. Res. 2016, 8, 1005–1015. [Google Scholar] [PubMed]
- Liu, T.; Ma, Q.; Zhang, Y.; Wang, X.; Xu, K.; Yan, K.; Dong, W.; Fan, Q.; Zhang, Y.; Qiu, X. Self-seeding circulating tumor cells promote the proliferation and metastasis of human osteosarcoma by upregulating interleukin-8. Cell Death Dis. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gvozdenovic, A.; Boro, A.; Meier, D.; Bode-Lesniewska, B.; Born, W.; Muff, R.; Fuchs, B. Targeting αvβ3 and αvβ5 integrins inhibits pulmonary metastasis in an intratibial xenograft osteosarcoma mouse model. Oncotarget 2016, 7, 55141–55154. [Google Scholar] [CrossRef] [PubMed]
- Trovato, R.; Canè, S.; Petrova, V.; Sartoris, S.; Ugel, S.; De Sanctis, F. The Engagement Between MDSCs and Metastases: Partners in Crime. Front. Oncol. 2020, 10, 165. [Google Scholar] [CrossRef]
- Sharma, S.K.; Chintala, N.K.; Vadrevu, S.K.; Patel, J.; Karbowniczek, M.; Markiewski, M.M. Pulmonary Alveolar Macrophages Contribute to the Premetastatic Niche by Suppressing Antitumor T Cell Responses in the Lungs. J. Immunol. 2015, 194, 5529–5538. [Google Scholar] [CrossRef]
- Wolf-Dennen, K.; Gordon, N.; Kleinerman, E.S. Exosomal communication by metastatic osteosarcoma cells modulates alveolar macrophages to an M2 tumor-promoting phenotype and inhibits tumoricidal functions. OncoImmunology 2020, 9, 1747677. [Google Scholar] [CrossRef]
- Behjati, S.; Tarpey, P.S.; Haase, K.; Ye, H.; Young, M.D.; Alexandrov, L.B.; Farndon, S.J.; Collord, G.; Wedge, D.C.; Martincorena, I.; et al. Recurrent mutation of IGF signalling genes and distinct patterns of genomic rearrangement in osteosarcoma. Nat. Commun. 2017, 8, 15936. [Google Scholar] [CrossRef]
- Morrow, J.J.; Bayles, I.; Funnell, A.P.W.; Miller, T.E.; Saiakhova, A.; Lizardo, M.M.; Bartels, C.F.; Kapteijn, M.Y.; Hung, S.; Mendoza, A.; et al. Positively selected enhancer elements endow osteosarcoma cells with metastatic competence. Nat. Med. 2018, 24, 176–185. [Google Scholar] [CrossRef]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed]
- Gvozdenovic, A.; Arlt, M.J.; Campanile, C.; Brennecke, P.; Husmann, K.; Li, Y.; Born, W.; Muff, R.; Fuchs, B. CD44 enhances tumor formation and lung metastasis in experimental osteosarcoma and is an additional predictor for poor patient outcome. J. Bone Miner. Res. 2013, 28, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Robl, B.; Botter, S.M.; Boro, A.; Meier, D.; Neri, D.; Fuchs, B. Evaluation of F8-TNF-α in Models of Early and Progressive Metastatic Osteosarcoma. Transl. Oncol. 2017, 10, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Husmann, K.; Arlt, M.J.; Jirkof, P.; Arras, M.; Born, W.; Fuchs, B. Primary tumour growth in an orthotopic osteosarcoma mouse model is not influenced by analgesic treatment with buprenorphine and meloxicam. Lab. Anim. 2015, 49, 284–293. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef]
- Arlt, M.J.; Banke, I.J.; Walters, D.K.; Puskas, G.J.; Steinmann, P.; Muff, R.; Born, W.; Fuchs, B. LacZ transgene expression in the subcutaneous Dunn/LM8 osteosarcoma mouse model allows for the identification of micrometastasis. J. Orthop. Res. 2011, 29, 938–946. [Google Scholar] [CrossRef]
- Urdinez, J.; Boro, A.; Mazumdar, A.; Arlt, M.J.; Muff, R.; Botter, S.M.; Bode-Lesniewska, B.; Fuchs, B.; Snedeker, J.G.; Gvozdenovic, A. The miR-143/145 Cluster, a Novel Diagnostic Biomarker in Chondrosarcoma, Acts as a Tumor Suppressor and Directly Inhibits Fascin-1. J. Bone Miner. Res. 2020, 35, 1077–1091. [Google Scholar] [CrossRef]
- Hatakeyama, M.; Opitz, L.; Russo, G.; Qi, W.; Schlapbach, R.; Rehrauer, H. SUSHI: An exquisite recipe for fully documented, reproducible and reusable NGS data analysis. BMC Bioinform. 2016, 17, 228. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Müllner, D. fastcluster: Fast hierarchical, agglomerative clustering routines for R and Python. J. Stat. Softw. 2013, 53, 1–18. [Google Scholar] [CrossRef]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference (SciPy), Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar]
- Waskom, M.; Botvinnik, O.; O’Kane, D.; Hobson, P.; Ostblom, J.; Lukauskas, S.; Gemperline, D.C.; Augspurger, T.; Halchenko, Y.; Cole, J.B. Mwaskom/Seaborn: V0. 9.0, version v0. 9.0; Zenodo: Meyrin, Switzerland, 2018. [Google Scholar]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.W.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Robl, B.; Botter, S.M.; Pellegrini, G.; Neklyudova, O.; Fuchs, B. Evaluation of intraarterial and intravenous cisplatin chemotherapy in the treatment of metastatic osteosarcoma using an orthotopic xenograft mouse model. J. Exp. Clin. Cancer Res. 2016, 35, 113. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazumdar, A.; Urdinez, J.; Boro, A.; Arlt, M.J.E.; Egli, F.E.; Niederöst, B.; Jaeger, P.K.; Moschini, G.; Muff, R.; Fuchs, B.; et al. Exploring the Role of Osteosarcoma-Derived Extracellular Vesicles in Pre-Metastatic Niche Formation and Metastasis in the 143-B Xenograft Mouse Osteosarcoma Model. Cancers 2020, 12, 3457. https://doi.org/10.3390/cancers12113457
Mazumdar A, Urdinez J, Boro A, Arlt MJE, Egli FE, Niederöst B, Jaeger PK, Moschini G, Muff R, Fuchs B, et al. Exploring the Role of Osteosarcoma-Derived Extracellular Vesicles in Pre-Metastatic Niche Formation and Metastasis in the 143-B Xenograft Mouse Osteosarcoma Model. Cancers. 2020; 12(11):3457. https://doi.org/10.3390/cancers12113457
Chicago/Turabian StyleMazumdar, Alekhya, Joaquin Urdinez, Aleksandar Boro, Matthias J. E. Arlt, Fabian E. Egli, Barbara Niederöst, Patrick K. Jaeger, Greta Moschini, Roman Muff, Bruno Fuchs, and et al. 2020. "Exploring the Role of Osteosarcoma-Derived Extracellular Vesicles in Pre-Metastatic Niche Formation and Metastasis in the 143-B Xenograft Mouse Osteosarcoma Model" Cancers 12, no. 11: 3457. https://doi.org/10.3390/cancers12113457
APA StyleMazumdar, A., Urdinez, J., Boro, A., Arlt, M. J. E., Egli, F. E., Niederöst, B., Jaeger, P. K., Moschini, G., Muff, R., Fuchs, B., Snedeker, J. G., & Gvozdenovic, A. (2020). Exploring the Role of Osteosarcoma-Derived Extracellular Vesicles in Pre-Metastatic Niche Formation and Metastasis in the 143-B Xenograft Mouse Osteosarcoma Model. Cancers, 12(11), 3457. https://doi.org/10.3390/cancers12113457






