Cancer and pH Dynamics: Transcriptional Regulation, Proteostasis, and the Need for New Molecular Tools
Abstract
Simple Summary
Abstract
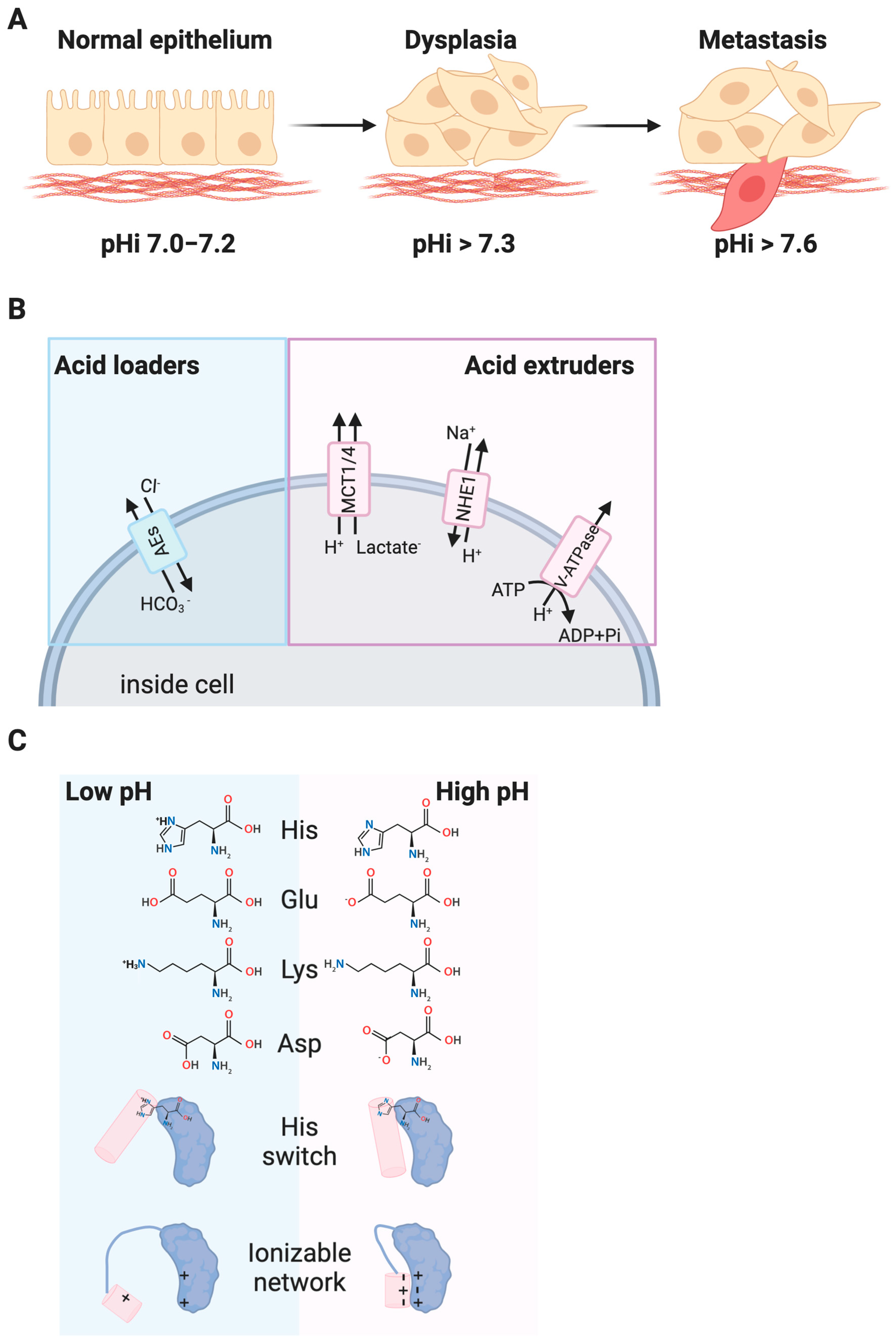
1. Introduction
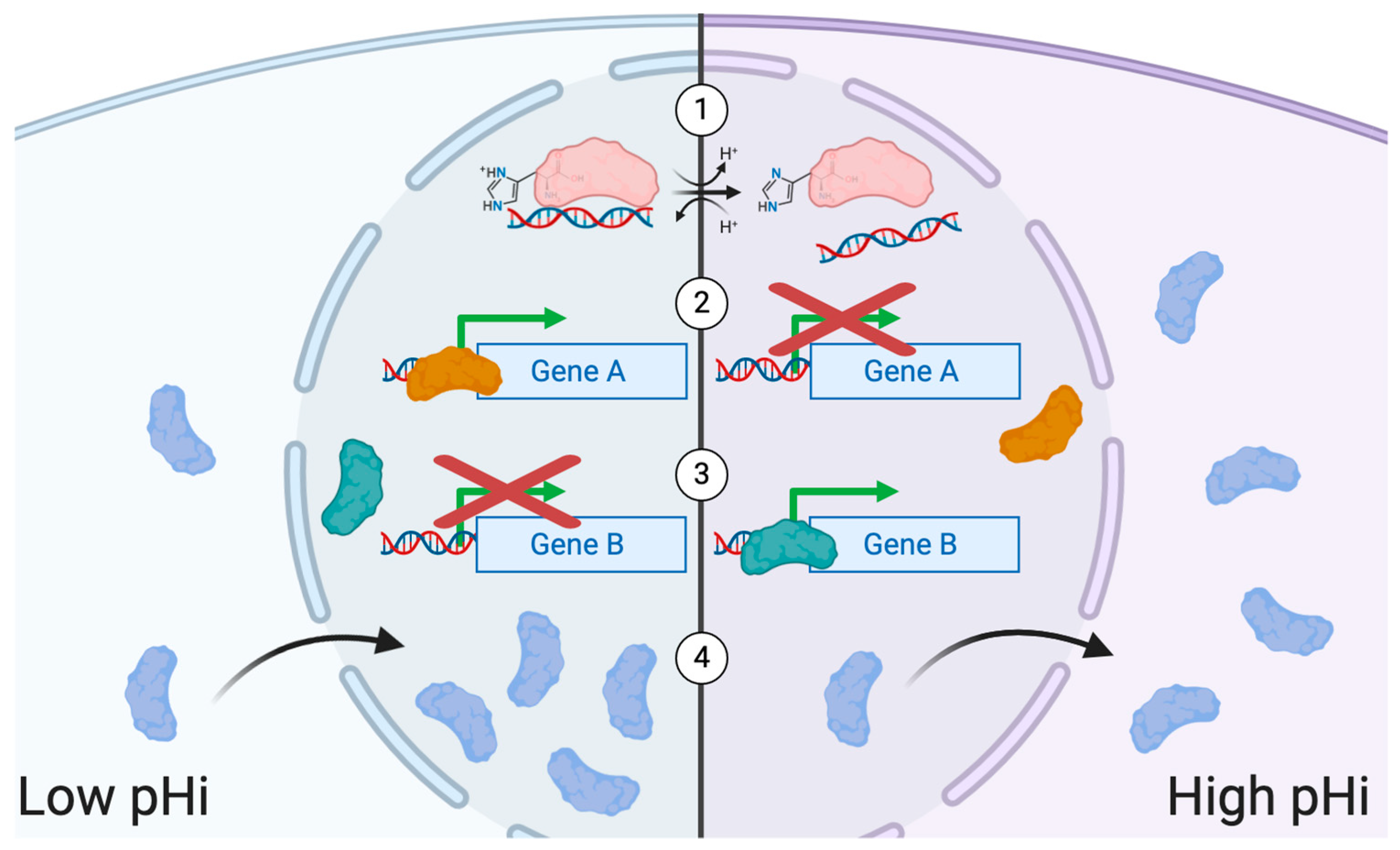
2. Transcriptional Regulation and pH
3. Heterogeneity and pHi
4. Relationship between Transcript and Protein Abundance
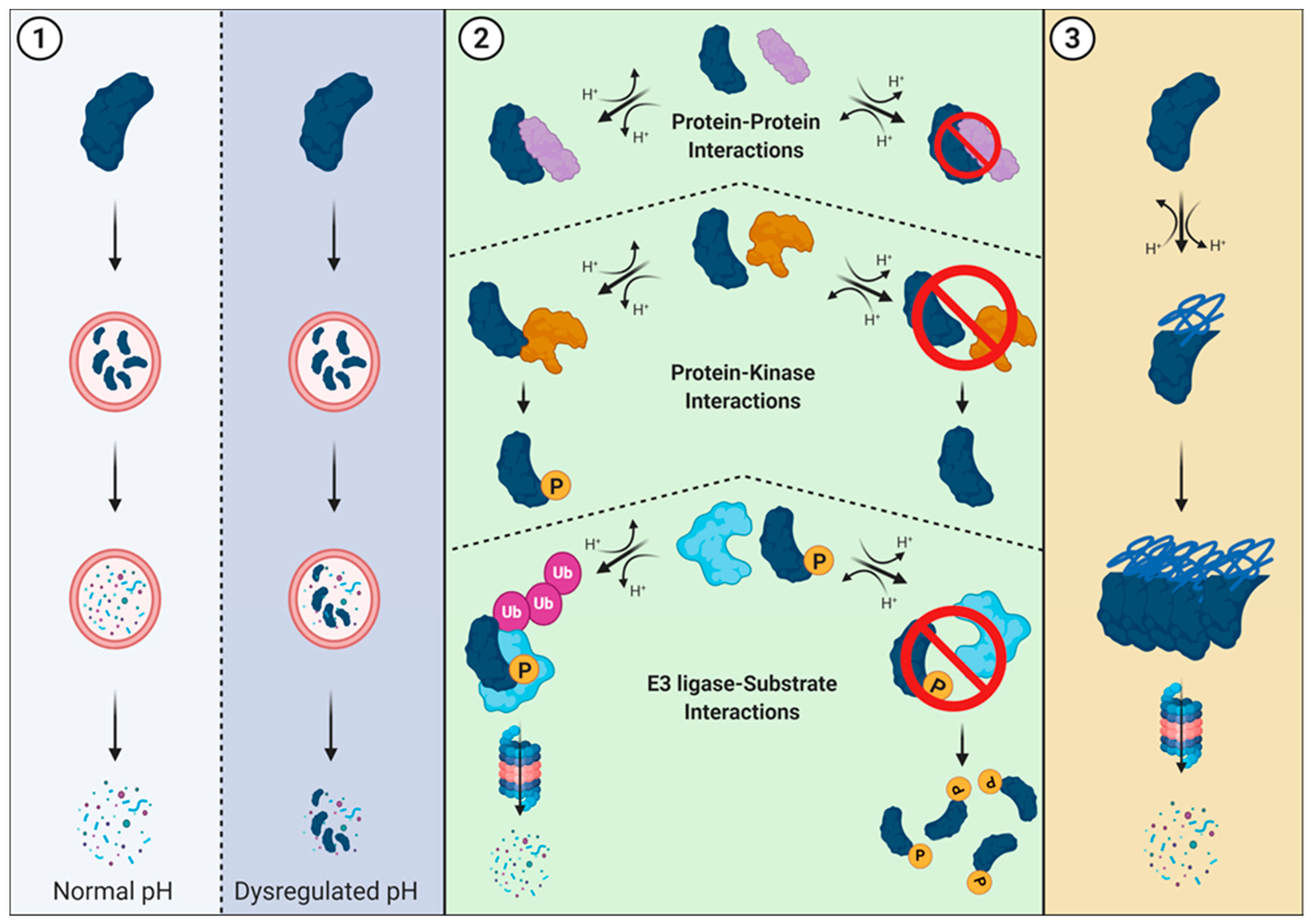
5. Tumorigenesis, Proteostasis, and pHi
6. Proteasome-Mediated Degradation and pHi
7. Roles for Proteostasis in Tumorigenesis
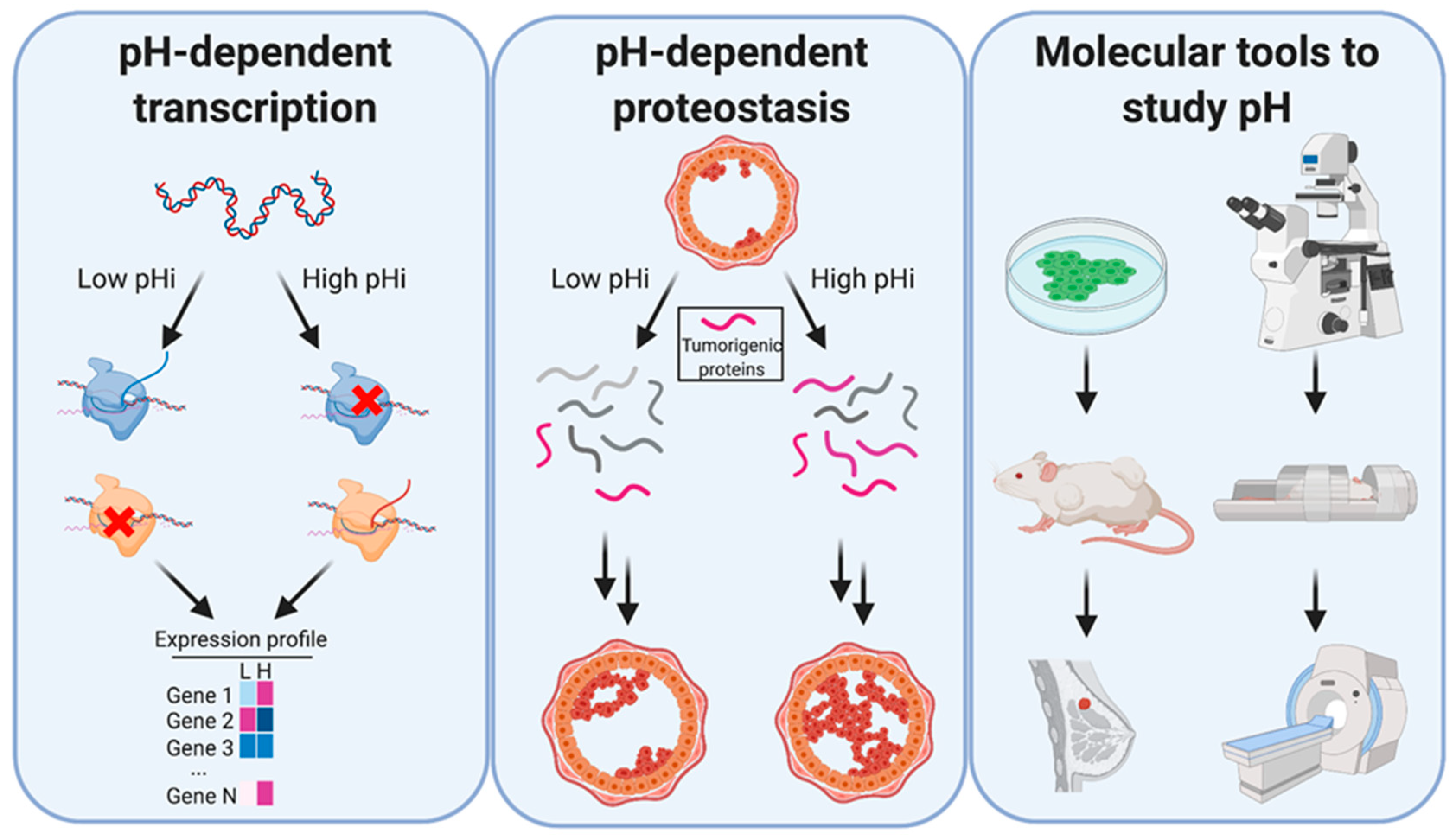
8. Tool Development and New Horizons
9. Tools to Measure pHi
10. Tools to Manipulate pHi
11. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef]
- Gillies, R.J.; Verduzco, D.; Gatenby, R.A. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat. Rev. Cancer 2012, 12, 487–493. [Google Scholar] [CrossRef]
- Ramachandran, S.; Ient, J.; Gottgens, E.L.; Krieg, A.J.; Hammond, E.M. Epigenetic Therapy for Solid Tumors: Highlighting the Impact of Tumor Hypoxia. Genes 2015, 6, 935–956. [Google Scholar] [CrossRef] [PubMed]
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85–95. [Google Scholar] [CrossRef]
- Brahimi-Horn, M.C.; Bellot, G.; Pouyssegur, J. Hypoxia and energetic tumour metabolism. Curr. Opin. Genet. Dev. 2011, 21, 67–72. [Google Scholar] [CrossRef]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef]
- Reshkin, S.J.; Bellizzi, A.; Caldeira, S.; Albarani, V.; Malanchi, I.; Poignee, M.; Alunni-Fabbroni, M.; Casavola, V.; Tommasino, M. Na+/H+ exchanger-dependent intracellular alkalinization is an early event in malignant transformation and plays an essential role in the development of subsequent transformation-associated phenotypes. FASEB J. 2000, 14, 2185–2197. [Google Scholar] [CrossRef]
- Grillo-Hill, B.K.; Choi, C.; Jimenez-Vidal, M.; Barber, D.L. Increased H(+) efflux is sufficient to induce dysplasia and necessary for viability with oncogene expression. Elife 2015, 4, e03270. [Google Scholar] [CrossRef]
- Gorbatenko, A.; Olesen, C.W.; Boedtkjer, E.; Pedersen, S.F. Regulation and roles of bicarbonate transporters in cancer. Front. Physiol. 2014, 5, 130. [Google Scholar] [CrossRef]
- Putney, L.K.; Denker, S.P.; Barber, D.L. The Changing Face of the Na+/H+ Exchanger, NHE1: Structure, Regulation, and Cellular Actions. Ann. Rev. Pharmacol. Toxicol. 2002, 42, 527–552. [Google Scholar] [CrossRef]
- De Saedeleer, C.J.; Porporato, P.E.; Copetti, T.; Perez-Escuredo, J.; Payen, V.L.; Brisson, L.; Feron, O.; Sonveaux, P. Glucose deprivation increases monocarboxylate transporter 1 (MCT1) expression and MCT1-dependent tumor cell migration. Oncogene 2014, 33, 4060–4068. [Google Scholar] [CrossRef] [PubMed]
- Stransky, L.; Cotter, K.; Forgac, M. The Function of V-ATPases in Cancer. Physiol. Rev. 2016, 96, 1071–1091. [Google Scholar] [CrossRef] [PubMed]
- Pamarthy, S.; Kulshrestha, A.; Katara, G.K.; Beaman, K.D. The curious case of vacuolar ATPase: Regulation of signaling pathways. Mol. Cancer 2018, 17, 41. [Google Scholar] [CrossRef] [PubMed]
- Hinton, A.; Bond, S.; Forgac, M. V-ATPase functions in normal and disease processes. Pflugers Arch. Eur. J. Physiol. 2009, 457, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, C.; Hauser, A.D.; Vucic, E.A.; Bar-Sagi, D. Plasma membrane V-ATPase controls oncogenic RAS-induced macropinocytosis. Nature 2019, 576, 477–481. [Google Scholar] [CrossRef]
- Webb, B.A.; Chimenti, M.; Jacobson, M.P.; Barber, D.L. Dysregulated pH: A perfect storm for cancer progression. Nat. Rev. Cancer 2011, 11, 671–677. [Google Scholar] [CrossRef]
- White, K.A.; Grillo-Hill, B.K.; Barber, D.L. Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J. Cell Sci. 2017, 130, 663–669. [Google Scholar] [CrossRef]
- Korenchan, D.E.; Flavell, R.R. Spatiotemporal pH Heterogeneity as a Promoter of Cancer Progression and Therapeutic Resistance. Cancers 2019, 11, 1026. [Google Scholar] [CrossRef]
- Alfarouk, K.O.; Ahmed, S.B.M.; Elliott, R.L.; Benoit, A.; Alqahtani, S.S.; Ibrahim, M.E.; Bashir, A.H.H.; Alhoufie, S.T.S.; Elhassan, G.O.; Wales, C.C.; et al. The Pentose Phosphate Pathway Dynamics in Cancer and Its Dependency on Intracellular pH. Metabolites 2020, 10, 285. [Google Scholar] [CrossRef]
- Cardone, R.A.; Alfarouk, K.O.; Elliott, R.L.; Alqahtani, S.S.; Ahmed, S.B.M.; Aljarbou, A.N.; Greco, M.R.; Cannone, S.; Reshkin, S.J. The Role of Sodium Hydrogen Exchanger 1 in Dysregulation of Proton Dynamics and Reprogramming of Cancer Metabolism as a Sequela. Int. J. Mol. Sci. 2019, 20, 3694. [Google Scholar] [CrossRef]
- Lyssiotis, C.A.; Vander-Heiden, M.G.; Heiden, M.G.V.; Muñoz-Pinedo, C.; Emerling, B.M. Emerging concepts: Linking hypoxic signaling and cancer metabolism. Cell Death Dis. 2012, 3, e303. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, J.; Barreiro, G.; Groscurth, S.; Gingras, A.R.; Goult, B.T.; Critchley, D.R.; Kelly, M.J.; Jacobson, M.P.; Barber, D.L. Structural model and functional significance of pH-dependent talin-actin binding for focal adhesion remodeling. Proc. Natl. Acad. Sci. USA 2008, 105, 14436–14441. [Google Scholar] [CrossRef] [PubMed]
- Frantz, C.; Barreiro, G.; Dominguez, L.; Chen, X.; Eddy, R.; Condeelis, J.; Kelly, M.J.; Jacobson, M.P.; Barber, D.L. Cofilin is a pH sensor for actin free barbed end formation: Role of phosphoinositide binding. J. Cell Biol. 2008, 183, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.H.; Webb, B.A.; Chimenti, M.S.; Jacobson, M.P.; Barber, D.L. pH sensing by FAK-His58 regulates focal adhesion remodeling. J. Cell Biol. 2013, 202, 849–859. [Google Scholar] [CrossRef]
- Isom, D.G.; Sridharan, V.; Baker, R.; Clement, S.T.; Smalley, D.M.; Dohlman, H.G. Protons as second messenger regulators of G protein signaling. Mol. Cell 2013, 51, 531–538. [Google Scholar] [CrossRef]
- Trivedi, B.; Danforth, W.H. Effect of pH on the kinetics of frog muscle phosphofructokinase. J. Biol. Chem. 1966, 241, 4110–4112. [Google Scholar]
- Castaneda, C.A.; Fitch, C.A.; Majumdar, A.; Khangulov, V.; Schlessman, J.L.; Garcia-Moreno, B.E. Molecular determinants of the pKa values of Asp and Glu residues in staphylococcal nuclease. Proteins 2009, 77, 570–588. [Google Scholar] [CrossRef]
- Isom, D.G.; Castaneda, C.A.; Cannon, B.R.; Garcia-Moreno, B. Large shifts in pKa values of lysine residues buried inside a protein. Proc. Natl. Acad. Sci. USA 2011, 108, 5260–5265. [Google Scholar] [CrossRef]
- Isom, D.G.; Dohlman, H.G. Buried ionizable networks are an ancient hallmark of G protein-coupled receptor activation. Proc. Natl. Acad. Sci. USA 2015, 112, 5702–5707. [Google Scholar] [CrossRef]
- Schlessman, J.L.; Abe, C.; Gittis, A.; Karp, D.A.; Dolan, M.A.; Garcia-Moreno, E.B. Crystallographic study of hydration of an internal cavity in engineered proteins with buried polar or ionizable groups. Biophys. J. 2008, 94, 3208–3216. [Google Scholar] [CrossRef][Green Version]
- Schonichen, A.; Webb, B.A.; Jacobson, M.P.; Barber, D.L. Considering protonation as a posttranslational modification regulating protein structure and function. Annu. Rev. Biophys. 2013, 42, 289–314. [Google Scholar] [CrossRef] [PubMed]
- Buccitelli, C.; Salgueiro, L.; Rowald, K.; Sotillo, R.; Mardin, B.R.; Korbel, J.O. Pan-cancer analysis distinguishes transcriptional changes of aneuploidy from proliferation. Genome Res. 2017, 27, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef] [PubMed]
- Casamassimi, A.; Ciccodicola, A. Transcriptional Regulation: Molecules, Involved Mechanisms, and Misregulation. Int. J. Mol. Sci. 2019, 20, 1281. [Google Scholar] [CrossRef]
- Eisen, T.J.; Eichhorn, S.W.; Subtelny, A.O.; Lin, K.S.; McGeary, S.E.; Gupta, S.; Bartel, D.P. The Dynamics of Cytoplasmic mRNA Metabolism. Mol. Cell 2020, 77, 786–799.e10. [Google Scholar] [CrossRef]
- Decker, C.; Parker, R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem. Sci. 1994, 19, 336–340. [Google Scholar] [CrossRef]
- Nakayama, K.; Kataoka, N. Regulation of Gene Expression under Hypoxic Conditions. Int. J. Mol. Sci. 2019, 20, 3278. [Google Scholar] [CrossRef]
- Huang, Q.; Hu, J.K.; Lohr, F.; Zhang, L.; Braun, R.; Lanzen, J.; Little, J.B.; Dewhirst, M.W.; Li, C.Y. Heat-induced gene expression as a novel targeted cancer gene therapy strategy. Cancer Res. 2000, 60, 3435–3439. [Google Scholar]
- Martín-Martín, N.; Carracedo, A.; Torrano, V. Metabolism and Transcription in Cancer: Merging Two Classic Tales. Front. Cell Dev. Biol. 2017, 5, 119. [Google Scholar] [CrossRef]
- Silberman, A.; Goldman, O.; Boukobza Assayag, O.; Jacob, A.; Rabinovich, S.; Adler, L.; Lee, J.S.; Keshet, R.; Sarver, A.; Frug, J.; et al. Acid-Induced Downregulation of ASS1 Contributes to the Maintenance of Intracellular pH in Cancer. Cancer Res. 2019, 79, 518–533. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Xu, H.; Shen, Q.; Schaefer, F.; Schmitt, C.P.; Chen, J.; Liu, H.; Liu, J.; Liu, J. pH-mediated upregulation of AQP1 gene expression through the Spi-B transcription factor. BMC Mol. Biol. 2018, 19, 4. [Google Scholar] [CrossRef] [PubMed]
- Duggan, S.P.; Gallagher, W.M.; Fox, E.J.P.; Abdel-Latif, M.M.; Reynolds, J.V.; Kelleher, D. Low pH induces co-ordinate regulation of gene expression in oesophageal cells. Carcinogenesis 2006, 27, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wang, J.; Jin, W.; Wang, L.; Li, H.; Ma, L.; Li, Q.; Pang, T. NHE1 mediates migration and invasion of HeLa cells via regulating the expression and localization of MT1-MMP. Cell Biochem. Funct. 2012, 30, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chang, G.; Wang, J.; Jin, W.; Wang, L.; Li, H.; Ma, L.; Li, Q.; Pang, T. NHE1 mediates MDA-MB-231 cells invasion through the regulation of MT1-MMP. Exp. Cell Res. 2011, 317, 2031–2040. [Google Scholar] [CrossRef]
- Valvona, C.J.; Fillmore, H.L.; Nunn, P.B.; Pilkington, G.J. The Regulation and Function of Lactate Dehydrogenase A: Therapeutic Potential in Brain Tumor. Brain Pathol. 2016, 26, 3–17. [Google Scholar] [CrossRef]
- Xie, H.; Valera, V.A.; Merino, M.J.; Amato, A.M.; Signoretti, S.; Linehan, W.M.; Sukhatme, V.P.; Seth, P. LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer. Mol. Cancer Ther. 2009, 8, 626–635. [Google Scholar] [CrossRef]
- Martin, T.A. Interleukin-8 and Angiogenesis. In Growth Factors and Their Receptors in Cancer Metastasis; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2001; pp. 51–65. [Google Scholar]
- Shibuya, M. Vascular Endothelial Growth Factor (VEGF) and Its Receptor (VEGFR) Signaling in Angiogenesis: A Crucial Target for Anti- and Pro-Angiogenic Therapies. Genes Cancer 2011, 2, 1097. [Google Scholar] [CrossRef]
- Xie, R.; Wang, H.; Jin, H.; Wen, G.; Tuo, B.; Xu, J. NHE1 is upregulated in gastric cancer and regulates gastric cancer cell proliferation, migration and invasion. Oncol. Rep. 2017, 37, 1451–1460. [Google Scholar] [CrossRef][Green Version]
- Amith, S.R.; Fliegel, L. Regulation of the Na+/H+ Exchanger (NHE1) in Breast Cancer Metastasis. Cancer Res. 2013, 73, 1259–1264. [Google Scholar] [CrossRef]
- Jiang, W.; Davies, G.; Martin, T.; Parr, C.; Watkins, G.; Mason, M.; Mansel, R. Expression of membrane type-1 matrix metalloproteinase, MT1-MMP in human breast cancer and its impact on invasiveness of breast cancer cells. Int. J. Mol. Med. 2006, 17, 583–590. [Google Scholar] [CrossRef]
- Harguindey, S.; Arranz, J.; Polo Orozco, J.; Rauch, C.; Fais, S.; Cardone, R.; Reshkin, S.J. Cariporide and other new and powerful NHE1 inhibitors as potentially selective anticancer drugs—An integral molecular/biochemical/metabolic/clinical approach after one hundred years of cancer research. J. Transl. Med. 2013, 11, 282. [Google Scholar] [CrossRef] [PubMed]
- Kondo, A.; Yamamoto, S.; Nakaki, R.; Shimamura, T.; Hamakubo, T.; Sakai, J.; Kodama, T.; Yoshida, T.; Aburatani, H.; Osawa, T. Extracellular Acidic pH Activates the Sterol Regulatory Element-Binding Protein 2 to Promote Tumor Progression. Cell Rep. 2017, 18, 2228–2242. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Guo, X.; Gui, Y. Acetyl-CoA Synthetase 2 Promotes Cell Migration and Invasion of Renal Cell Carcinoma by Upregulating Lysosomal-Associated Membrane Protein 1 Expression. Cell. Physiol. Biochem. 2018, 45, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Fukuma, Y.; Matsui, H.; Koike, H.; Sekine, Y.; Shechter, I.; Ohtake, N.; Nakata, S.; Ito, K.; Suzuki, K. Role of squalene synthase in prostate cancer risk and the biological aggressiveness of human prostate cancer. Prostate Cancer Prostatic Dis. 2012, 15, 339–345. [Google Scholar] [CrossRef]
- Ashida, S.; Kawada, C.; Inoue, K. Stromal regulation of prostate cancer cell growth by mevalonate pathway enzymes HMGCS1 and HMGCR. Oncol. Lett. 2017, 14, 6533–6542. [Google Scholar] [CrossRef]
- Wang, C.; Li, P.; Xuan, J.; Zhu, C.; Liu, J.; Shan, L.; Du, Q.; Ren, Y.; Ye, J. Cholesterol Enhances Colorectal Cancer Progression via ROS Elevation and MAPK Signaling Pathway Activation. Cell. Physiol. Biochem. 2017, 42, 729–742. [Google Scholar] [CrossRef]
- Fang, Y.; Liu, Z.; Chen, Z.; Xu, X.; Xiao, M.; Yu, Y.; Zhang, Y.; Zhang, X.; Du, Y.; Jiang, C.; et al. Smad5 acts as an intracellular pH messenger and maintains bioenergetic homeostasis. Cell Res. 2017, 27, 1083–1099. [Google Scholar] [CrossRef]
- Miccoli, L.; Oudard, S.; Sureau, F.; Poirson, F.; Dutrillaux, B.; Poupon, M.F. Intracellular pH governs the subcellular distribution of hexokinase in a glioma cell line. Biochem. J. 1996, 313 Pt 3, 957–962. [Google Scholar] [CrossRef]
- Persi, E.; Duran-Frigola, M.; Damaghi, M.; Roush, W.R.; Aloy, P.; Cleveland, J.L.; Gillies, R.J.; Ruppin, E. Systems analysis of intracellular pH vulnerabilities for cancer therapy. Nat. Commun. 2018, 9, 2997. [Google Scholar] [CrossRef]
- White, K.A.; Kisor, K.; Barber, D.L. Intracellular pH dynamics and charge-changing somatic mutations in cancer. Cancer Metastasis Rev. 2019, 38, 17–24. [Google Scholar] [CrossRef]
- Szpiech, Z.A.; Strauli, N.B.; White, K.A.; Ruiz, D.G.; Jacobson, M.P.; Barber, D.L.; Hernandez, R.D. Prominent features of the amino acid mutation landscape in cancer. PLoS ONE 2017, 12, e0183273. [Google Scholar] [CrossRef]
- Joerger, A.C.; Fersht, A.R. The tumor suppressor p53: From structures to drug discovery. Cold Spring Harb. Perspect. Biol. 2010, 2, a000919. [Google Scholar] [CrossRef] [PubMed]
- White, K.A.; Garrido Ruiz, D.; Szpiech, Z.A.; Strauli, N.B.; Hernandez, R.D.; Jacobson, M.P.; Barber, D.L. Cancer-associated arginine-to-histidine mutations confer a gain in pH sensing to mutant proteins. Sci. Signal. 2017, 10, eaam9931. [Google Scholar] [CrossRef] [PubMed]
- Munro, D.; Ghersi, D.; Singh, M. Two critical positions in zinc finger domains are heavily mutated in three human cancer types. PLoS Comput. Biol. 2018, 14, e1006290. [Google Scholar] [CrossRef] [PubMed]
- Aynaud, M.-M.; Mirabeau, O.; Gruel, N.; Grossetête, S.; Boeva, V.; Durand, S.; Surdez, D.; Saulnier, O.; Zaïdi, S.; Gribkova, S.; et al. Transcriptional Programs Define Intratumoral Heterogeneity of Ewing Sarcoma at Single-Cell Resolution. Cell Rep. 2020, 30, 1767–1779.e6. [Google Scholar] [CrossRef]
- Powell, A.A.; Talasaz, A.H.; Zhang, H.; Coram, M.A.; Reddy, A.; Deng, G.; Telli, M.L.; Advani, R.H.; Carlson, R.W.; Mollick, J.A.; et al. Single Cell Profiling of Circulating Tumor Cells: Transcriptional Heterogeneity and Diversity from Breast Cancer Cell Lines. PLoS ONE 2012, 7, e33788. [Google Scholar] [CrossRef]
- Riemann, A.; Reime, S.; Thews, O. Acidic extracellular environment affects miRNA expression in tumors in vitro and in vivo. Int. J. Cancer 2019, 144, 1609–1618. [Google Scholar] [CrossRef]
- González-Silva, L.; Quevedo, L.; Varela, I. Tumor Functional Heterogeneity Unraveled by scRNA-seq Technologies. Trends Cancer 2020, 6, 13–19. [Google Scholar] [CrossRef]
- Dengler, V.L.; Galbraith, M.D.; Espinosa, J.M. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 1–15. [Google Scholar] [CrossRef]
- van der Knaap, J.A.; Verrijzer, C.P. Undercover: Gene control by metabolites and metabolic enzymes. Genes Dev. 2016, 30, 2345–2369. [Google Scholar] [CrossRef]
- Chen, L.Q.; Pagel, M.D. Evaluating pH in the Extracellular Tumor Microenvironment Using CEST MRI and Other Imaging Methods. Adv. Radiol. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Ozawa, S.; Miyamoto, C.; Maehata, Y.; Suzuki, A.; Maeda, T.; Baba, Y. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Stock, C.; Schwab, A. Ion channels and transporters in metastasis. Biochim. Biophys. Acta 2015, 1848, 2638–2646. [Google Scholar] [CrossRef] [PubMed]
- Parks, S.K.; Cormerais, Y.; Pouysségur, J. Hypoxia and cellular metabolism in tumour pathophysiology: Hypoxia and tumour pathophysiology. J. Physiol. 2017, 595, 2439–2450. [Google Scholar] [CrossRef]
- Lee, A.H.; Tannock, I.F. Heterogeneity of Intracellular pH and of Mechanisms That Regulate Intracellular pH in Populations of Cultured Cells. Cancer Res. 1998, 58, 1901–1908. [Google Scholar]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.; McDermott, J.E.; Zhou, J.-Y.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, J.; Wang, X.; Zhu, J.; Liu, Q.; Shi, Z.; Chambers, M.C.; Zimmerman, L.J.; Shaddox, K.F.; Kim, S.; et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014, 513, 382–387. [Google Scholar] [CrossRef]
- Sinha, A.; Huang, V.; Livingstone, J.; Wang, J.; Fox, N.S.; Kurganovs, N.; Ignatchenko, V.; Fritsch, K.; Donmez, N.; Heisler, L.E.; et al. The Proteogenomic Landscape of Curable Prostate Cancer. Cancer Cell 2019, 35, 414–427.e6. [Google Scholar] [CrossRef]
- Lakatos, E.; Salehi-Reyhani, A.; Barclay, M.; Stumpf, M.P.H.; Klug, D.R. Protein degradation rate is the dominant mechanism accounting for the differences in protein abundance of basal p53 in a human breast and colorectal cancer cell line. PLoS ONE 2017, 12, e0177336. [Google Scholar] [CrossRef]
- Arceci, A.; Bonacci, T.; Wang, X.; Stewart, K.; Damrauer, J.S.; Hoadley, K.A.; Emanuele, M.J. FOXM1 Deubiquitination by USP21 Regulates Cell Cycle Progression and Paclitaxel Sensitivity in Basal-like Breast Cancer. Cell Rep. 2019, 26, 3076–3086.e6. [Google Scholar] [CrossRef]
- Senft, D.; Qi, J.; Ronai, Z.A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 2018, 18, 69–88. [Google Scholar] [CrossRef] [PubMed]
- Urra, H.; Dufey, E.; Avril, T.; Chevet, E.; Hetz, C. Endoplasmic Reticulum Stress and the Hallmarks of Cancer. Trends Cancer 2016, 2, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Qian, T.; Liu, C.; Ding, Y.; Guo, C.; Cai, R.; Wang, X.; Wang, R.; Zhang, K.; Zhou, L.; Deng, Y.; et al. PINCH-1 interacts with myoferlin to promote breast cancer progression and metastasis. Oncogene 2020, 39, 2069–2087. [Google Scholar] [CrossRef]
- Fahmy, K.; Gonzalez, A.; Arafa, M.; Peixoto, P.; Bellahcène, A.; Turtoi, A.; Delvenne, P.; Thiry, M.; Castronovo, V.; Peulen, O. Myoferlin plays a key role in VEGFA secretion and impacts tumor-associated angiogenesis in human pancreas cancer: Myoferlin, a key regulator of VEGFA secretion and neoangiogenesis in PDAC. Int. J. Cancer 2016, 138, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Turtoi, A.; Blomme, A.; Bellahcène, A.; Gilles, C.; Hennequière, V.; Peixoto, P.; Bianchi, E.; Noel, A.; De Pauw, E.; Lifrange, E.; et al. Myoferlin is a key regulator of EGFR activity in breast cancer. Cancer Res. 2013, 73, 5438–5448. [Google Scholar] [CrossRef] [PubMed]
- Rademaker, G.; Costanza, B.; Anania, S.; Agirman, F.; Maloujahmoum, N.; Di Valentin, E.; Goval, J.J.; Bellahcène, A.; Castronovo, V.; Peulen, O. Myoferlin Contributes to the Metastatic Phenotype of Pancreatic Cancer Cells by Enhancing Their Migratory Capacity through the Control of Oxidative Phosphorylation. Cancers 2019, 11, 853. [Google Scholar] [CrossRef]
- Qin, Z.-Y.; Wang, T.; Su, S.; Shen, L.-T.; Zhu, G.-X.; Liu, Q.; Zhang, L.; Liu, K.-W.; Zhang, Y.; Zhou, Z.-H.; et al. BRD4 Promotes Gastric Cancer Progression and Metastasis through Acetylation-Dependent Stabilization of Snail. Cancer Res. 2019, 79, 4869–4881. [Google Scholar] [CrossRef]
- Martínez-Jiménez, F.; Muiños, F.; Sentís, I.; Deu-Pons, J.; Reyes-Salazar, I.; Arnedo-Pac, C.; Mularoni, L.; Pich, O.; Bonet, J.; Kranas, H.; et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 2020, 20, 555–572. [Google Scholar] [CrossRef]
- White, K.A.; Grillo-Hill, B.K.; Esquivel, M.; Peralta, J.; Bui, V.N.; Chire, I.; Barber, D.L. β-Catenin is a pH sensor with decreased stability at higher intracellular pH. J. Cell Biol. 2018, 217, 3965–3976. [Google Scholar] [CrossRef]
- Hao, B.; Oehlmann, S.; Sowa, M.E.; Harper, J.W.; Pavletich, N.P. Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 2007, 26, 131–143. [Google Scholar] [CrossRef]
- Akhoondi, S.; Sun, D.; von der Lehr, N.; Apostolidou, S.; Klotz, K.; Maljukova, A.; Cepeda, D.; Fiegl, H.; Dafou, D.; Marth, C.; et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007, 67, 9006–9012. [Google Scholar] [CrossRef] [PubMed]
- Korphaisarn, K.; Morris, V.K.; Overman, M.J.; Fogelman, D.R.; Kee, B.K.; Raghav, K.P.S.; Manuel, S.; Shureiqi, I.; Wolff, R.A.; Eng, C.; et al. FBXW7 missense mutation: A novel negative prognostic factor in metastatic colorectal adenocarcinoma. Oncotarget 2017, 8, 39268–39279. [Google Scholar] [CrossRef] [PubMed]
- Minella, A.C.; Welcker, M.; Clurman, B.E. Ras activity regulates cyclin E degradation by the Fbw7 pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 9649–9654. [Google Scholar] [CrossRef] [PubMed]
- Karki, P.; Li, X.; Schrama, D.; Fliegel, L. B-Raf associates with and activates the NHE1 isoform of the Na+/H+ exchanger. J. Biol. Chem. 2011, 286, 13096–13105. [Google Scholar] [CrossRef]
- Thomas, R.C.; Pagnotta, S.E.; Nistri, A. Whole-cell recording of intracellular pH with silanized and oiled patch-type single or double-barreled microelectrodes. Pflugers Arch. 2003, 447, 259–265. [Google Scholar] [CrossRef]
- Ozkan, P.; Mutharasan, R. A rapid method for measuring intracellular pH using BCECF-AM. Biochim. Biophys. Acta 2002, 1572, 143–148. [Google Scholar] [CrossRef]
- Wieder, E.D.; Hang, H.; Fox, M.H. Measurement of intracellular pH using flow cytometry with carboxy-SNARF-1. Cytometry 1993, 14, 916–921. [Google Scholar] [CrossRef]
- Sun, C.; Du, W.; Wang, B.; Dong, B.; Wang, B. Research progress of near-infrared fluorescence probes based on indole heptamethine cyanine dyes in vivo and in vitro. BMC Chem. 2020, 14, 21. [Google Scholar] [CrossRef]
- Gao, L.; Lin, X.; Zheng, A.; Shuang, E.; Wang, J.; Chen, X. Real-time monitoring of intracellular pH in live cells with fluorescent ionic liquid. Anal. Chim. Acta 2020, 1111, 132–138. [Google Scholar] [CrossRef]
- Miesenböck, G.; De Angelis, D.A.; Rothman, J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998, 394, 192–195. [Google Scholar] [CrossRef]
- Schotthöfer, S.K.; Bohrmann, J. Analysing bioelectrical phenomena in the Drosophila ovary with genetic tools: Tissue-specific expression of sensors for membrane potential and intracellular pH, and RNAi-knockdown of mechanisms involved in ion exchange. BMC Dev. Biol. 2020, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Koivusalo, M.; Welch, C.; Hayashi, H.; Scott, C.C.; Kim, M.; Alexander, T.; Touret, N.; Hahn, K.M.; Grinstein, S. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J. Cell Biol. 2010, 188, 547–563. [Google Scholar] [CrossRef]
- Mahon, M.J. pHluorin2: An enhanced, ratiometric, pH-sensitive green florescent protein. Adv. Biosci. Biotechnol. 2011, 2, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Tantama, M.; Hung, Y.P.; Yellen, G. Imaging Intracellular pH in Live Cells with a Genetically Encoded Red Fluorescent Protein Sensor. J. Am. Chem. Soc. 2011, 133, 10034–10037. [Google Scholar] [CrossRef] [PubMed]
- Zanotelli, M.R.; Goldblatt, Z.E.; Miller, J.P.; Bordeleau, F.; Li, J.; VanderBurgh, J.A.; Lampi, M.C.; King, M.R.; Reinhart-King, C.A. Regulation of ATP utilization during metastatic cell migration by collagen architecture. Mol. Biol. Cell 2018, 29, 1–9. [Google Scholar] [CrossRef]
- Shen, Y.; Rosendale, M.; Campbell, R.E.; Perrais, D. pHuji, a pH-sensitive red fluorescent protein for imaging of exo- and endocytosis. J. Cell Biol. 2014, 207, 419–432. [Google Scholar] [CrossRef]
- Rajendran, M.; Claywell, B.; Haynes, E.P.; Scales, U.; Henning, C.K.; Tantama, M. Imaging pH Dynamics Simultaneously in Two Cellular Compartments Using a Ratiometric pH-Sensitive Mutant of mCherry. ACS Omega 2018, 3, 9476–9486. [Google Scholar] [CrossRef]
- Grillo-Hill, B.K.; Webb, B.A.; Barber, D.L. Ratiometric imaging of pH probes. Methods Cell Biol. 2014, 123, 429–448. [Google Scholar] [CrossRef]
- Martynov, V.I.; Pakhomov, A.A.; Deyev, I.E.; Petrenko, A.G. Genetically encoded fluorescent indicators for live cell pH imaging. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2018, 1862, 2924–2939. [Google Scholar] [CrossRef] [PubMed]
- Canton, J.; Grinstein, S. Chapter 5-Measuring lysosomal pH by fluorescence microscopy. In Methods in Cell Biology; Platt, F., Platt, N., Eds.; Lysosomes and Lysosomal Diseases; Academic Press: Cambridge, MA, USA, 2015; Volume 126, pp. 85–99. [Google Scholar]
- Takahashi, A.; Zhang, Y.; Centonze, V.E.; Herman, B. Measurement of Mitochondrial pH In Situ. BioTechniques 2001, 30, 804–815. [Google Scholar] [CrossRef] [PubMed]
- Sarkisyan, K.S.; Goryashchenko, A.S.; Lidsky, P.V.; Gorbachev, D.A.; Bozhanova, N.G.; Gorokhovatsky, A.Y.; Pereverzeva, A.R.; Ryumina, A.P.; Zherdeva, V.V.; Savitsky, A.P.; et al. Green Fluorescent Protein with Anionic Tryptophan-Based Chromophore and Long Fluorescence Lifetime. Biophys. J. 2015, 109, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Rost, B.R.; Schneider, F.; Grauel, M.K.; Wozny, C.; Bentz, C.; Blessing, A.; Rosenmund, T.; Jentsch, T.J.; Schmitz, D.; Hegemann, P.; et al. Optogenetic Acidification of Synaptic Vesicles and Lysosomes. Nat. Neurosci. 2015, 18, 1845–1852. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, A.; Colinet, A.-S.; Zimmermannova, O.; Sychrova, H.; Morsomme, P. A new pH sensor localized in the Golgi apparatus of Saccharomyces cerevisiae reveals unexpected roles of Vph1p and Stv1p isoforms. Sci. Rep. 2020, 10, 1881. [Google Scholar] [CrossRef]
- Reifenrath, M.; Boles, E. A superfolder variant of pH-sensitive pHluorin for in vivo pH measurements in the endoplasmic reticulum. Sci. Rep. 2018, 8, 11985. [Google Scholar] [CrossRef]
- Rieger, B.; Shalaeva, D.N.; Söhnel, A.-C.; Kohl, W.; Duwe, P.; Mulkidjanian, A.Y.; Busch, K.B. Lifetime imaging of GFP at CoxVIIIa reports respiratory supercomplex assembly in live cells. Sci. Rep. 2017, 7, 46055. [Google Scholar] [CrossRef] [PubMed]
- Goudge, M.C.; Kuo, J.C.-H.; Metzloff, A.E.; Huang, L.-T.; Colville, M.J.; Park, S.; Zipfel, W.R.; Paszek, M.J. Litmus-Body: A Molecularly Targeted Sensor for Cell-Surface pH Measurements. ACS Sens. 2020, 5, 1555–1566. [Google Scholar] [CrossRef]
- Haynes, E.; Rajendran, M.; Lyon, A.; Noinaj, N.; Day, R.; Tantama, M. Fluorescence lifetime imaging of compartmental pH dynamics using red fluorescent protein sensors in live cells. FASEB J. 2018, 32, 657-14. [Google Scholar] [CrossRef]
- Dey, S.; Bielytskyi, P.; Gräsing, D.; Das, A.; Kundu, R.; Matysik, J.; Maiti, S.; Madhu, P.K. Precise in situ photo-induced pH modulation during NMR spectrometry. Chem. Phys. Lett. 2018, 706, 665–668. [Google Scholar] [CrossRef]
- Chakraborty, S.; Nandi, S.; Bhattacharyya, K.; Mukherjee, S. Time Evolution of Local pH Around a Photo-Acid in Water and a Polymer Hydrogel: Time Resolved Fluorescence Spectroscopy of Pyranine. ChemPhysChem 2019, 20, 3221–3227. [Google Scholar] [CrossRef]
- Wu, L.; Dong, A.; Dong, L.; Wang, S.-Q.; Li, Y. PARIS, an optogenetic method for functionally mapping gap junctions. Elife 2019, 8, e43366. [Google Scholar] [CrossRef]
- Adams, D.S.; Tseng, A.-S.; Levin, M. Light-activation of the Archaerhodopsin H(+)-pump reverses age-dependent loss of vertebrate regeneration: Sparking system-level controls in vivo. Biol. Open 2013, 2, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Chaudhuri, P.K.; Ramalingam, N.; Tan, D.S.; Lim, C.T.; Warkiani, M.E. Single-cell profiling approaches to probing tumor heterogeneity. Int. J. Cancer 2016, 139, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Teng, L.; He, B.; Wang, J.; Tan, K. 4DGenome: A comprehensive database of chromatin interactions. Bioinformatics 2015, 31, 2560–2564. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Zou, Q.; Liu, Y.; Yang, X. Genome-wide assessment of differential translations with ribosome profiling data. Nat. Commun. 2016, 7, 11194. [Google Scholar] [CrossRef]
- Muthuswamy, S.K. Organoid Models of Cancer Explode with Possibilities. Cell Stem Cell 2018, 22, 290–291. [Google Scholar] [CrossRef]
- Hochhaus, A.; Larson, R.A.; Guilhot, F.; Radich, J.P.; Branford, S.; Hughes, T.P.; Baccarani, M.; Deininger, M.W.; Cervantes, F.; Fujihara, S.; et al. Long-Term Outcomes of Imatinib Treatment for Chronic Myeloid Leukemia. N. Engl. J. Med. 2017, 376, 917–927. [Google Scholar] [CrossRef]
- Maeda, H.; Khatami, M. Analyses of repeated failures in cancer therapy for solid tumors: Poor tumor-selective drug delivery, low therapeutic efficacy and unsustainable costs. Clin. Transl. Med. 2018, 7, 11. [Google Scholar] [CrossRef]
- Rolver, M.G.; Elingaard-Larsen, L.O.; Andersen, A.P.; Counillon, L.; Pedersen, S.F. Pyrazine ring-based Na+/H+ exchanger (NHE) inhibitors potently inhibit cancer cell growth in 3D culture, independent of NHE1. Sci. Rep. 2020, 10, 5800. [Google Scholar] [CrossRef]
- Mauvezin, C.; Nagy, P.; Juhász, G.; Neufeld, T.P. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 2015, 6, 7007. [Google Scholar] [CrossRef]
| Tool | In Vivo Compatibility | Cytotoxicity | Long-Term Measurements | Spatial Resolution | Brightness | Quantitative | Requires Standardization |
|---|---|---|---|---|---|---|---|
| Patch Clamp [96] | Incompatible | high | minutes | Single Cell | NA | Yes | No |
| BCECF [97] | cell-based | mild | minutes-hours | Subcellular | mid | Yes | Yes |
| SNARF [98] | cell-based | mild | minutes-hours | Subcellular | mid | Yes | Yes |
| Indole Heptamethine Cyanine Dyes [99] | cell-based | mild | minutes | Subcellular | low | Yes | Yes |
| Ionic Liquids [100] | cell-based | low | hours | Subcellular | low | Yes | Yes |
| pHluorin [101] | cell-based, some tissue | low | hours | Subcellular, targetable | low | Yes | Yes |
| SuperEclipticpHluorin [101,102] | cell-based, some tissue | low | hours | Subcellular, targetable | high | No | Yes |
| pHluorin-mCherry [103] | cell-based, some tissue | low | hours | Subcellular, targetable | high | Yes | Yes |
| pHluorin 2 [104] | cell-based, some tissue | low | hours | Subcellular, targetable | mid | Yes | Yes |
| pHred [105,106] | cell-based, deeper tissue | low-mild | hours | Subcellular, targetable | low | Yes | Yes |
| pHuji [107] | cell-based, deeper tissue | low-mild | hours | Subcellular, targetable | low | No | Yes |
| mCherry EA-mutant [108] | cell-based, deeper tissue | low-mild | hours | Subcellular, targetable | low | Yes | Yes |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czowski, B.J.; Romero-Moreno, R.; Trull, K.J.; White, K.A. Cancer and pH Dynamics: Transcriptional Regulation, Proteostasis, and the Need for New Molecular Tools. Cancers 2020, 12, 2760. https://doi.org/10.3390/cancers12102760
Czowski BJ, Romero-Moreno R, Trull KJ, White KA. Cancer and pH Dynamics: Transcriptional Regulation, Proteostasis, and the Need for New Molecular Tools. Cancers. 2020; 12(10):2760. https://doi.org/10.3390/cancers12102760
Chicago/Turabian StyleCzowski, Brandon J., Ricardo Romero-Moreno, Keelan J. Trull, and Katharine A. White. 2020. "Cancer and pH Dynamics: Transcriptional Regulation, Proteostasis, and the Need for New Molecular Tools" Cancers 12, no. 10: 2760. https://doi.org/10.3390/cancers12102760
APA StyleCzowski, B. J., Romero-Moreno, R., Trull, K. J., & White, K. A. (2020). Cancer and pH Dynamics: Transcriptional Regulation, Proteostasis, and the Need for New Molecular Tools. Cancers, 12(10), 2760. https://doi.org/10.3390/cancers12102760






