In-Depth Mapping of the Urinary N-Glycoproteome: Distinct Signatures of ccRCC-related Progression
Abstract
1. Introduction
2. Results
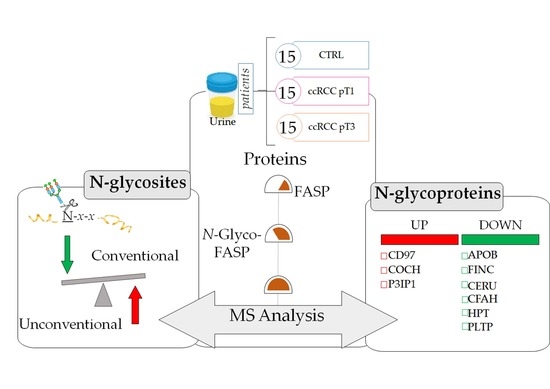
2.1. Clinical Data and Study Design
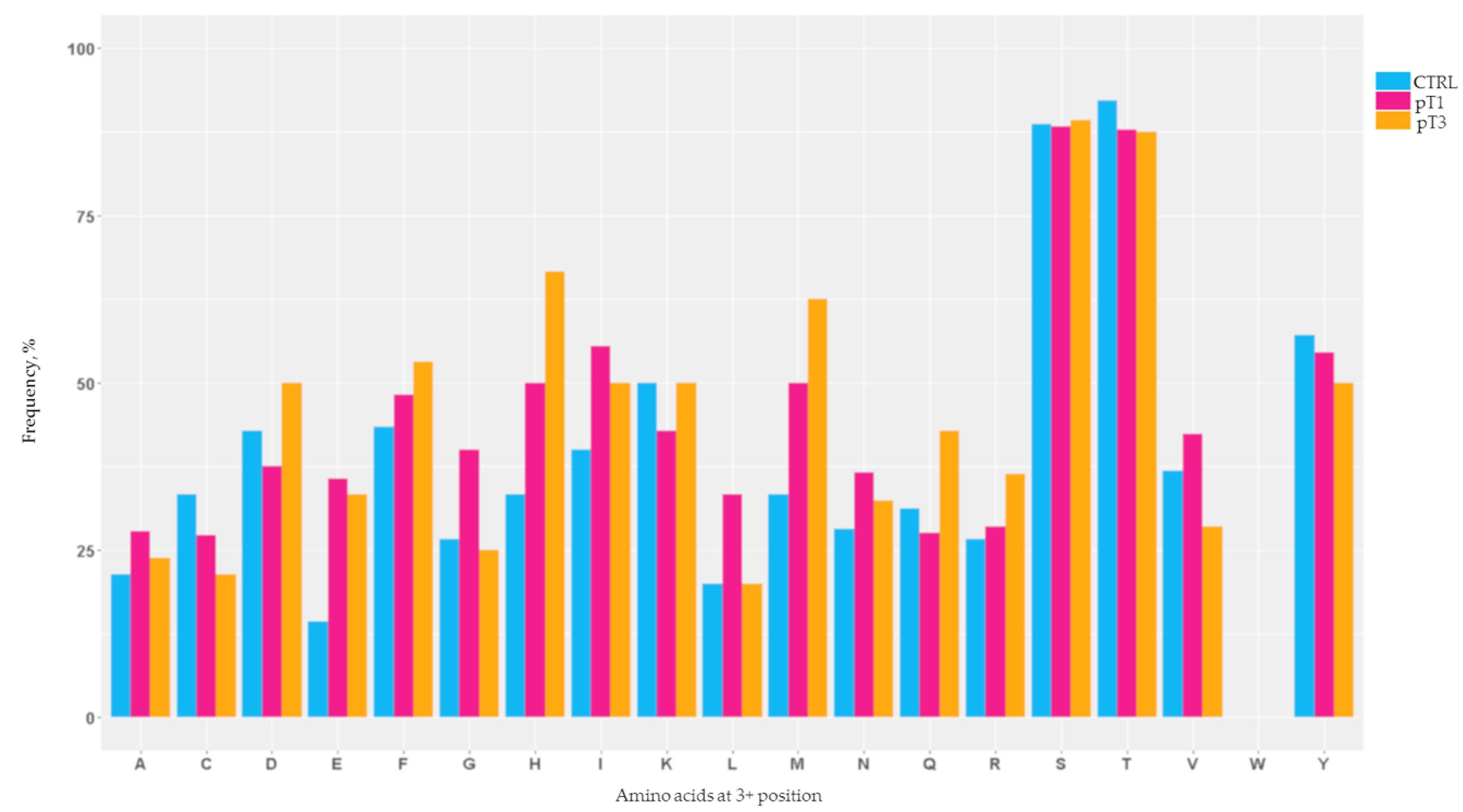
2.2. Mapping of the N-Glycosylation Sites
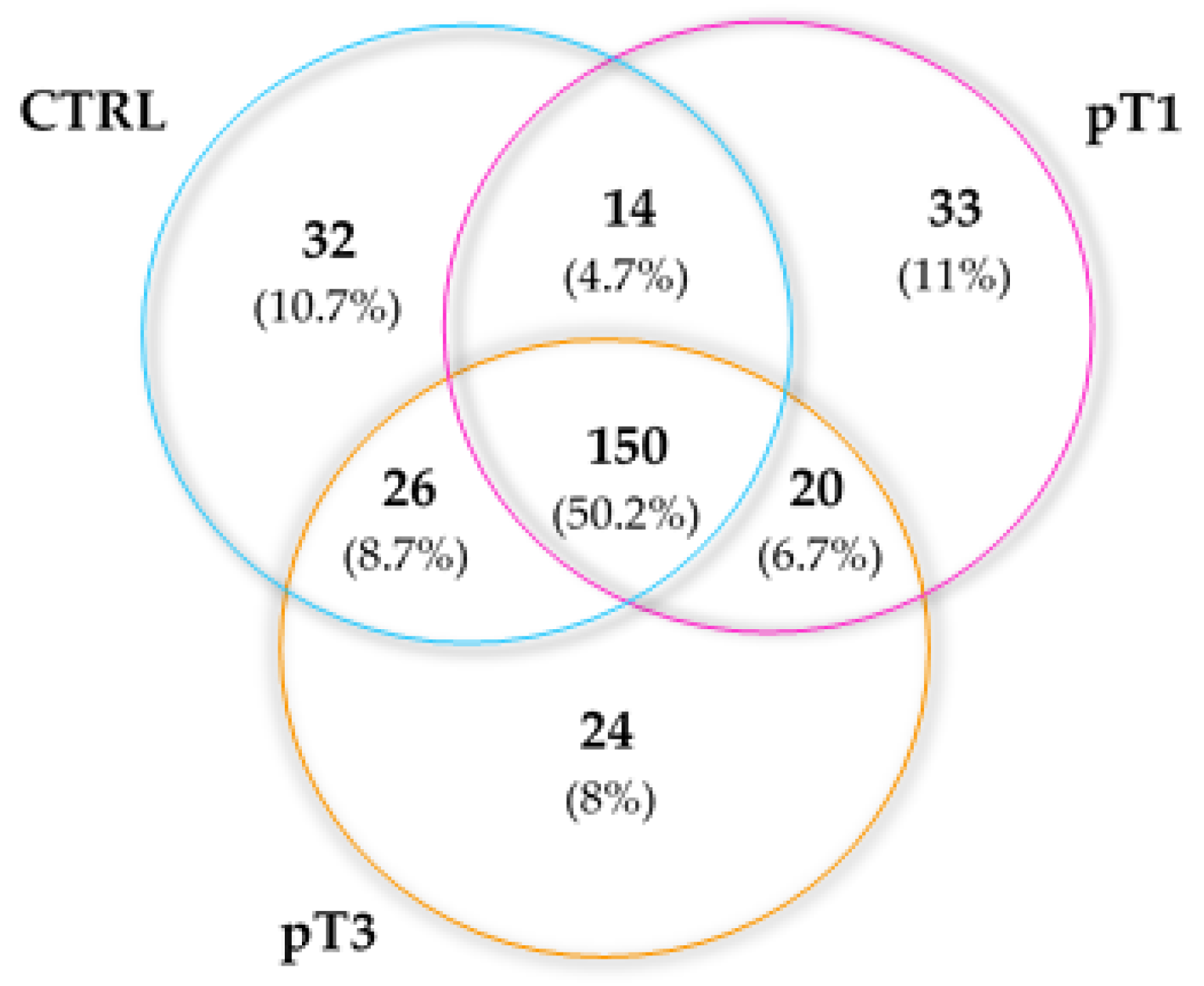
2.3. Characterisation of the Urinary N-Glycoproteins: Identification
2.4. Characterisation of the Urinary N-Glycoproteins: Functional Analysis
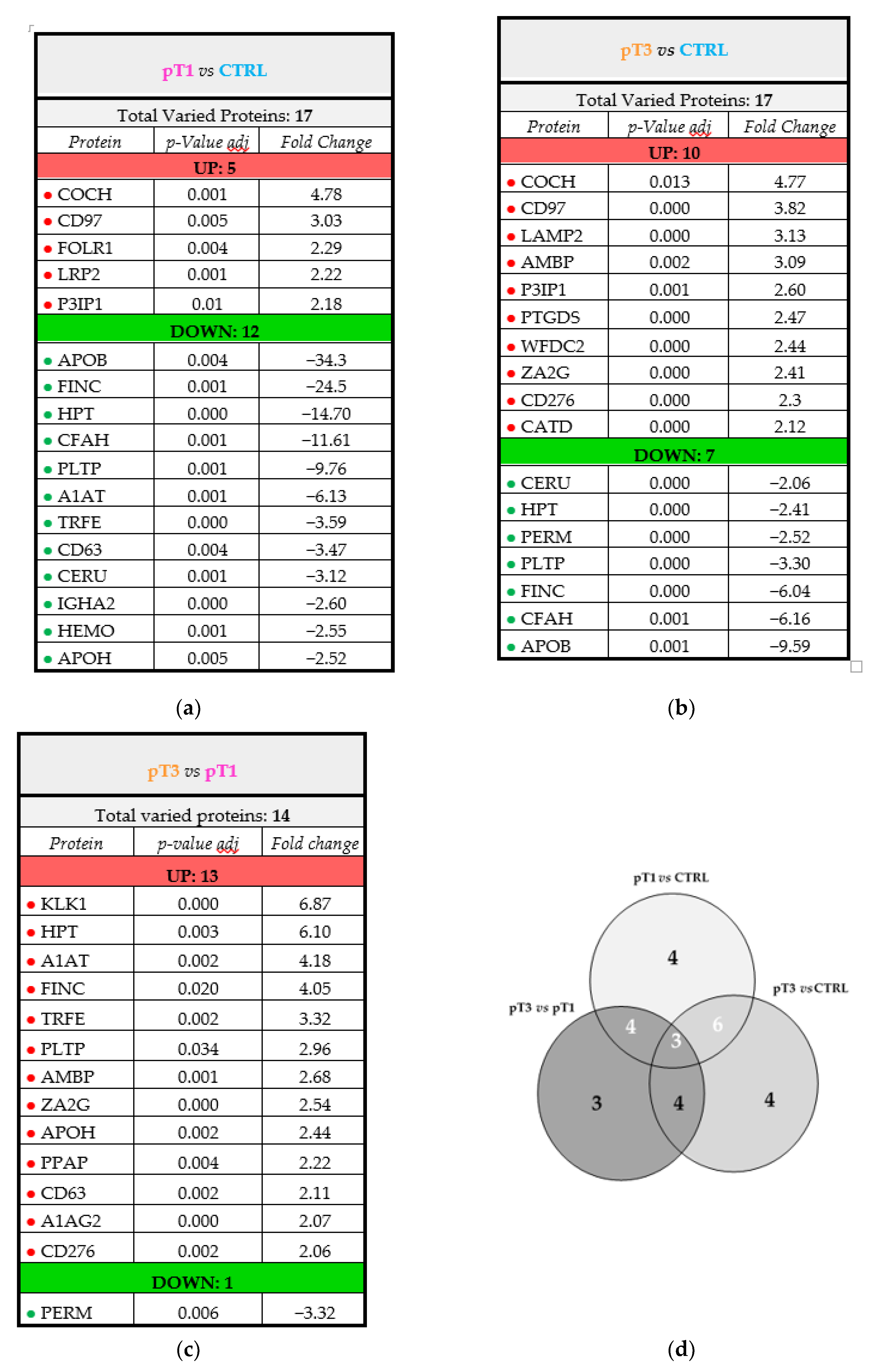
2.5. Characterisation of the Urinary N-Glycoproteins: Quantitative Analysis
3. Discussion
4. Materials and Methods
4.1. Urine Collection and Processing
4.2. Sample Population
4.3. Urinary Proteins Digestion
4.4. Urinary N-Glycopeptides Enrichment and Deglycosylation
4.5. Mass Spectrometric Analysis
4.6. Data Processing
4.6.1. Protein Identification
4.6.2. Bioinformatics and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chandler, K.B.; Costello, C.E.; Rahimi, N. Glycosylation in the Tumor Microenvironment: Implications for Tumor Angiogenesis and Metastasis. Cells 2019, 8, 544. [Google Scholar] [CrossRef] [PubMed]
- Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 2430–2437. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Marth, J.D. Glycosylation in Cellular Mechanisms of Health and Disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, N.; Kizuka, Y. Glycans and cancer: Role of N-Glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv. Cancer Res. 2015. [Google Scholar] [CrossRef]
- Lau, K.S.; Dennis, J.W. N-Glycans in cancer progression. Glycobiology 2008, 18, 750–760. [Google Scholar] [CrossRef]
- Häuselmann, I.; Borsig, L. Altered tumor-cell glycosylation promotes metastasis. Front. Oncol. 2014, 4, 28. [Google Scholar] [CrossRef]
- Oliveira-Ferrer, L.; Legler, K.; Milde-Langosch, K. Role of protein glycosylation in cancer metastasis. Semin. Cancer Biol. 2017, 44, 141–152. [Google Scholar] [CrossRef]
- Gilgunn, S.; Conroy, P.J.; Saldova, R.; Rudd, P.M.; O’Kennedy, R.J. Aberrant PSA glycosylation-A sweet predictor of prostate cancer. Nat. Rev. Urol. 2013, 10, 99. [Google Scholar] [CrossRef]
- Ilic, D.; Neuberger, M.M.; Djulbegovic, M.; Dahm, P. Screening for prostate cancer. Cochrane database Syst. Rev. 2013, CD004720. [Google Scholar] [CrossRef]
- Drake, R.R.; Jones, E.E.; Powers, T.W.; Nyalwidhe, J.O. Altered Glycosylation in Prostate Cancer. Adv. Cancer Res. 2015, 345–382. [Google Scholar] [CrossRef]
- Duffy, M.J.; Shering, S.; Sherry, F.; McDermott, E.; O’Higgins, N. CA 15-3: A prognostic marker in breast cancer. Int. J. Biol. Markers 2000, 15, 330–333. [Google Scholar] [CrossRef]
- Choi, J.W.; Moon, B.I.; Lee, J.W.; Kim, H.J.; Jin, Y.; Kim, H.J. Use of CA15-3 for screening breast cancer: An antibody-lectin sandwich assay for detecting glycosylation of CA15-3 in sera. Oncol. Rep. 2018, 40, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.W.; Lloyd, K.O. Molecular cloning of the CA125 ovarian cancer antigen: Identification as a new mucin, MUC16. J. Biol. Chem. 2001, 276, 27371–27375. [Google Scholar] [CrossRef]
- Duffy, M.J. Carcinoembryonic antigen as a marker for colorectal cancer: Is it clinically useful? Clin. Chem. 2001, 47, 624–630. [Google Scholar] [PubMed]
- Saito, G.; Sadahiro, S.; Okada, K.; Tanaka, A.; Suzuki, T.; Kamijo, A. Relation between Carcinoembryonic Antigen Levels in Colon Cancer Tissue and Serum Carcinoembryonic Antigen Levels at Initial Surgery and Recurrence. Oncology 2016, 91, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Moertel, C.G.; Fleming, T.R.; Macdonald, J.S.; Haller, D.G.; Laurie, J.A.; Tangen, C. An Evaluation of the Carcinoembryonic Antigen (CEA) Test for Monitoring Patients with Resected Colon Cancer. JAMA J. Am. Med. Assoc. 1993, 270, 943–947. [Google Scholar] [CrossRef]
- Hammarstrom, S. The carcinoembryonic antigen (CEA) family: Structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999, 9, 67–81. [Google Scholar] [CrossRef]
- Davis, M.T.; Spahr, C.S.; McGinley, M.D.; Robinson, J.H.; Bures, E.J.; Beierle, J.; Mort, J.; Yu, W.; Luethy, R.; Patterson, S.D. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. II. Limitations of complex mixture analyses. Proteomics 2001, 1, 108–117. [Google Scholar] [CrossRef]
- Sun, W.; Li, F.; Wu, S.; Wang, X.; Zheng, D.; Wang, J.; Gao, Y. Human urine proteome analysis by three separation approaches. Proteomics 2005, 5, 4994–5001. [Google Scholar] [CrossRef]
- Arivusudar Marimuthu A Comprehensive Map of the Human Urinary Proteome. J. Proteome Res. 2011, 23, 1–7. [CrossRef]
- Desiere, F.; Deutsch, E.W.; King, N.L.; Nesvizhskii, A.I.; Mallick, P.; Eng, J.; Chen, S.; Eddes, J.; Loevenich, S.N.; Aebersold, R. The PeptideAtlas project. Nucleic Acids Res. 2006, 34, D655–D658. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, R.; Saad, J.; Angeli, C.B.; Palmisano, G. Site-specific characterization of N-linked glycosylation in human urinary glycoproteins and endogenous glycopeptides. Glycoconj. J. 2016, 33, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Different collaborators Comprehensive Molecular Characterization of clear cell renal cell carcinoma. Nature 2014, 499, 43–49. [CrossRef]
- Zielinska, D.F.; Gnad, F.; Wiśniewski, J.R.; Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 2010, 141, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Wang, X.-J.; Xue, Y.; Liu, M.-Q.; Zeng, W.-F.; Zhang, Y.; Zhang, L.; Gao, X.; Yan, G.-Q.; Yao, J.; et al. In-depth mapping of the mouse brain N-glycoproteome reveals widespread N-glycosylation of diverse brain proteins. Oncotarget 2016, 7, 38796–38809. [Google Scholar] [CrossRef]
- Geng, F.; Wang, J.; Liu, D.; Jin, Y.; Ma, M. Identification of N-Glycosites in Chicken Egg White Proteins Using an Omics Strategy. J. Agric. Food Chem. 2017, 65, 5357–5364. [Google Scholar] [CrossRef]
- Song, W.; Mentink, R.A.; Henquet, M.G.L.; Cordewener, J.H.G.; van Dijk, A.D.J.; Bosch, D.; America, A.H.P.; van der Krol, A.R. N-glycan occupancy of Arabidopsis N-glycoproteins. J. Proteomics 2013, 93, 343–355. [Google Scholar] [CrossRef]
- Xu, Y.; Bailey, U.-M.; Punyadeera, C.; Schulz, B.L. Identification of salivary N-glycoproteins and measurement of glycosylation site occupancy by boronate glycoprotein enrichment and liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2014, 28, 471–482. [Google Scholar] [CrossRef]
- Malerod, H.; Graham, R.L.J.; Sweredoski, M.J.; Hess, S. Comprehensive profiling of N-linked glycosylation sites in HeLa cells using hydrazide enrichment. J. Proteome Res. 2013, 12, 248–259. [Google Scholar] [CrossRef][Green Version]
- Hollander, N.; Haimovich, J. Altered N-Linked Glycosylation in Follicular Lymphoma and Chronic Lymphocytic Leukemia: Involvement in Pathogenesis and Potential Therapeutic Targeting. Front. Immunol. 2017, 8, 912. [Google Scholar] [CrossRef] [PubMed]
- Mamessier, E.; Drevet, C.; Broussais-Guillaumot, F.; Mollichella, M.-L.; Garciaz, S.; Roulland, S.; Benchetrit, M.; Nadel, B.; Xerri, L. Contiguous follicular lymphoma and follicular lymphoma in situ harboring N-glycosylated sites. Haematologica 2015, 100, e155–e157. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Chen, R.; Tamura, Y.; Crispin, D.A.; Lai, L.A.; May, D.H.; McIntosh, M.W.; Goodlett, D.R.; Brentnall, T.A. Quantitative glycoproteomics analysis reveals changes in N-glycosylation level associated with pancreatic ductal adenocarcinoma. J. Proteome Res. 2014, 13, 1293–1306. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Ruhaak, L.R.; Stroble, C.; Salemi, M.R.; Phinney, B.; Lebrilla, C.B.; Leiserowitz, G.S. Glycoproteomic Analysis of Malignant Ovarian Cancer Ascites Fluid Identifies Unusual Glycopeptides. J. Proteome Res. 2016, 15, 3358–3376. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Qin, H.; Li, T.; Cheng, K.; Dong, J.; Tian, M.; Chai, N.; Guo, H.; Li, J.; You, X.; et al. Characterization of site-specific glycosylation of secreted proteins associated with multi-drug resistance of gastric cancer. Oncotarget 2016, 7, 25315–25327. [Google Scholar] [CrossRef]
- Valliere-Douglass, J.F.; Kodama, P.; Mujacic, M.; Brady, L.J.; Wang, W.; Wallace, A.; Yan, B.; Reddy, P.; Treuheit, M.J.; Balland, A. Asparagine-linked oligosaccharides present on a non-consensus amino acid sequence in the CH1 domain of human antibodies. J. Biol. Chem. 2009, 284, 32493–32506. [Google Scholar] [CrossRef]
- Matsumoto, S.; Taguchi, Y.; Shimada, A.; Igura, M.; Kohda, D. Tethering an N-Glycosylation Sequon-Containing Peptide Creates a Catalytically Competent Oligosaccharyltransferase Complex. Biochemistry 2017, 56, 602–611. [Google Scholar] [CrossRef]
- Trinidad, J.C.; Schoepfer, R.; Burlingame, A.L.; Medzihradszky, K.F. N-and O-glycosylation in the murine synaptosome. Mol. Cell. Proteomics 2013, 12, 3474–3488. [Google Scholar] [CrossRef]
- Valliere-Douglass, J.F.; Eakin, C.M.; Wallace, A.; Ketchem, R.R.; Wang, W.; Treuheit, M.J.; Balland, A. Glutamine-linked and non-consensus asparagine-linked oligosaccharides present in human recombinant antibodies define novel protein glycosylation motifs. J. Biol. Chem. 2010, 285, 16012–16022. [Google Scholar] [CrossRef]
- Vance, B.A.; Wu, W.; Ribaudo, R.K.; Segal, D.M.; Kearse, K.P. Multiple dimeric forms of human CD69 result from differential addition of N-glycans to typical (Asn-X-Ser/Thr) and atypical (Asn-X-cys) glycosylation motifs. J. Biol. Chem. 1997, 272, 23117–23122. [Google Scholar] [CrossRef]
- Mooser, V.; Seabra, M.C.; Abedin, M.; Landschulz, K.T.; Marcovina, S.; Hobbs, H.H. Apolipoprotein (a) kringle 4-containing fragments in human urine. Relationship to plasma levels of lipoprotein (a). J. Clin. Investig. 1996, 97, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Bolenz, C.; Schröppel, B.; Eisenhardt, A.; Schmitz-Dräger, B.J.; Grimm, M.-O. Abklärung der Hämaturie. Dtsch. Aerzteblatt Online 2018, 25, 127–135. [Google Scholar] [CrossRef]
- Tian, Z.-H.; Yuan, C.; Yang, K.; Gao, X.-L. Systematic identification of key genes and pathways in clear cell renal cell carcinoma on bioinformatics analysis. Ann. Transl. Med. 2019, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Dobryszycka, W. Biological functions of haptoglobin--new pieces to an old puzzle. Eur. J. Clin. Chem. Clin. Biochem. 1997, 35, 647–654. [Google Scholar]
- Liu, T.; Qian, W.J.; Gritsenko, M.A.; Camp, D.G.; Monroe, M.E.; Moore, R.J.; Smith, R.D. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 2005, 4, 2070–2080. [Google Scholar] [CrossRef]
- Zhu, J.; Lin, Z.; Wu, J.; Yin, H.; Dai, J.; Feng, Z.; Marrero, J.; Lubman, D.M. Analysis of serum haptoglobin fucosylation in hepatocellular carcinoma and liver cirrhosis of different etiologies. J. Proteome Res. 2014, 13, 2986–2997. [Google Scholar] [CrossRef]
- Park, S.Y.; Yoon, S.J.; Jeong, Y.T.; Kim, J.M.; Kim, J.Y.; Bernert, B.; Ullman, T.; Itzkowitz, S.H.; Kim, J.H.; Hakomori, S.I. N-glycosylation status of β-haptoglobin in sera of patients with colon cancer, chronic inflammatory diseases and normal subjects. Int. J. Cancer 2010, 126, 142–155. [Google Scholar] [CrossRef]
- Lin, Z.; Simeone, D.M.; Anderson, M.A.; Brand, R.E.; Xie, X.; Shedden, K.A.; Ruffin, M.T.; Lubman, D.M. Mass spectrometric assay for analysis of haptoglobin fucosylation in pancreatic cancer. J. Proteome Res. 2011, 10, 2602–2611. [Google Scholar] [CrossRef]
- Sandim, V.; de Abreu Pereira, D.; Kalume, D.E.; Oliveira-Carvalho, A.L.; Ornellas, A.A.; Soares, M.R.; Alves, G.; Zingali, R.B. Proteomic analysis reveals differentially secreted proteins in the urine from patients with clear cell renal cell carcinoma. Urol. Oncol. Semin. Orig. Investig. 2016, 34, e11–e15. [Google Scholar] [CrossRef]
- Bruneel, A.; Habarou, F.; Stojkovic, T.; Plouviez, G.; Bougas, L.; Guillemet, F.; Brient, N.; Henry, D.; Dupre, T.; Vuillaumier-Barrot, S.; et al. Two-dimensional electrophoresis highlights haptoglobin beta chain as an additional biomarker of congenital disorders of glycosylation. Clin. Chim. Acta. 2017, 470, 70–74. [Google Scholar] [CrossRef]
- To, W.S.; Midwood, K.S. Plasma and cellular fibronectin: Distinct and independent functions during tissue repair. Fibrogenesis Tissue Repair 2011, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Kumra, H.; Reinhardt, D.P. Fibronectin-targeted drug delivery in cancer. Adv. Drug Deliv. Rev. 2016, 97, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.T.; Cheng, H.W.; Huang, C.M.; Li, H.R.; Ou, M.H.; Huang, J.R.; Khoo, K.H.; Yu, H.W.; Chen, Y.Q.; Wang, Y.K.; et al. Fibronectin in cell adhesion and migration via N-glycosylation. Oncotarget 2017, 8, 70653–70668. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Cheng, H.; Parvani, J.G.; Zhou, Z.; Lu, Z.R. Magnetic resonance molecular imaging of metastatic breast cancer by targeting extradomain-B fibronectin in the tumor microenvironment. Magn. Reson. Med. 2018, 79, 3135–3143. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, K.F.; Du, W.; Sorrelle, N.B.; Wnuk-Lipinska, K.; Topalovski, M.; Toombs, J.E.; Cruz, V.H.; Yabuuchi, S.; Rajeshkumar, N.V.; Maitra, A.; et al. Small-Molecule Inhibition of Axl Targets Tumor Immune Suppression and Enhances Chemotherapy in Pancreatic Cancer. Cancer Res. 2018, 78, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.J.; Lee, S.W.; Lin, L.C.; Chen, T.J.; Chang, I.W.; Hsu, H.P.; Chang, K.Y.; Huang, H.Y.; Li, C.F. Fibronectin overexpression is associated with latentmembrane protein 1 expression and has independent prognostic value for nasopharyngeal carcinoma. Tumor Biol. 2014, 35, 1703–1712. [Google Scholar] [CrossRef]
- Gopal, S.; Veracini, L.; Grall, D.; Butori, C.; Schaub, S.; Audebert, S.; Camoin, L.; Baudelet, E.; Adwanska, A.; Beghelli-De La Forest Divonne, S.; et al. Fibronectin-guided migration of carcinoma collectives. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Ou, Y.C.; Li, J.R.; Wang, J.D.; Chang, C.Y.; Wu, C.C.; Chen, W.Y.; Kuan, Y.H.; Liao, S.L.; Lu, H.C.; Chen, C.J. Fibronectin Promotes Cell Growth and Migration in Human Renal Cell Carcinoma Cells. Int. J. Mol. Sci. 2019, 20, 2792. [Google Scholar] [CrossRef]
- Kondisetty, S.; Menon, K.N.; Pooleri, G.K. Fibronectin protein expression in renal cell carcinoma in correlation with clinical stage of tumour. Biomark. Res. 2018, 6, 1–6. [Google Scholar] [CrossRef]
- Hellman, N.E.; Gitlin, J.D. Ceruloplasmin metabolism and function. Annu. Rev. Nutr. 2002, 22, 439–458. [Google Scholar] [CrossRef]
- Delmonico, L.; Bravo, M.; Silvestre, R.T.; Ornellas, M.H.F.; De Azevedo, C.M.; Alves, G. Proteomic profile of saliva and plasma from women with impalpable breast lesions. Oncol. Lett. 2016, 12, 2145–2152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Han, I.W.; Jang, J.-Y.; Kwon, W.; Park, T.; Kim, Y.; Lee, K.B.; Kim, S.-W. Ceruloplasmin as a prognostic marker in patients with bile duct cancer. Oncotarget 2017, 8, 29028–29037. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.H.; Venkatesh, R.; More, C.B. Determination of role of ceruloplasmin in oral potentially malignant disorders and oral malignancy-A cross-sectional study. Oral Dis. 2017, 23, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Boz, A.; Evliyaoglu, O.; Yildirim, M.; Erkan, N.; Karaca, B. The value of serum zinc, copper, ceruloplasmin levels in patients with gastrointestinal tract cancers. Turk. J. Gastroenterol. 2005, 16, 81–84. [Google Scholar]
- Gajbhiye, A.; Dabhi, R.; Taunk, K.; Vannuruswamy, G.; RoyChoudhury, S.; Adhav, R.; Seal, S.; Mane, A.; Bayatigeri, S.; Santra, M.K.; et al. Urinary proteome alterations in HER2 enriched breast cancer revealed by multipronged quantitative proteomics. Proteomics 2016, 16, 2403–2418. [Google Scholar] [CrossRef]
- Bleu, M.; Gaulis, S.; Lopes, R.; Sprouffske, K.; Apfel, V.; Holwerda, S.; Pregnolato, M.; Yildiz, U.; Cordo’, V.; Dost, A.F.M.; et al. PAX8 activates metabolic genes via enhancer elements in Renal Cell Carcinoma. Nat. Commun. 2019, 10, 3739. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, H.; Xu, G.; Chu, N.; Xu, N.; Wen, H.; Gu, B.; Liu, J.; Mao, S.; Na, R.; et al. iTRAQ-based quantitative proteomic analysis reveals potential early diagnostic markers of clear-cell Renal cell carcinoma. Biosci. Trends 2016, 10, 210–219. [Google Scholar] [CrossRef][Green Version]
- Raimondo, F.; Morosi, L.; Corbetta, S.; Chinello, C.; Brambilla, P.; Della Mina, P.; Villa, A.; Albo, G.; Battaglia, C.; Bosari, S.; et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol. Biosyst. 2013, 9, 1220–1233. [Google Scholar] [CrossRef]
- Yan, X.; Yao, M.; Wen, X.; Zhu, Y.; Zhao, E.; Qian, X.; Chen, X.; Lu, W.; Lv, Q.; Zhang, L.; et al. Elevated apolipoprotein B predicts poor postsurgery prognosis in patients with hepatocellular carcinoma. Onco Targets Ther. 2019, 12, 1957–1964. [Google Scholar] [CrossRef]
- Liu, J.-X.; Yuan, Q.; Min, Y.-L.; He, Y.; Xu, Q.-H.; Li, B.; Shi, W.-Q.; Lin, Q.; Li, Q.-H.; Zhu, P.-W.; et al. Apolipoprotein A1 and B as risk factors for development of intraocular metastasis in patients with breast cancer. Cancer Manag. Res. 2019, 11, 2881–2888. [Google Scholar] [CrossRef]
- Chen, C.-L.; Lin, T.-S.; Tsai, C.-H.; Wu, C.-C.; Chung, T.; Chien, K.-Y.; Wu, M.; Chang, Y.-S.; Yu, J.-S.; Chen, Y.-T. Identification of potential bladder cancer markers in urine by abundant-protein depletion coupled with quantitative proteomics. J. Proteomics 2013, 85, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Velagapudi, S.; Schraml, P.; Yalcinkaya, M.; Bolck, H.A.; Rohrer, L.; Moch, H.; von Eckardstein, A. Scavenger receptor BI promotes cytoplasmic accumulation of lipoproteins in clear-cell renal cell carcinoma. J. Lipid Res. 2018, 59, 2188–2201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Xie, Y.; Ma, X.; Gu, L.; Li, H.; Li, X.; Guo, G.; Zhang, X. Preoperative apolipoprotein B/A1 ratio is an independent prognostic factor in metastatic renal cell carcinoma. Urol. Oncol. 2019, 37, e9–e184. [Google Scholar] [CrossRef] [PubMed]
- van den Boogert, M.A.W.; Larsen, L.E.; Ali, L.; Kuil, S.D.; Chong, P.L.W.; Loregger, A.; Kroon, J.; Schnitzler, J.G.; Schimmel, A.W.M.; Peter, J.; et al. N-Glycosylation Defects in Humans Lower Low-Density Lipoprotein Cholesterol Through Increased Low-Density Lipoprotein Receptor Expression. Circulation 2019, 140, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.-C. Phospholipid transfer protein: its impact on lipoprotein homeostasis and atherosclerosis. J. Lipid Res. 2018, 59, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Sirwi, A.; Hussain, M.M. Lipid transfer proteins in the assembly of apoB-containing lipoproteins. J. Lipid Res. 2018, 59, 1094–1102. [Google Scholar] [CrossRef]
- Dong, W.; Gong, H.; Zhang, G.; Vuletic, S.; Albers, J.; Zhang, J.; Liang, H.; Sui, Y.; Zheng, J. Lipoprotein lipase and phospholipid transfer protein overexpression in human glioma cells and their effect on cell growth, apoptosis, and migration. Acta Biochim. Et Biophys. Sin. 2017, 49, 62–73. [Google Scholar] [CrossRef]
- Cserhalmi, M.; Papp, A.; Brandus, B.; Uzonyi, B.; Jozsi, M. Regulation of regulators: Role of the complement factor H-related proteins. Semin. Immunol. 2019, 101341. [Google Scholar] [CrossRef]
- Ajona, D.; Castano, Z.; Garayoa, M.; Zudaire, E.; Pajares, M.J.; Martinez, A.; Cuttitta, F.; Montuenga, L.M.; Pio, R. Expression of complement factor H by lung cancer cells: effects on the activation of the alternative pathway of complement. Cancer Res. 2004, 64, 6310–6318. [Google Scholar] [CrossRef]
- Bjorge, L.; Hakulinen, J.; Vintermyr, O.K.; Jarva, H.; Jensen, T.S.; Iversen, O.E.; Meri, S. Ascitic complement system in ovarian cancer. Br. J. Cancer 2005, 92, 895–905. [Google Scholar] [CrossRef]
- Wilczek, E.; Rzepko, R.; Nowis, D.; Legat, M.; Golab, J.; Glab, M.; Gorlewicz, A.; Konopacki, F.; Mazurkiewicz, M.; Sladowski, D.; et al. The possible role of factor H in colon cancer resistance to complement attack. Int. J. Cancer 2008, 122, 2030–2037. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.-H.; Hwang, H.-J.; Sung, H.-J.; Heo, S.-H.; Kim, D.-S.; Hong, S.-H.; Lee, K.-H.; Cho, J.-Y. Upregulation of Complement Factor H by SOCS-1/3(-)STAT4 in Lung Cancer. Cancers 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; He, X.; Johnson, C.; Stoops, J.; Eaker, A.E.; Stoffer, D.S.; Bell, A.; Zarnegar, R.; DeFrances, M.C. PI3K is negatively regulated by PIK3IP1, a novel p110 interacting protein. Biochem. Biophys. Res. Commun. 2007, 358, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.C.; Martincorena, I.; Rust, A.G.; Rashid, M.; Alifrangis, C.; Alexandrov, L.B.; Tiffen, J.C.; Kober, C.; Green, A.R.; Massie, C.E.; et al. Inactivating CUX1 mutations promote tumorigenesis. Nat. Genet. 2014, 46, 33–38. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhu, Z.; Johnson, C.; Stoops, J.; Eaker, A.E.; Bowen, W.; DeFrances, M.C. PIK3IP1, a negative regulator of PI3K, suppresses the development of hepatocellular carcinoma. Cancer Res. 2008, 68, 5591–5598. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.H.; Roy, N.; Chakraborty, S.; Desrichard, A.; Chung, S.S.; Woolthuis, C.M.; Hu, W.; Berezniuk, I.; Garrett-Bakelman, F.E.; Hamann, J.; et al. CD97 is a critical regulator of acute myeloid leukemia stem cell function. J. Exp. Med. 2019, 216, 2362–2377. [Google Scholar] [CrossRef]
- Chance, M.R.; Chang, J.; Liu, S.; Gokulrangan, G.; Chen, D.H.-C.; Lindsay, A.; Geng, R.; Zheng, Q.Y.; Alagramam, K. Proteomics, bioinformatics and targeted gene expression analysis reveals up-regulation of cochlin and identifies other potential biomarkers in the mouse model for deafness in Usher syndrome type 1F. Hum. Mol. Genet. 2010, 19, 1515–1527. [Google Scholar] [CrossRef]
- Lee, R.S.; Monigatti, F.; Briscoe, A.C.; Waldon, Z.; Freeman, M.R.; Steen, H. Optimizing sample handling for urinary proteomics. J. Proteome Res. 2008, 7, 4022–4030. [Google Scholar] [CrossRef]
- Chinello, C.; Stella, M.; Piga, I.; Smith, A.J.; Bovo, G.; Varallo, M.; Ivanova, M.; Denti, V.; Grasso, M.; Grasso, A.; et al. Proteomics of liquid biopsies: Depicting RCC infiltration into the renal vein by MS analysis of urine and plasma. J. Proteomics 2019, 191, 29–37. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Severi, L.; Losi, L.; Fonda, S.; Taddia, L.; Gozzi, G.; Marverti, G.; Magni, F.; Chinello, C.; Stella, M.; Sheouli, J.; et al. Proteomic and Bioinformatic Studies for the Characterization of Response to Pemetrexed in Platinum Drug Resistant Ovarian Cancer. Front. Pharmacol. 2018, 9, 454. [Google Scholar] [CrossRef] [PubMed]

| Group | # of Patients | Gender (Male-Female) | Age Mean (Range) | Tumour Dimension (cm) |
|---|---|---|---|---|
| CTRL | 15 | 10–5 | 57.9 (39–77) | / |
| pT1 | 15 | 8–7 | 67.8 (42–82) | 4 |
| pT3 | 15 | 12–3 | 68.9 (45–81) | 12 |
| N-glycosylated Asparagine Distribution | % | |
|---|---|---|
| CTRL | ccRCC | |
| N/N tot | 21 | 21 |
| N in ncSeq/N ncSeq tot | 5.6 | 6.8 |
| N in cSeq/N cSeq tot | 81 | 77 |
| N-x-T/N-x-T tot | 88 | 82 |
| N-x-S/N-x-S tot | 71 | 69 |
| % | % | |||||
|---|---|---|---|---|---|---|
| CTRL | pT1 | Variation | CTRL | pT3 | Variation | |
| 21 | 19 | −10 | N/N tot | 21 | 23 | 9 |
| 5.6 | 6.4 | 14 | N in ncSeq/N ncSeq tot | 5.6 | 7.2 | 29 |
| 81 | 75 | −7 | N in cSeq/N cSeq tot | 81 | 79 | −2 |
| 88 | 81 | −8 | N-x-T/N-x-T tot | 88 | 82 | −7 |
| 71 | 66 | 7 | N-x-S/N-x-S tot | 71 | 73 | 3 |
| % | % | |||||
|---|---|---|---|---|---|---|
| CTRL | pT1 | Variation | CTRL | pT3 | Variation | |
| 66 | 66 | 0 | N/N tot | 66 | 66 | 0 |
| 31 | 37 | 19 | N in ncSeq/N ncSeq tot | 31 | 37 | 19 |
| 91 | 88 | −3 | N in cSeq/N cSeq tot | 91 | 88 | −3 |
| 92 | 88 | −4 | N-x-T/N-x-T tot | 92 | 88 | −4 |
| 89 | 88 | −1 | N-x-S/N-x-S tot | 89 | 89 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santorelli, L.; Capitoli, G.; Chinello, C.; Piga, I.; Clerici, F.; Denti, V.; Smith, A.; Grasso, A.; Raimondo, F.; Grasso, M.; et al. In-Depth Mapping of the Urinary N-Glycoproteome: Distinct Signatures of ccRCC-related Progression. Cancers 2020, 12, 239. https://doi.org/10.3390/cancers12010239
Santorelli L, Capitoli G, Chinello C, Piga I, Clerici F, Denti V, Smith A, Grasso A, Raimondo F, Grasso M, et al. In-Depth Mapping of the Urinary N-Glycoproteome: Distinct Signatures of ccRCC-related Progression. Cancers. 2020; 12(1):239. https://doi.org/10.3390/cancers12010239
Chicago/Turabian StyleSantorelli, Lucia, Giulia Capitoli, Clizia Chinello, Isabella Piga, Francesca Clerici, Vanna Denti, Andrew Smith, Angelica Grasso, Francesca Raimondo, Marco Grasso, and et al. 2020. "In-Depth Mapping of the Urinary N-Glycoproteome: Distinct Signatures of ccRCC-related Progression" Cancers 12, no. 1: 239. https://doi.org/10.3390/cancers12010239
APA StyleSantorelli, L., Capitoli, G., Chinello, C., Piga, I., Clerici, F., Denti, V., Smith, A., Grasso, A., Raimondo, F., Grasso, M., & Magni, F. (2020). In-Depth Mapping of the Urinary N-Glycoproteome: Distinct Signatures of ccRCC-related Progression. Cancers, 12(1), 239. https://doi.org/10.3390/cancers12010239