TRAIL Induces Nuclear Translocation and Chromatin Localization of TRAIL Death Receptors
Abstract
1. Introduction
2. Results
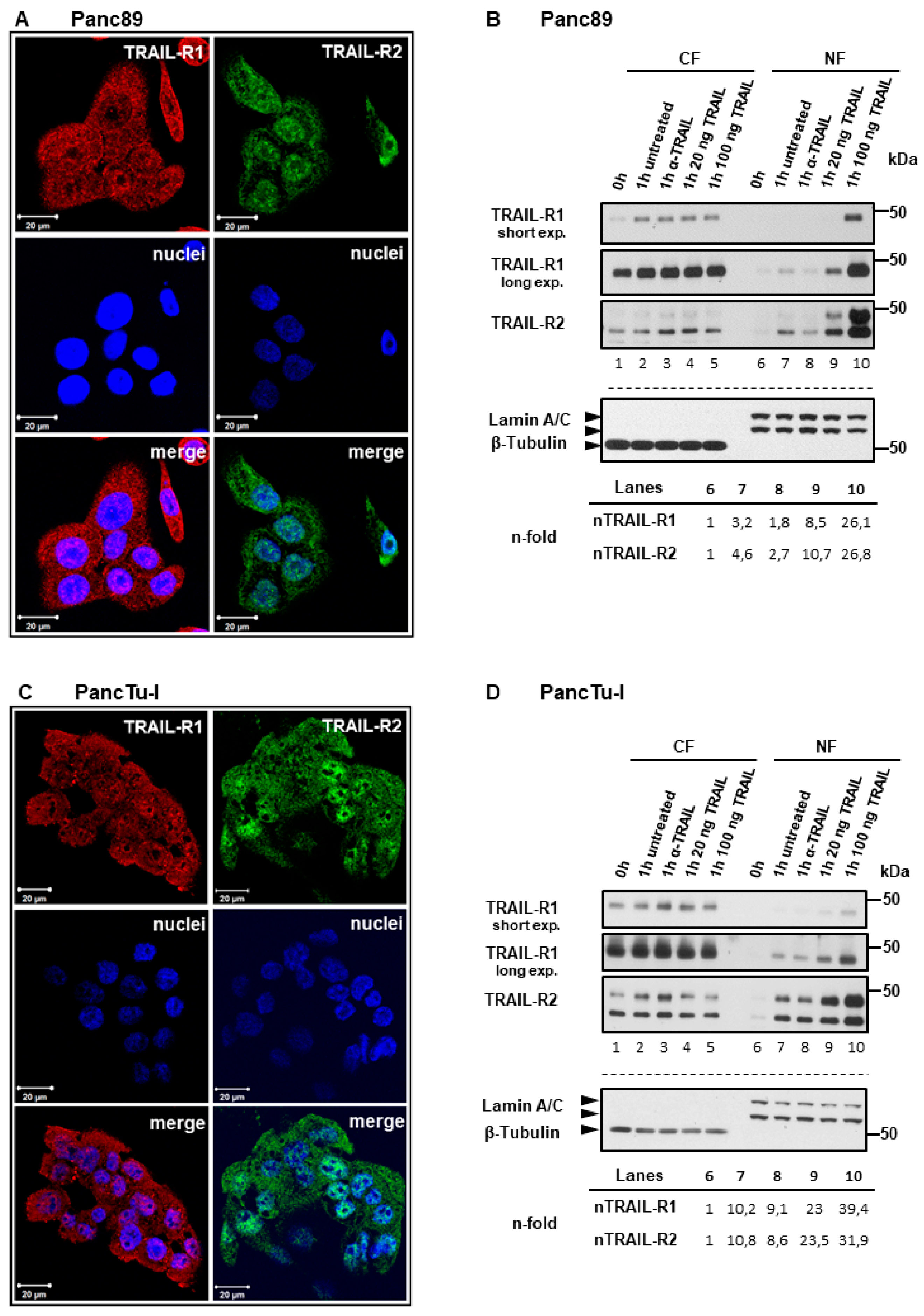
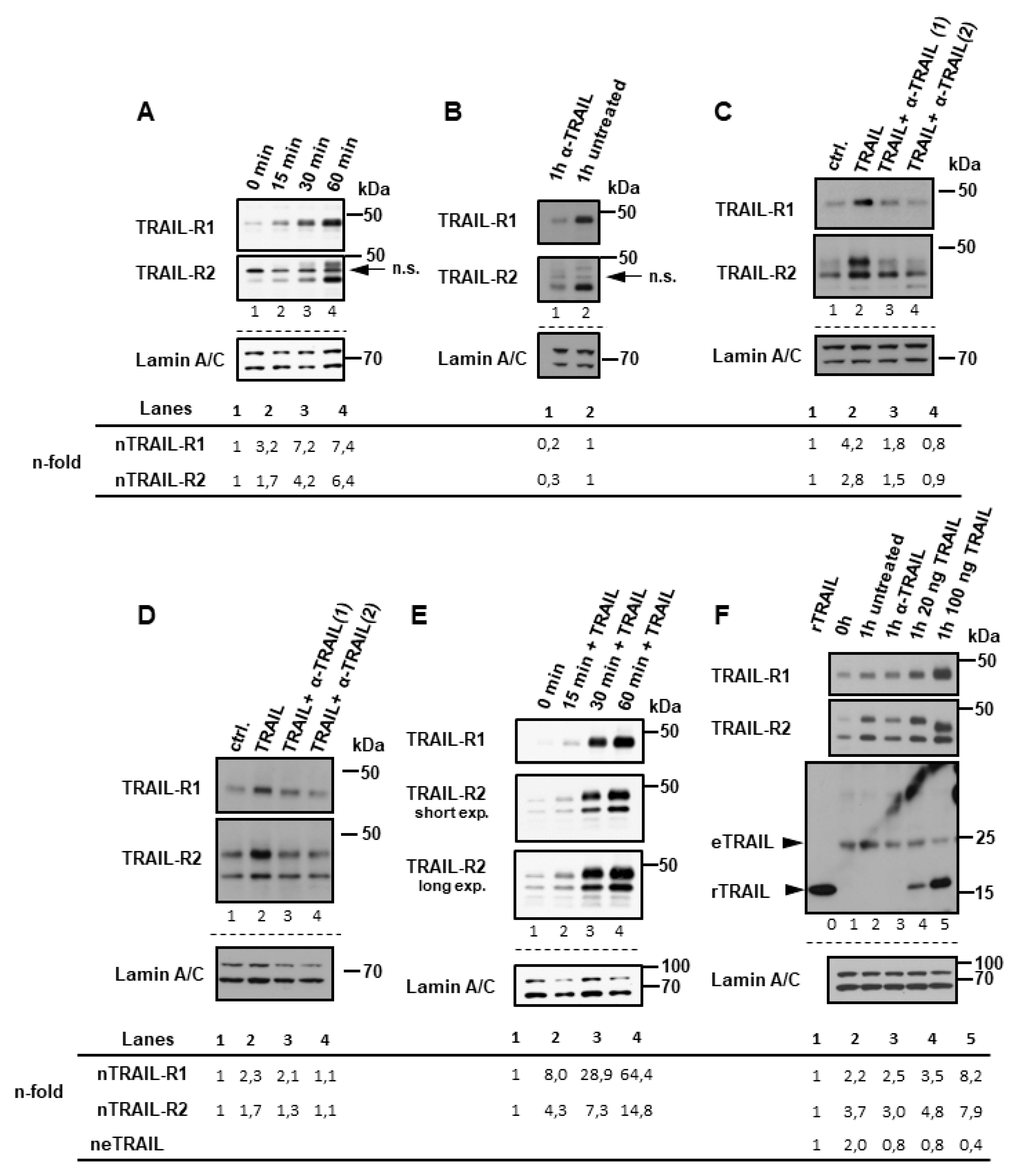
2.1. Plasma Membrane TRAIL-R1 and TRAIL-R2 Translocate to the Nucleus in a TRAIL-Dependent Manner
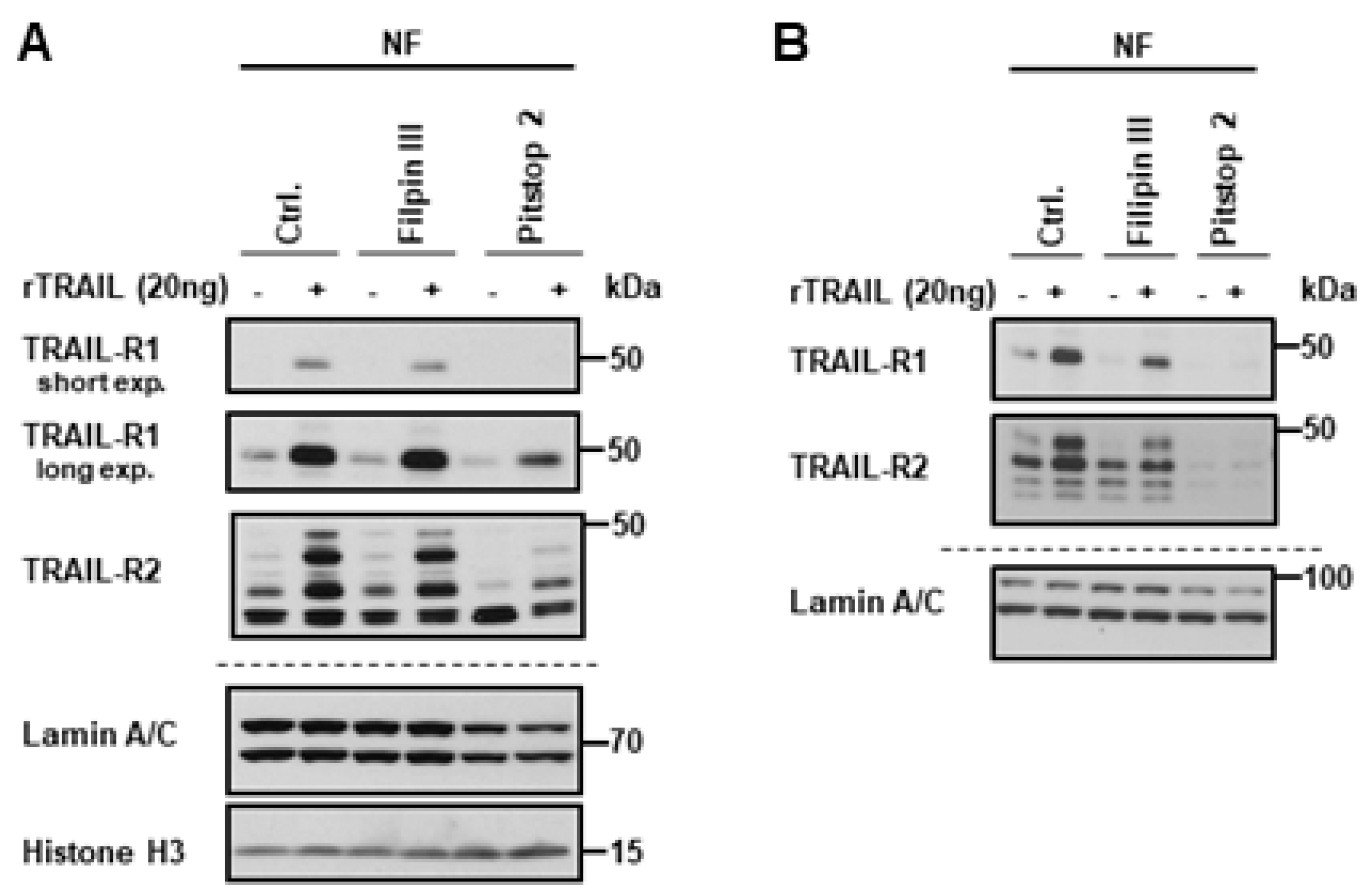
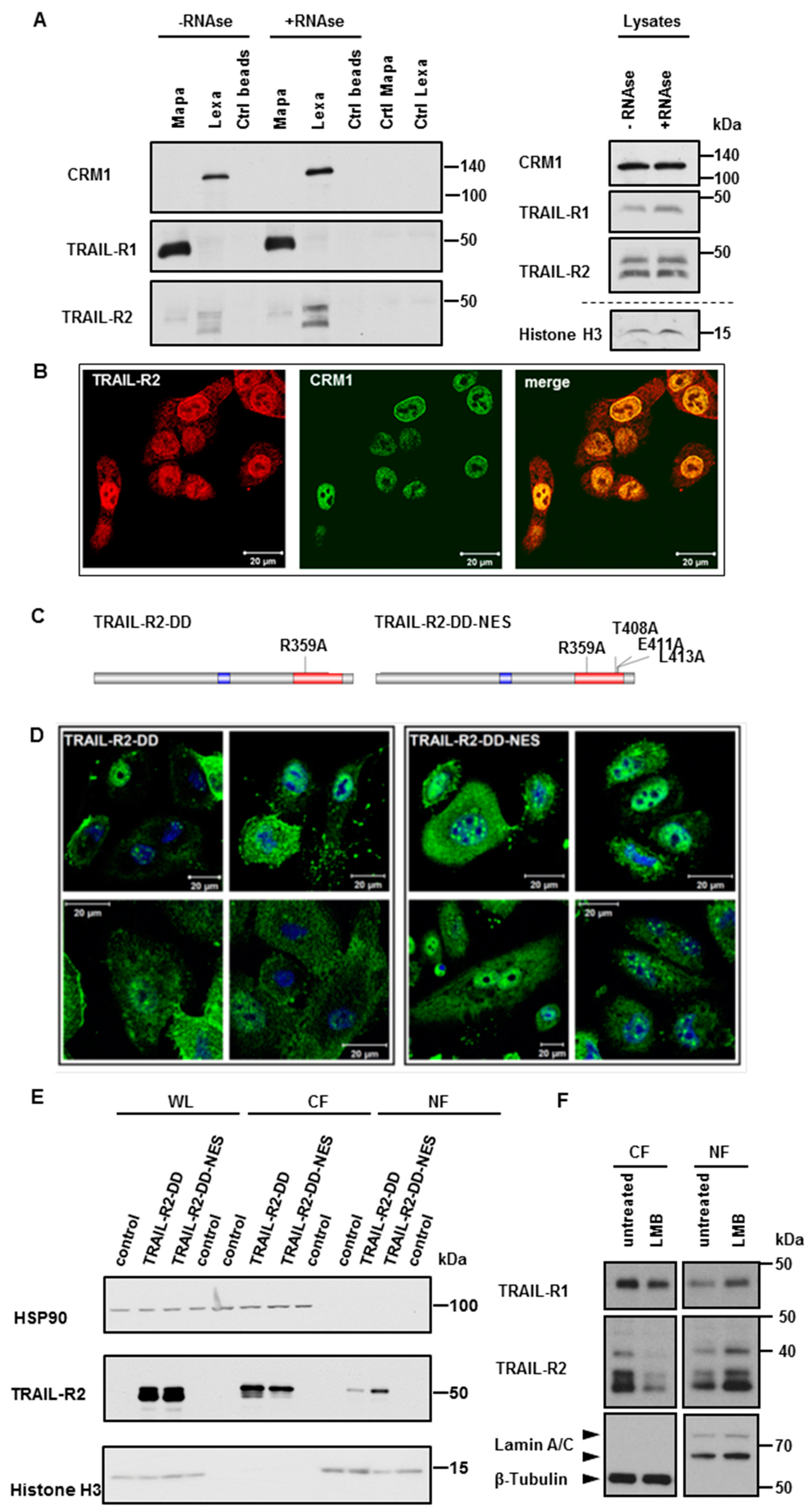
2.2. Exportin-1/CRM-1 Mediates the Nuclear Export of TRAIL Death Receptors
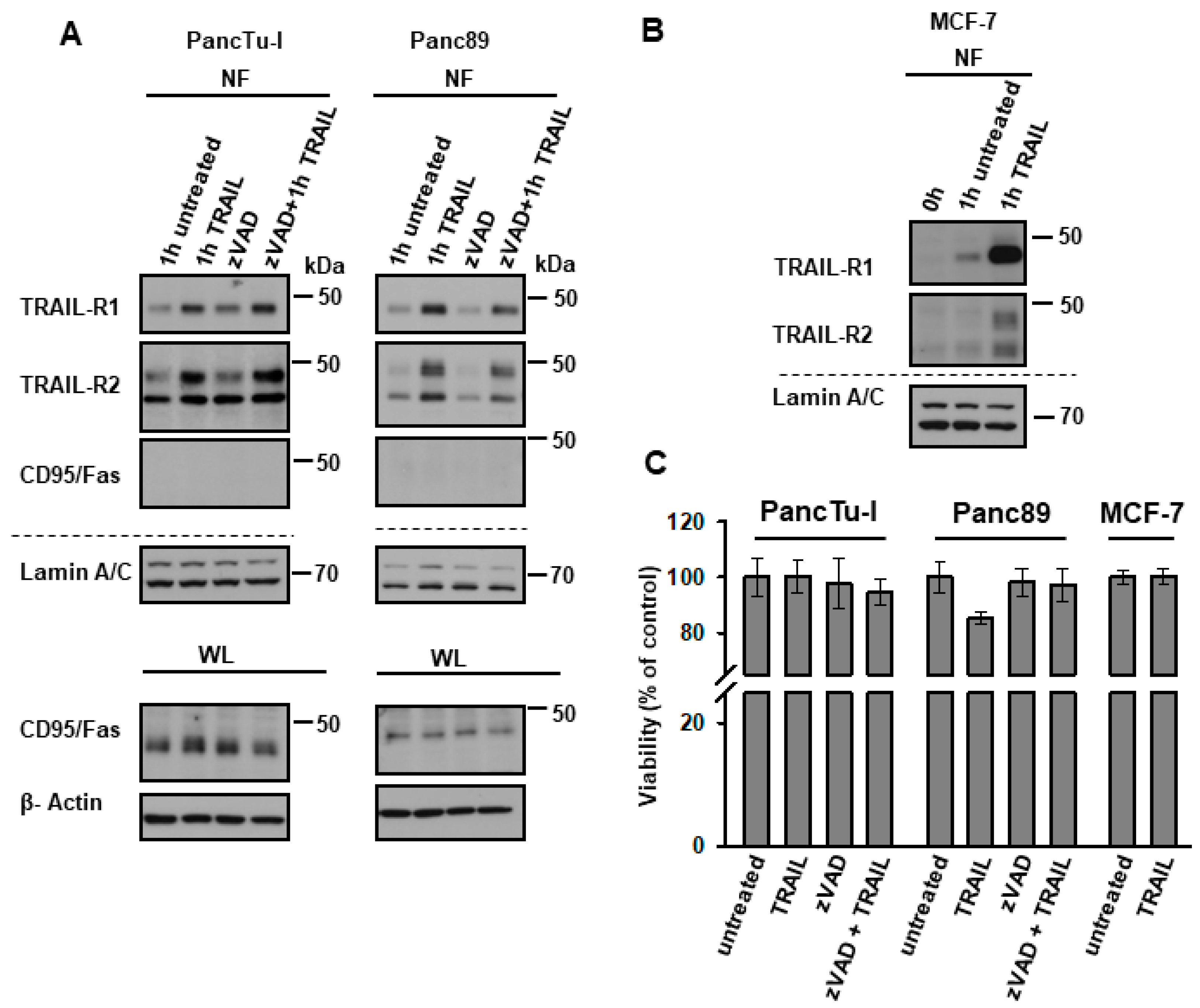
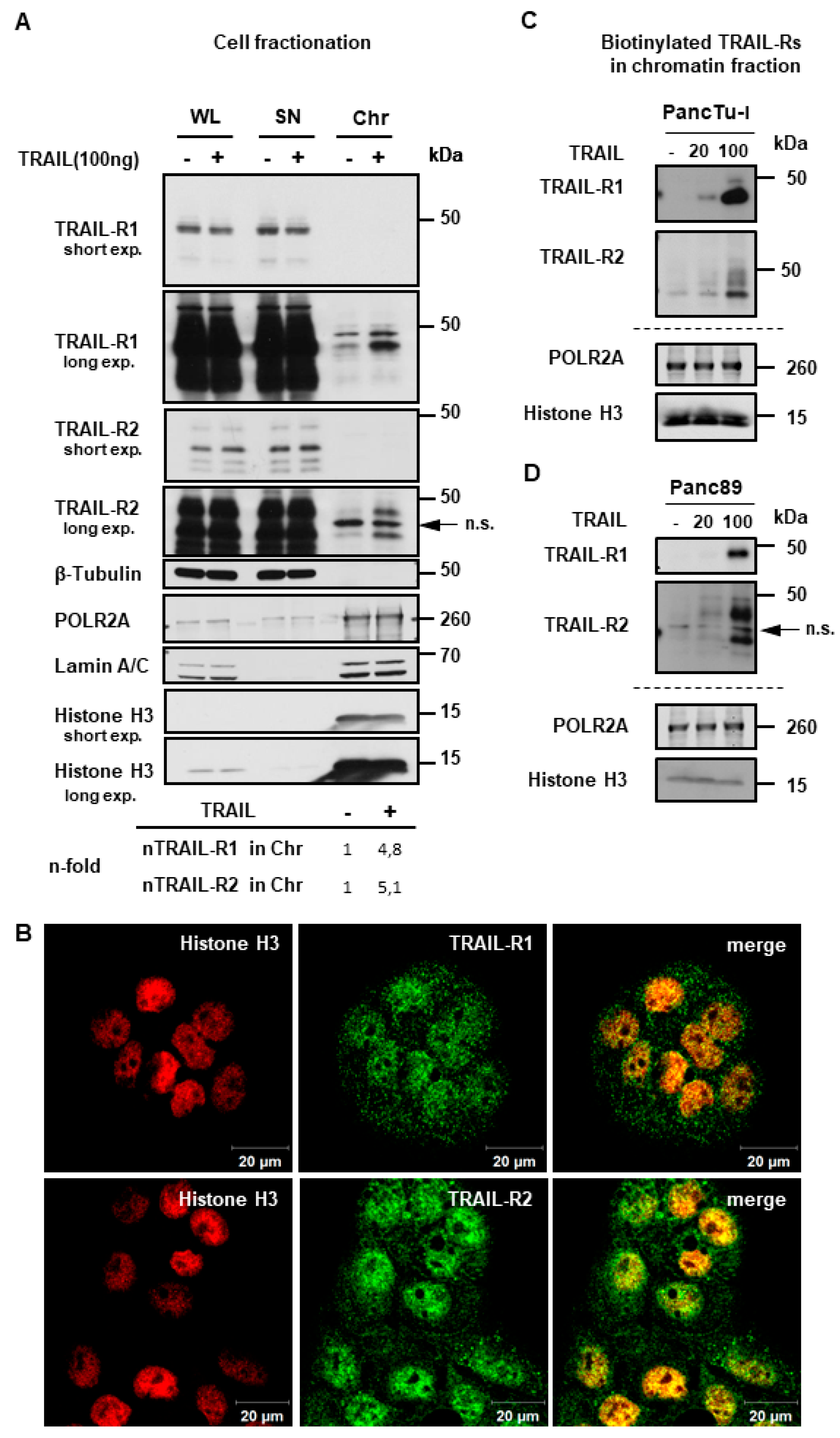
2.3. TRAIL-R1 and TRAIL-R2 Translocate into Chromatin-Rich Fractions in a TRAIL-Dependent Manner
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Nuclear Fractionation
4.3. Chromatin Fractionation
4.4. Cell Surface Protein Labeling
4.5. Flow Cytometric Analyses of Cell Surface Expression of TRAIL Receptors
4.6. TRAIL Receptors Internalization Assay
4.7. Immunofluorescence Staining
4.8. Western Blot Analysis
4.9. Immunoprecipitation
4.10. Generation and Expression of TRAIL-R2 Constructs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pan, G.; O’Rourke, K.; Chinnaiyan, A.M.; Gentz, R.; Ebner, R.; Ni, J.; Dixit, V.M. The receptor for the cytotoxic ligand TRAIL. Science 1997, 276, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Walczak, H.; Degli-Esposti, M.A.; Johnson, R.S.; Smolak, P.J.; Waugh, J.Y.; Boiani, N.; Timour, M.S.; Gerhart, M.J.; Schooley, K.A.; Smith, C.A.; et al. TRAIL-R2: A novel apoptosis-mediating receptor for TRAIL. EMBO J. 1997, 16, 5386–5397. [Google Scholar] [CrossRef] [PubMed]
- Screaton, G.R.; Mongkolsapaya, J.; Xu, X.N.; Cowper, A.E.; McMichael, A.J.; Bell, J.I. TRICK2, a new alternatively spliced receptor that transduces the cytotoxic signal from TRAIL. Curr. Biol. 1997, 7, 693–696. [Google Scholar] [CrossRef]
- Kischkel, F.C.; Lawrence, D.A.; Chuntharapai, A.; Schow, P.; Kim, K.J.; Ashkenazi, A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 2000, 12, 611–620. [Google Scholar] [CrossRef]
- Sprick, M.R.; Weigand, M.A.; Rieser, E.; Rauch, C.T.; Juo, P.; Blenis, J.; Krammer, P.H.; Walczak, H. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 2000, 12, 599–609. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, C.; Zheng, Y.; Zhang, J.; Tao, X.; Liu, S.; Zheng, D.; Liu, Y. A Novel Anti-human DR5 Monoclonal Antibody with Tumoricidal Activity Induces Caspase-dependent and Caspase-independent Cell Death. J. Biol. Chem. 2005, 280, 41940–41952. [Google Scholar] [CrossRef]
- Voigt, S.; Philipp, S.; Davarnia, P.; Winoto-Morbach, S.; Röder, C.; Arenz, C.; Trauzold, A.; Kabelitz, D.; Schütze, S.; Kalthoff, H.; et al. TRAIL-induced programmed necrosis as a novel approach to eliminate tumor cells. BMC Cancer 2014, 14, 74. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Degli-Esposti, M.A.; Smolak, P.J.; Walczak, H.; Waugh, J.; Huang, C.P.; DuBose, R.F.; Goodwin, R.G.; Smith, C.A. Cloning and characterization of TRAIL-R3, a novel member of the emerging TRAIL receptor family. J. Exp. Med. 1997, 186, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Ni, J.; Wei, Y.F.; Yu, G.; Gentz, R.; Dixit, V.M. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 1997, 277, 815–818. [Google Scholar] [CrossRef]
- Degli-Esposti, M.A.; Dougall, W.C.; Smolak, P.J.; Waugh, J.Y.; Smith, C.A.; Goodwin, R.G. The novel receptor TRAIL-R4 induces NF-kappaB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity 1997, 7, 813–820. [Google Scholar] [CrossRef]
- Mérino, D.; Lalaoui, N.; Morizot, A.; Schneider, P.; Solary, E.; Micheau, O. Differential inhibition of TRAIL-mediated DR5-DISC formation by decoy receptors 1 and 2. Mol. Cell. Biol. 2006, 26, 7046–7055. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Hasenauer, J.; Pollak, N.; Scheurich, P. Dominant negative effects of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) receptor 4 on TRAIL receptor 1 signaling by formation of heteromeric complexes. J. Biol. Chem. 2014, 289, 16576–16587. [Google Scholar] [CrossRef] [PubMed]
- Walczak, H.; Miller, R.E.; Ariail, K.; Gliniak, B.; Griffith, T.S.; Kubin, M.; Chin, W.; Jones, J.; Woodward, A.; Le, T.; et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 1999, 5, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazi, A.; Pai, R.C.; Fong, S.; Leung, S.; Lawrence, D.A.; Marsters, S.A.; Blackie, C.; Chang, L.; McMurtrey, A.E.; Hebert, A.; et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Investig. 1999, 104, 155–162. [Google Scholar] [CrossRef] [PubMed]
- de Miguel, D.; Lemke, J.; Anel, A.; Walczak, H.; Martinez-Lostao, L. Onto better TRAILs for cancer treatment. Cell Death Differ. 2016, 23, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Secchiero, P.; Melloni, E.; Heikinheimo, M.; Mannisto, S.; Di Pietro, R.; Iacone, A.; Zauli, G. TRAIL regulates normal erythroid maturation through an ERK-dependent pathway. Blood 2004, 103, 517–522. [Google Scholar] [CrossRef]
- Siegmund, D.; Klose, S.; Zhou, D.; Baumann, B.; Röder, C.; Kalthoff, H.; Wajant, H.; Trauzold, A. Role of caspases in CD95L- and TRAIL-induced non-apoptotic signalling in pancreatic tumour cells. Cell Signal. 2007, 19, 1172–1184. [Google Scholar] [CrossRef]
- Trauzold, A.; Wermann, H.; Arlt, A.; Schütze, S.; Schäfer, H.; Oestern, S.; Röder, C.; Ungefroren, H.; Lampe, E.; Heinrich, M.; et al. CD95 and TRAIL receptor-mediated activation of protein kinase C and NF-kappaB contributes to apoptosis resistance in ductal pancreatic adenocarcinoma cells. Oncogene 2001, 20, 4258–4269. [Google Scholar] [CrossRef]
- Zauli, G.; Sancilio, S.; Cataldi, A.; Sabatini, N.; Bosco, D.; Di Pietro, R. PI-3K/Akt and NF-kappaB/IkappaBalpha pathways are activated in Jurkat T cells in response to TRAIL treatment. J. Cell. Physiol. 2005, 202, 900–911. [Google Scholar] [CrossRef]
- Azijli, K.; Yuvaraj, S.; Peppelenbosch, M.P.; Würdinger, T.; Dekker, H.; Joore, J.; van Dijk, E.; Quax, W.J.; Peters, G.J.; de Jong, S.; et al. Kinome profiling of non-canonical TRAIL signaling reveals RIP1-Src-STAT3-dependent invasion in resistant non-small cell lung cancer cells. J. Cell Sci. 2012, 125, 4651–4661. [Google Scholar] [CrossRef]
- Azijli, K.; Weyhenmeyer, B.; Peters, G.J.; de Jong, S.; Kruyt, F.A.E. Non-canonical kinase signaling by the death ligand TRAIL in cancer cells: Discord in the death receptor family. Cell Death Differ. 2013, 20, 858–868. [Google Scholar] [CrossRef]
- Trauzold, A.; Siegmund, D.; Schniewind, B.; Sipos, B.; Egberts, J.; Zorenkov, D.; Emme, D.; Röder, C.; Kalthoff, H.; Wajant, H. TRAIL promotes metastasis of human pancreatic ductal adenocarcinoma. Oncogene 2006, 25, 7434–7439. [Google Scholar] [CrossRef]
- Ishimura, N.; Isomoto, H.; Bronk, S.F.; Gores, G.J. Trail induces cell migration and invasion in apoptosis-resistant cholangiocarcinoma cells. Am. J. Physiol. Liver Physiol. 2006, 290, G129–G136. [Google Scholar] [CrossRef]
- Hoogwater, F.J.H.; Nijkamp, M.W.; Smakman, N.; Steller, E.J.A.; Emmink, B.L.; Westendorp, B.F.; Raats, D.A.E.; Sprick, M.R.; Schaefer, U.; Van Houdt, W.J.; et al. Oncogenic K-Ras Turns Death Receptors Into Metastasis-Promoting Receptors in Human and Mouse Colorectal Cancer Cells. Gastroenterology 2010, 138, 2357–2367. [Google Scholar] [CrossRef]
- Hartwig, T.; Montinaro, A.; von Karstedt, S.; Sevko, A.; Surinova, S.; Chakravarthy, A.; Taraborrelli, L.; Draber, P.; Lafont, E.; Arce Vargas, F.; et al. The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2. Mol. Cell 2017, 65, 730–742. [Google Scholar] [CrossRef]
- von Karstedt, S.; Conti, A.; Nobis, M.; Montinaro, A.; Hartwig, T.; Lemke, J.; Legler, K.; Annewanter, F.; Campbell, A.D.; Taraborrelli, L.; et al. Cancer Cell-Autonomous TRAIL-R Signaling Promotes KRAS-Driven Cancer Progression, Invasion, and Metastasis. Cancer Cell 2015, 27, 561–573. [Google Scholar] [CrossRef]
- Bertsch, U.; Röder, C.; Kalthoff, H.; Trauzold, A. Compartmentalization of TNF-related apoptosis-inducing ligand (TRAIL) death receptor functions: Emerging role of nuclear TRAIL-R2. Cell Death Dis. 2014, 5, e1390. [Google Scholar] [CrossRef]
- Kojima, Y.; Nakayama, M.; Nishina, T.; Nakano, H.; Koyanagi, M.; Takeda, K.; Okumura, K.; Yagita, H. Importin β1 protein-mediated nuclear localization of death receptor 5 (DR5) limits DR5/tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-induced cell death of human tumor cells. J. Biol. Chem. 2011, 286, 43383–43393. [Google Scholar] [CrossRef]
- Reis, C.R.; Chen, P.-H.; Bendris, N.; Schmid, S.L. TRAIL-death receptor endocytosis and apoptosis are selectively regulated by dynamin-1 activation. Proc. Natl. Acad. Sci. USA 2017, 114, 504–509. [Google Scholar] [CrossRef]
- Chen, J.-J.; Shen, H.-C.J.; Rivera Rosado, L.A.; Zhang, Y.; Di, X.; Zhang, B. Mislocalization of death receptors correlates with cellular resistance to their cognate ligands in human breast cancer cells. Oncotarget 2012, 3, 833–842. [Google Scholar]
- Di, X.; Zhang, G.; Zhang, Y.; Takeda, K.; Rivera Rosado, L.A.; Zhang, B. Accumulation of autophagosomes in breast cancer cells induces TRAIL resistance through downregulation of surface expression of death receptors 4 and 5. Oncotarget 2013, 4, 1349–1364. [Google Scholar] [CrossRef]
- Dufour, F.; Rattier, T.; Constantinescu, A.A.; Zischler, L.; Morlé, A.; Ben Mabrouk, H.; Humblin, E.; Jacquemin, G.; Szegezdi, E.; Delacote, F.; et al. TRAIL receptor gene editing unveils TRAIL-R1 as a master player of apoptosis induced by TRAIL and ER stress. Oncotarget 2017, 8, 9974–9985. [Google Scholar] [CrossRef]
- Lu, M.; Lawrence, D.A.; Marsters, S.; Acosta-Alvear, D.; Kimmig, P.; Mendez, A.S.; Paton, A.W.; Paton, J.C.; Walter, P.; Ashkenazi, A. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 2014, 345, 98–101. [Google Scholar] [CrossRef]
- Haselmann, V.; Kurz, A.; Bertsch, U.; Hübner, S.; Olempska-Müller, M.; Fritsch, J.; Häsler, R.; Pickl, A.; Fritsche, H.; Annewanter, F.; et al. Nuclear death receptor TRAIL-R2 inhibits maturation of let-7 and promotes proliferation of pancreatic and other tumor cells. Gastroenterology 2014, 146, 278–290. [Google Scholar] [CrossRef]
- Kohlhaas, S.L.; Craxton, A.; Sun, X.-M.; Pinkoski, M.J.; Cohen, G.M. Receptor-mediated Endocytosis Is Not Required for Tumor Necrosis Factor-related Apoptosis-inducing Ligand (TRAIL)-induced Apoptosis. J. Biol. Chem. 2007, 282, 12831–12841. [Google Scholar] [CrossRef]
- Zhang, Y.; Yoshida, T.; Zhang, B. TRAIL induces endocytosis of its death receptors in MDA-MB-231 breast cancer cells. Cancer Biol. Ther. 2009, 8, 917–922. [Google Scholar] [CrossRef]
- Akazawa, Y.; Mott, J.L.; Bronk, S.F.; Werneburg, N.W.; Kahraman, A.; Guicciardi, M.E.; Meng, X.W.; Kohno, S.; Shah, V.H.; Kaufmann, S.H.; et al. Death receptor 5 internalization is required for lysosomal permeabilization by TRAIL in malignant liver cell lines. Gastroenterology 2009, 136, 2365–2376. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, B. TRAIL resistance of breast cancer cells is associated with constitutive endocytosis of death receptors 4 and 5. Mol. Cancer Res. 2008, 6, 1861–1871. [Google Scholar] [CrossRef]
- Monleón, I.; Martínez-Lorenzo, M.J.; Monteagudo, L.; Lasierra, P.; Taulés, M.; Iturralde, M.; Piñeiro, A.; Larrad, L.; Alava, M.A.; Naval, J.; et al. Differential secretion of Fas ligand- or APO2 ligand/TNF-related apoptosis-inducing ligand-carrying microvesicles during activation-induced death of human T cells. J. Immunol. 2001, 167, 6736–6744. [Google Scholar] [CrossRef]
- Koga, Y.; Matsuzaki, A.; Suminoe, A.; Hattori, H.; Hara, T. Neutrophil-derived TNF-related apoptosis-inducing ligand (TRAIL): A novel mechanism of antitumor effect by neutrophils. Cancer Res. 2004, 64, 1037–1043. [Google Scholar] [CrossRef]
- Lemke, J.; Noack, A.; Adam, D.; Tchikov, V.; Bertsch, U.; Röder, C.; Schütze, S.; Wajant, H.; Kalthoff, H.; Trauzold, A. TRAIL signaling is mediated by DR4 in pancreatic tumor cells despite the expression of functional DR5. J. Mol. Med. 2010, 88, 729–740. [Google Scholar] [CrossRef]
- Turner, J.G.; Sullivan, D.M. CRM1-mediated nuclear export of proteins and drug resistance in cancer. Curr. Med. Chem. 2008, 15, 2648–2655. [Google Scholar] [CrossRef]
- la Cour, T.; Kiemer, L.; Mølgaard, A.; Gupta, R.; Skriver, K.; Brunak, S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef]
- Humphreys, R.C.; Halpern, W. Trail receptors: Targets for cancer therapy. Adv. Exp. Med. Biol. 2008, 615, 127–158. [Google Scholar]
- Kudo, N.; Matsumori, N.; Taoka, H.; Fujiwara, D.; Schreiner, E.P.; Wolff, B.; Yoshida, M.; Horinouchi, S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 1999, 96, 9112–9117. [Google Scholar] [CrossRef]
- Brand, T.M.; Iida, M.; Li, C.; Wheeler, D.L. The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discov. Med. 2011, 12, 419–432. [Google Scholar]
- Hsu, S.-C.; Miller, S.A.; Wang, Y.; Hung, M.-C. Nuclear EGFR is required for cisplatin resistance and DNA repair. Am. J. Transl. Res. 2009, 1, 249–258. [Google Scholar]
- Gundlach, J.-P.; Hauser, C.; Schlegel, F.M.; Böger, C.; Röder, C.; Röcken, C.; Becker, T.; Egberts, J.-H.; Kalthoff, H.; Trauzold, A. Cytoplasmic TRAIL-R1 is a positive prognostic marker in PDAC. BMC Cancer 2018, 18, 777. [Google Scholar] [CrossRef]
- Bryant, D.M.; Stow, J.L. Nuclear Translocation of Cell-Surface Receptors: Lessons from Fibroblast Growth Factor. Traffic 2005, 6, 947–953. [Google Scholar] [CrossRef]
- Liao, H.-J.; Carpenter, G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol. Biol. Cell 2007, 18, 1064–1072. [Google Scholar] [CrossRef]
- Giri, D.K.; Ali-Seyed, M.; Li, L.-Y.; Lee, D.-F.; Ling, P.; Bartholomeusz, G.; Wang, S.-C.; Hung, M.-C. Endosomal transport of ErbB-2: Mechanism for nuclear entry of the cell surface receptor. Mol. Cell. Biol. 2005, 25, 11005–11018. [Google Scholar] [CrossRef]
- Kutay, U.; Güttinger, S. Leucine-rich nuclear-export signals: Born to be weak. Trends Cell Biol. 2005, 15, 121–124. [Google Scholar] [CrossRef]
- Cook, A.; Bono, F.; Jinek, M.; Conti, E. Structural Biology of Nucleocytoplasmic Transport. Annu. Rev. Biochem. 2007, 76, 647–671. [Google Scholar] [CrossRef]
- Engelsma, D.; Bernad, R.; Calafat, J.; Fornerod, M. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 2004, 23, 3643–3652. [Google Scholar] [CrossRef]
- Nguyen, K.T.; Holloway, M.P.; Altura, R.A. The CRM1 nuclear export protein in normal development and disease. Int. J. Biochem. Mol. Biol. 2012, 3, 137–151. [Google Scholar]
- Wang, Y.-N.; Hung, M.-C. Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2012, 2, 13. [Google Scholar] [CrossRef]
- Hung, L.-Y.; Tseng, J.T.; Lee, Y.-C.; Xia, W.; Wang, Y.-N.; Wu, M.-L.; Chuang, Y.-H.; Lai, C.-H.; Chang, W.-C. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008, 36, 4337–4351. [Google Scholar] [CrossRef]
- Lo, H.-W.; Cao, X.; Zhu, H.; Ali-Osman, F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol. Cancer Res. 2010, 8, 232–245. [Google Scholar] [CrossRef]
- Lo, H.-W.; Hsu, S.-C.; Ali-Seyed, M.; Gunduz, M.; Xia, W.; Wei, Y.; Bartholomeusz, G.; Shih, J.-Y.; Hung, M.-C. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell 2005, 7, 575–589. [Google Scholar] [CrossRef]
- Hanada, N.; Lo, H.-W.; Day, C.-P.; Pan, Y.; Nakajima, Y.; Hung, M.-C. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol. Carcinog. 2006, 45, 10–17. [Google Scholar] [CrossRef]
- Huo, L.; Wang, Y.-N.; Xia, W.; Hsu, S.-C.; Lai, C.-C.; Li, L.-Y.; Chang, W.-C.; Wang, Y.; Hsu, M.-C.; Yu, Y.-L.; et al. RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. USA 2010, 107, 16125–16130. [Google Scholar] [CrossRef]
- Zwaagstra, J.C.; Guimond, A.; O’Connor-McCourt, M.D. Predominant intracellular localization of the type I transforming growth factor-beta receptor and increased nuclear accumulation after growth arrest. Exp. Cell Res. 2000, 258, 121–134. [Google Scholar] [CrossRef]
- Chandra, M.; Zang, S.; Li, H.; Zimmerman, L.J.; Champer, J.; Tsuyada, A.; Chow, A.; Zhou, W.; Yu, Y.; Gao, H.; et al. Nuclear translocation of type I transforming growth factor β receptor confers a novel function in RNA processing. Mol. Cell. Biol. 2012, 32, 2183–2195. [Google Scholar] [CrossRef]
- Zhou, H.-J.; Pham, L.V.; Tamayo, A.T.; Lin-Lee, Y.-C.; Fu, L.; Yoshimura, L.C.; Ford, R.J. Nuclear CD40 interacts with c-Rel and enhances proliferation in aggressive B-cell lymphoma. Blood 2007, 110, 2121–2127. [Google Scholar] [CrossRef]
- Fu, L.; Lin-Lee, Y.-C.; Pham, L.V.; Tamayo, A.T.; Yoshimura, L.C.; Ford, R.J. BAFF-R promotes cell proliferation and survival through interaction with IKKbeta and NF-kappaB/c-Rel in the nucleus of normal and neoplastic B-lymphoid cells. Blood 2009, 113, 4627–4636. [Google Scholar] [CrossRef]
- Gómez-Angelats, M.; Cidlowski, J.A. Molecular evidence for the nuclear localization of FADD. Cell Death Differ. 2003, 10, 791–797. [Google Scholar] [CrossRef]
- Benchoua, A.; Couriaud, C.; Guégan, C.; Tartier, L.; Couvert, P.; Friocourt, G.; Chelly, J.; Ménissier-de Murcia, J.; Onténiente, B. Active caspase-8 translocates into the nucleus of apoptotic cells to inactivate poly(ADP-ribose) polymerase-2. J. Biol. Chem. 2002, 277, 34217–34222. [Google Scholar] [CrossRef]
- Koschny, R.; Brost, S.; Hinz, U.; Sykora, J.; Batke, E.M.; Singer, S.; Breuhahn, K.; Stremmel, W.; Walczak, H.; Schemmer, P.; et al. Cytosolic and nuclear caspase-8 have opposite impact on survival after liver resection for hepatocellular carcinoma. BMC Cancer 2013, 13, 532. [Google Scholar] [CrossRef]
- Boege, Y.; Malehmir, M.; Healy, M.E.; Bettermann, K.; Lorentzen, A.; Vucur, M.; Ahuja, A.K.; Böhm, F.; Mertens, J.C.; Shimizu, Y.; et al. A Dual Role of Caspase-8 in Triggering and Sensing Proliferation-Associated DNA Damage, a Key Determinant of Liver Cancer Development. Cancer Cell 2017, 32, 342–359. [Google Scholar] [CrossRef]
- Yoon, S.; Bogdanov, K.; Kovalenko, A.; Wallach, D. Necroptosis is preceded by nuclear translocation of the signaling proteins that induce it. Cell Death Differ. 2016, 23, 253–260. [Google Scholar] [CrossRef]
- Henry, C.M.; Martin, S.J. Caspase-8 Acts in a Non-enzymatic Role as a Scaffold for Assembly of a Pro-inflammatory “FADDosome” Complex upon TRAIL Stimulation. Mol. Cell 2017, 65, 715–729. [Google Scholar] [CrossRef]
- Wang, S.-C.; Nakajima, Y.; Yu, Y.-L.; Xia, W.; Chen, C.-T.; Yang, C.-C.; McIntush, E.W.; Li, L.-Y.; Hawke, D.H.; Kobayashi, R.; et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 2006, 8, 1359–1368. [Google Scholar] [CrossRef]
- Dittmann, K.; Mayer, C.; Fehrenbacher, B.; Schaller, M.; Raju, U.; Milas, L.; Chen, D.J.; Kehlbach, R.; Rodemann, H.P. Radiation-induced Epidermal Growth Factor Receptor Nuclear Import Is Linked to Activation of DNA-dependent Protein Kinase. J. Biol. Chem. 2005, 280, 31182–31189. [Google Scholar] [CrossRef]
- Liccardi, G.; Hartley, J.A.; Hochhauser, D. EGFR Nuclear Translocation Modulates DNA Repair following Cisplatin and Ionizing Radiation Treatment. Cancer Res. 2011, 71, 1103–1114. [Google Scholar] [CrossRef]
- von Karstedt, S.; Montinaro, A.; Walczak, H. Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat. Rev. Cancer 2017, 17, 352–366. [Google Scholar] [CrossRef]
- Ledoux, S.; Yang, R.; Friedlander, G.; Laouari, D. Glucose depletion enhances P-glycoprotein expression in hepatoma cells: Role of endoplasmic reticulum stress response. Cancer Res. 2003, 63, 7284–7290. [Google Scholar]
- Montes de Oca, R.; Andreassen, P.R.; Margossian, S.P.; Gregory, R.C.; Taniguchi, T.; Wang, X.; Houghtaling, S.; Grompe, M.; D’Andrea, A.D. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood 2004, 105, 1003–1009. [Google Scholar] [CrossRef]
- McDonald, E.R.; Chui, P.C.; Martelli, P.F.; Dicker, D.T.; El-Deiry, W.S. Death domain mutagenesis of KILLER/DR5 reveals residues critical for apoptotic signaling. J. Biol. Chem. 2001, 276, 14939–14945. [Google Scholar] [CrossRef]
- Narita, M.; Narita, M.; Krizhanovsky, V.; Nuñez, S.; Chicas, A.; Hearn, S.A.; Myers, M.P.; Lowe, S.W. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 2006, 126, 503–514. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mert, U.; Adawy, A.; Scharff, E.; Teichmann, P.; Willms, A.; Haselmann, V.; Colmorgen, C.; Lemke, J.; von Karstedt, S.; Fritsch, J.; et al. TRAIL Induces Nuclear Translocation and Chromatin Localization of TRAIL Death Receptors. Cancers 2019, 11, 1167. https://doi.org/10.3390/cancers11081167
Mert U, Adawy A, Scharff E, Teichmann P, Willms A, Haselmann V, Colmorgen C, Lemke J, von Karstedt S, Fritsch J, et al. TRAIL Induces Nuclear Translocation and Chromatin Localization of TRAIL Death Receptors. Cancers. 2019; 11(8):1167. https://doi.org/10.3390/cancers11081167
Chicago/Turabian StyleMert, Ufuk, Alshaimaa Adawy, Elisabeth Scharff, Pierre Teichmann, Anna Willms, Verena Haselmann, Cynthia Colmorgen, Johannes Lemke, Silvia von Karstedt, Jürgen Fritsch, and et al. 2019. "TRAIL Induces Nuclear Translocation and Chromatin Localization of TRAIL Death Receptors" Cancers 11, no. 8: 1167. https://doi.org/10.3390/cancers11081167
APA StyleMert, U., Adawy, A., Scharff, E., Teichmann, P., Willms, A., Haselmann, V., Colmorgen, C., Lemke, J., von Karstedt, S., Fritsch, J., & Trauzold, A. (2019). TRAIL Induces Nuclear Translocation and Chromatin Localization of TRAIL Death Receptors. Cancers, 11(8), 1167. https://doi.org/10.3390/cancers11081167




