miR551b Regulates Colorectal Cancer Progression by Targeting the ZEB1 Signaling Axis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. miR551b Construct and Infection
2.3. Transfection with miR-551b Mimics
2.4. Cell Proliferation Assay (MTT Assay)
2.5. Migration and Invasion Assay
2.6. Wound Healing Assay
2.7. RNA Extraction and Real Time qPCR (RT-qPCR)
2.8. Western Blotting Analysis
2.9. Dual Luciferase Reporter Assay
2.10. Xenograft Mouse Model
2.11. Microarray Data Analysis
2.12. Survival Analysis of ZEB1
2.13. Information of Tissue Specimens
2.14. Statistical Analysis
3. Results
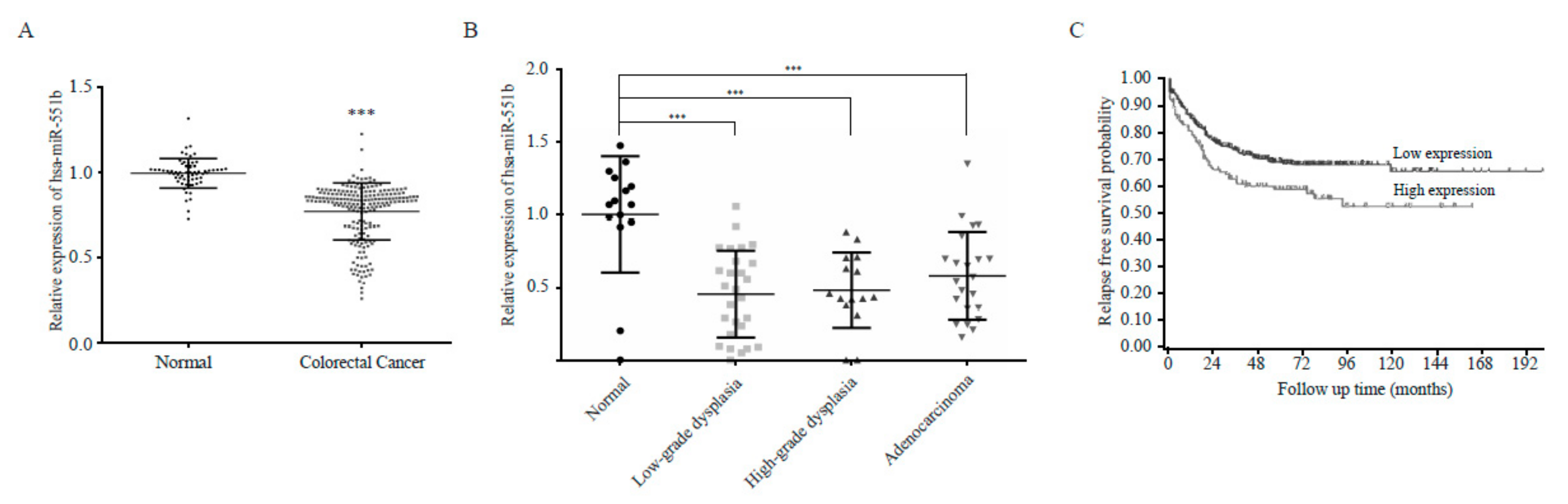
3.1. miR551b-3p is Downregulated in CRC Cell Lines and Patient-Derived Samples
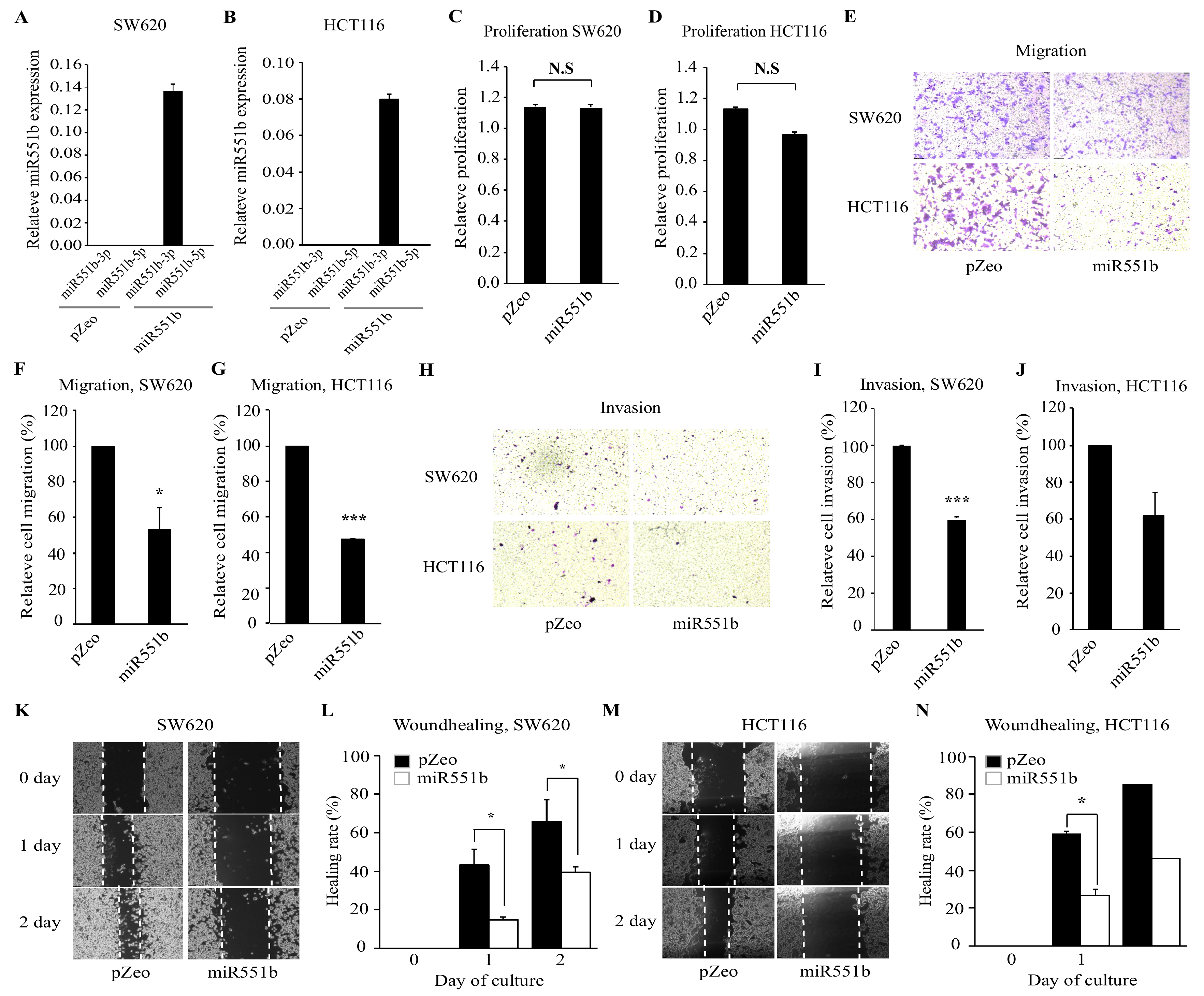
3.2. Ectopic Expression of miR551b Inhibits the Migration and Invasion of CRC Cell Lines In Vitro
3.3. miR551b-3p Mimic Inhibits Motility of CRC Cell Lines In Vitro
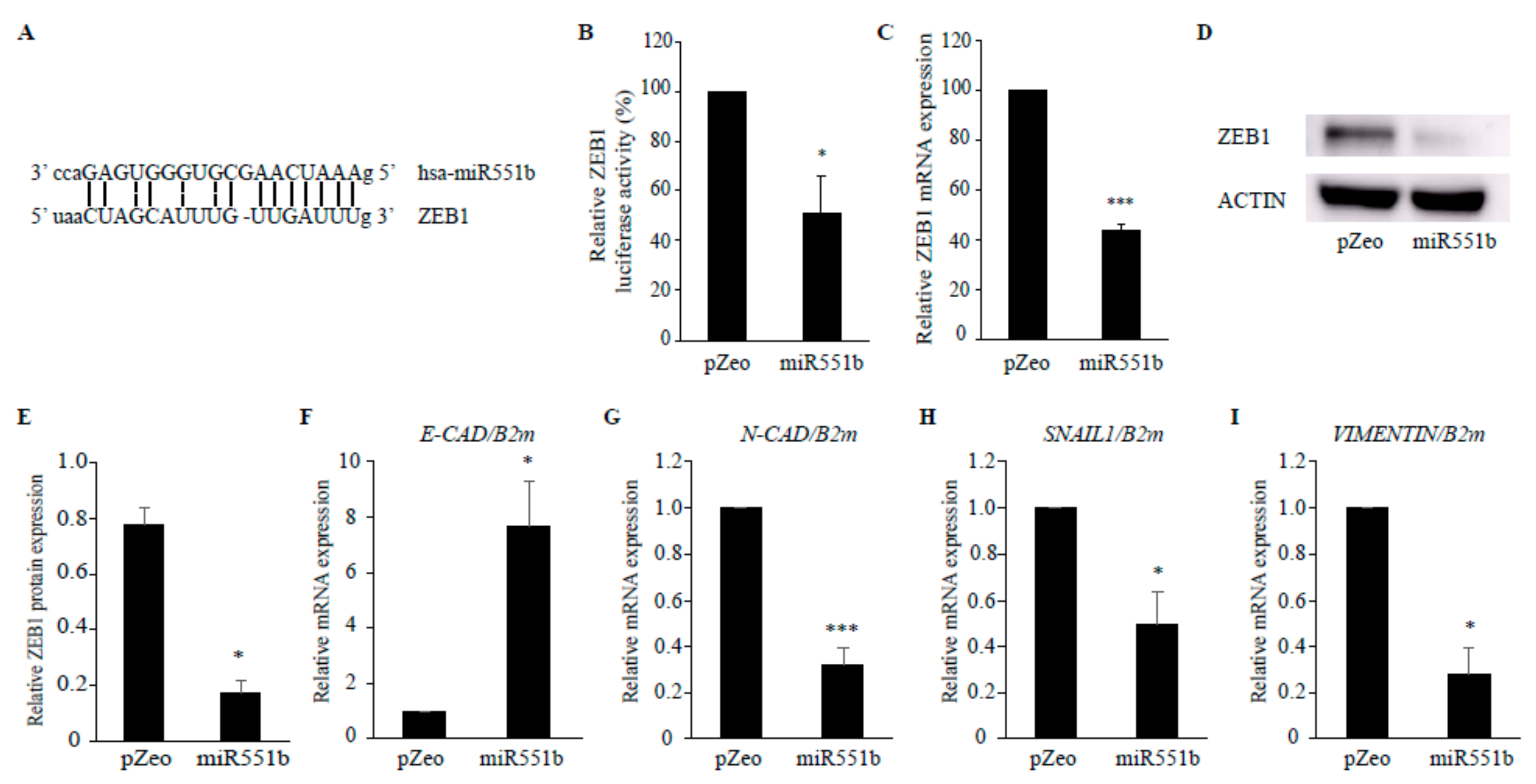
3.4. miR551b Targets ZEB1, Leading to the Dysregulation of the EMT Signaling Pathway
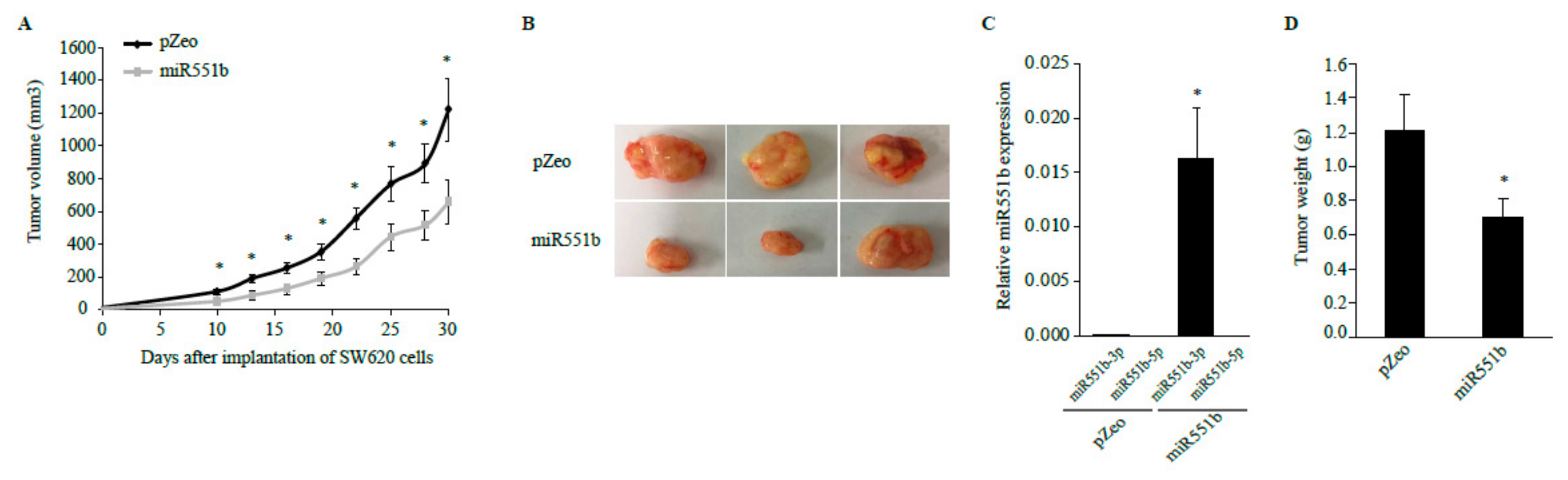
3.5. miR551b Overexpression Suppressed Tumor Growth in a Mouse Xenograft Model of CRC Cells In Vivo
3.6. The Inverse Correlation Between miR551b and ZEB1 in the Prognosis of CRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Ait Ouakrim, D.; Pizot, C.; Boniol, M.; Malvezzi, M.; Boniol, M.; Negri, E.; Bota, M.; Jenkins, M.A.; Bleiberg, H.; Autier, P. Trends in colorectal cancer mortality in Europe: Retrospective analysis of the WHO mortality database. BMJ 2015, 351, h4970. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Gao, W.; Lu, Y.; Lan, H.; Teng, L.; Cao, F. Mechanisms regulating colorectal cancer cell metastasis into liver (Review). Oncol. Lett. 2012, 3, 11–15. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Agarwal, E.; Robb, C.M.; Smith, L.M.; Brattain, M.G.; Wang, J.; Black, J.D.; Chowdhury, S. Role of Akt2 in regulation of metastasis suppressor 1 expression and colorectal cancer metastasis. Oncogene 2017, 36, 3104. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Weinberg, R.A. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Jou, J.; Diehl, A.M. Epithelial-mesenchymal transitions and hepatocarcinogenesis. J. Clin. Investig. 2010, 120, 1031–1034. [Google Scholar] [CrossRef]
- Hur, K.; Toiyama, Y.; Takahashi, M.; Balaguer, F.; Nagasaka, T.; Koike, J.; Hemmi, H.; Koi, M.; Boland, C.R.; Goel, A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut 2013, 62, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Spaderna, S.; Schmalhofer, O.; Hlubek, F.; Berx, G.; Eger, A.; Merkel, S.; Jung, A.; Kirchner, T.; Brabletz, T. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology 2006, 131, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Roseweir, A.K.; Kong, C.Y.; Park, J.H.; Bennett, L.; Powell, A.G.M.T.; Quinn, J.; van Wyk, H.C.; Horgan, P.G.; McMillan, D.C.; Edwards, J.; et al. A novel tumor-based epithelial-to-mesenchymal transition score that associates with prognosis and metastasis in patients with Stage II/III colorectal cancer. Int. J. Cancer 2019, 144, 150–159. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P.K. Regulation of EMT in colorectal cancer: A culprit in metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.P.; Jopling, C.L. Targeting viral infection by microRNA inhibition. Genome Biol. 2010, 11, 201. [Google Scholar] [CrossRef]
- Sayed, D.; Abdellatif, M. MicroRNAs in development and disease. Physiol. Rev. 2011, 91, 827–887. [Google Scholar] [CrossRef]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Hobert, O. Gene regulation by transcription factors and microRNAs. Science 2008, 319, 1785–1786. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Lovat, F.; Valeri, N.; Croce, C.M. MicroRNAs in the pathogenesis of cancer. Semin. Oncol. 2011, 38, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug. Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed]
- Chaluvally-Raghavan, P.; Jeong, K.J.; Pradeep, S.; Silva, A.M.; Yu, S.; Liu, W.; Moss, T.; Rodriguez-Aguayo, C.; Zhang, D.; Ram, P.; et al. Direct upregulation of STAT3 by microRNA-551b-3p deregulates growth and metastasis of ovarian cancer. Cell Rep. 2016, 15, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Karanam, N.K.; Ding, L.-H.; Hwang, T.H.; Tang, H.; Story, M.D. miR-551a and miR-551b-3p target GLIPR2 and promote tumor growth in High-Risk Head and Neck Cancer by modulating autophagy. bioRxiv 2015. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, Y.; Wang, Y.; Zhang, Y.; Luo, Q.; Man, X.; Wei, F.; Yu, X. Downregulation of Foxo3 and TRIM31 by miR-551b in side population promotes cell proliferation, invasion, and drug resistance of ovarian cancer. Med. Oncol. 2016, 33, 126. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, X.; Liu, M.; Liu, X.; Jia, J.; Ji, R.; Guo, Q.; Zhou, Y. Expression of miR-551b-3p in gastric cancer cell lines and tissues and its clinical significance. Zhonghua Zhong Liu Za Zhi 2014, 36, 903–904. [Google Scholar] [PubMed]
- Song, G.; Zhang, H.; Chen, C.; Gong, L.; Chen, B.; Zhao, S.; Shi, J.; Xu, J.; Ye, Z. miR-551b regulates epithelial-mesenchymal transition and metastasis of gastric cancer by inhibiting ERBB4 expression. Oncotarget 2017, 8, 45725–45735. [Google Scholar] [CrossRef]
- Ryu, S.; Joshi, N.; McDonnell, K.; Woo, J.; Choi, H.; Gao, D.; McCombie, W.R.; Mittal, V. Discovery of novel human breast cancer microRNAs from deep sequencing data by analysis of pri-microRNA secondary structures. PLoS ONE 2011, 6, e16403. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.G.; Koh, Y.W.; Sari, I.N.; Jun, N.; Lee, S.; Phi, L.T.H.; Kim, K.S.; Wijaya, Y.T.; Lee, S.H.; Baek, M.J.; et al. Interferon-induced transmembrane protein 1-mediated EGFR/SOX2 signaling axis is essential for progression of non-small cell lung cancer. Int. J. Cancer 2019, 144, 2020–2032. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sun, Y.; Ma, L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle 2015, 14, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Tillo, E.; Lazaro, A.; Torrent, R.; Cuatrecasas, M.; Vaquero, E.C.; Castells, A.; Engel, P.; Postigo, A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 2010, 29, 3490–3500. [Google Scholar] [CrossRef]
- Eger, A.; Aigner, K.; Sonderegger, S.; Dampier, B.; Oehler, S.; Schreiber, M.; Berx, G.; Cano, A.; Beug, H.; Foisner, R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005, 24, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Sawada, J.; Sui, G.; Affar el, B.; Whetstine, J.R.; Lan, F.; Ogawa, H.; Luke, M.P.; Nakatani, Y.; Shi, Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 2003, 422, 735–738. [Google Scholar] [CrossRef]
- Title, A.C.; Hong, S.-J.; Pires, N.D.; Hasenöhrl, L.; Godbersen, S.; Stokar-Regenscheit, N.; Bartel, D.P.; Stoffel, M. Genetic dissection of the miR-200–Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat. Commun. 2018, 9, 4671. [Google Scholar] [CrossRef]
- Diepenbruck, M.; Tiede, S.; Saxena, M.; Ivanek, R.; Kalathur, R.K.R.; Lüönd, F.; Meyer-Schaller, N.; Christofori, G. miR-1199-5p and Zeb1 function in a double-negative feedback loop potentially coordinating EMT and tumour metastasis. Nat. Commun. 2017, 8, 1168. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, R.; Xu, S.; Li, Y.; Zhao, P.; Dong, W.; Liu, Z.; Zhao, Q.; Tan, B. Tumor suppressor miR-128-3p inhibits metastasis and epithelial–mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim. Biophys. Sin. 2018, 50, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Postigo, A.A.; Depp, J.L.; Taylor, J.J.; Kroll, K.L. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003, 22, 2453–2462. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.S.; Jeong, D.; Sari, I.N.; Wijaya, Y.T.; Jun, N.; Lee, S.; Yang, Y.-G.; Lee, S.H.; Kwon, H.Y. miR551b Regulates Colorectal Cancer Progression by Targeting the ZEB1 Signaling Axis. Cancers 2019, 11, 735. https://doi.org/10.3390/cancers11050735
Kim KS, Jeong D, Sari IN, Wijaya YT, Jun N, Lee S, Yang Y-G, Lee SH, Kwon HY. miR551b Regulates Colorectal Cancer Progression by Targeting the ZEB1 Signaling Axis. Cancers. 2019; 11(5):735. https://doi.org/10.3390/cancers11050735
Chicago/Turabian StyleKim, Kwang Seock, Dongjun Jeong, Ita Novita Sari, Yoseph Toni Wijaya, Nayoung Jun, Sanghyun Lee, Ying-Gui Yang, Sae Hwan Lee, and Hyog Young Kwon. 2019. "miR551b Regulates Colorectal Cancer Progression by Targeting the ZEB1 Signaling Axis" Cancers 11, no. 5: 735. https://doi.org/10.3390/cancers11050735
APA StyleKim, K. S., Jeong, D., Sari, I. N., Wijaya, Y. T., Jun, N., Lee, S., Yang, Y.-G., Lee, S. H., & Kwon, H. Y. (2019). miR551b Regulates Colorectal Cancer Progression by Targeting the ZEB1 Signaling Axis. Cancers, 11(5), 735. https://doi.org/10.3390/cancers11050735




