Detecting and Tracking Circulating Tumour DNA Copy Number Profiles during First Line Chemotherapy in Oesophagogastric Adenocarcinoma
Abstract
1. Introduction
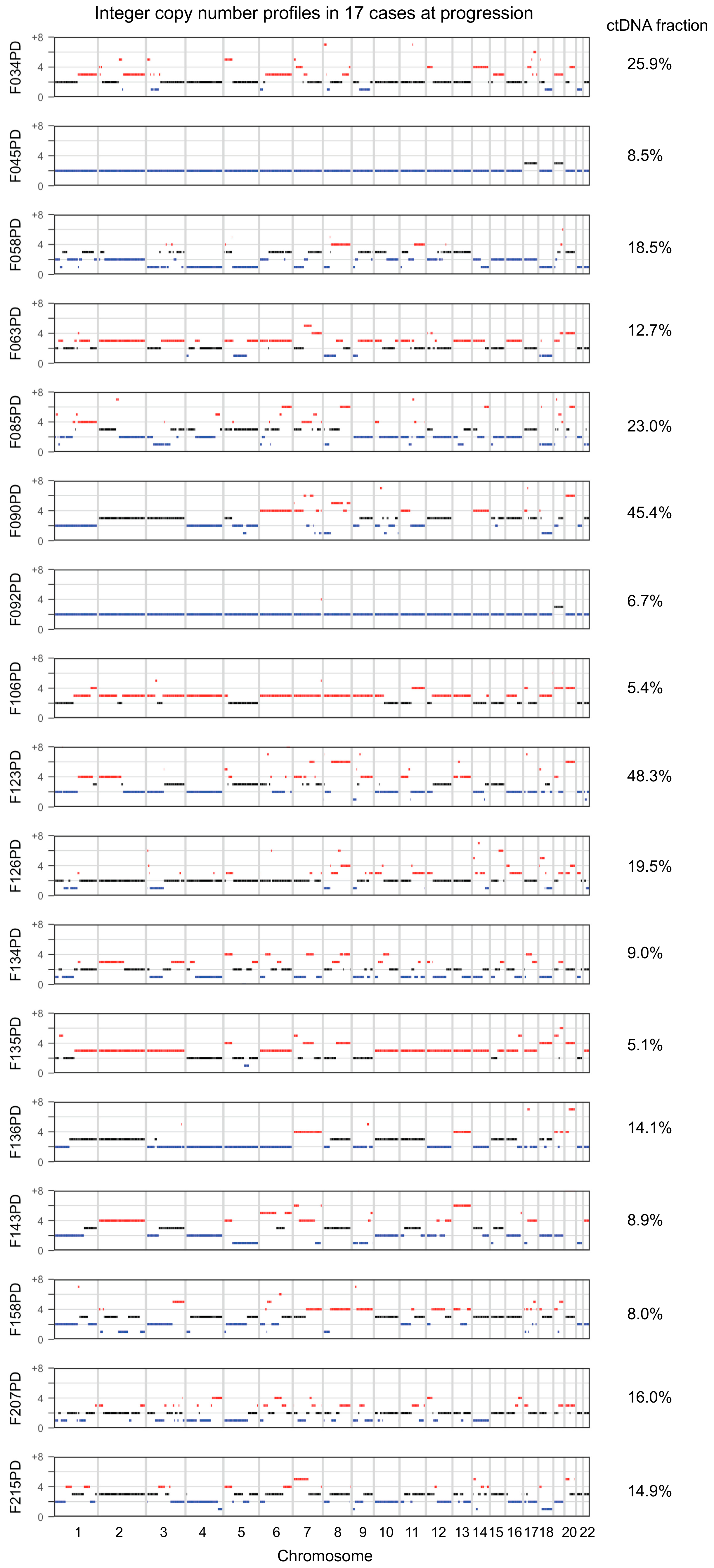
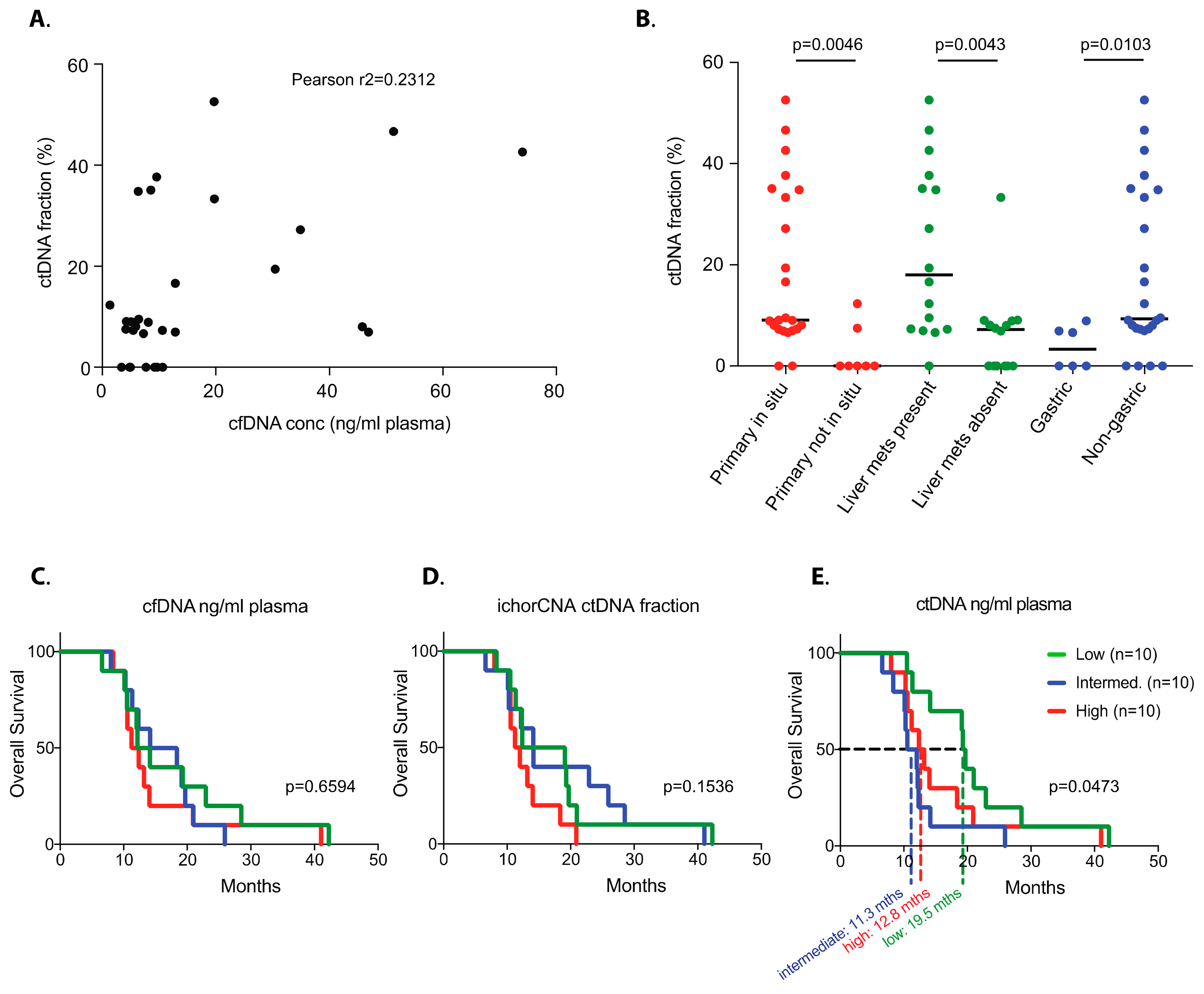
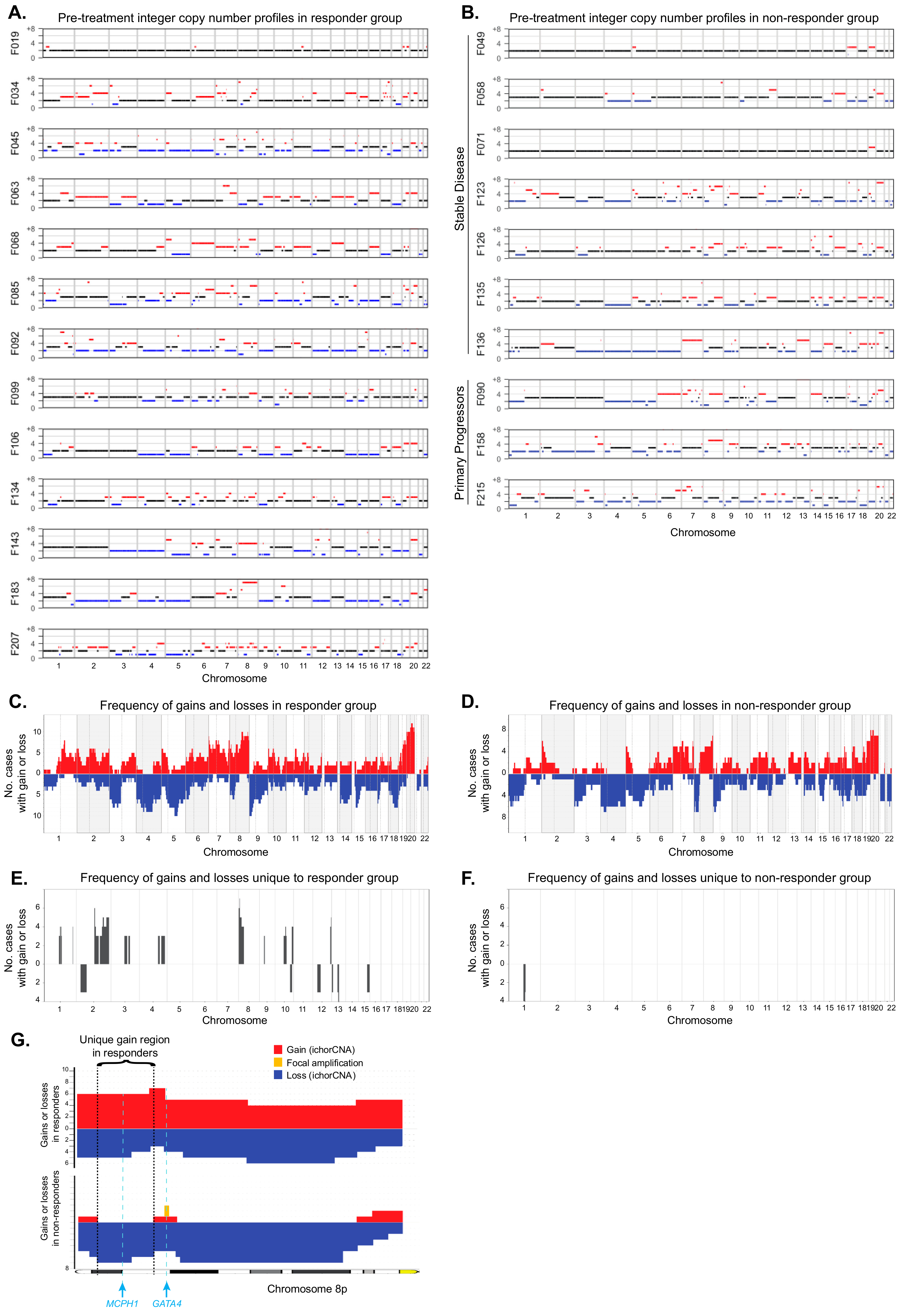
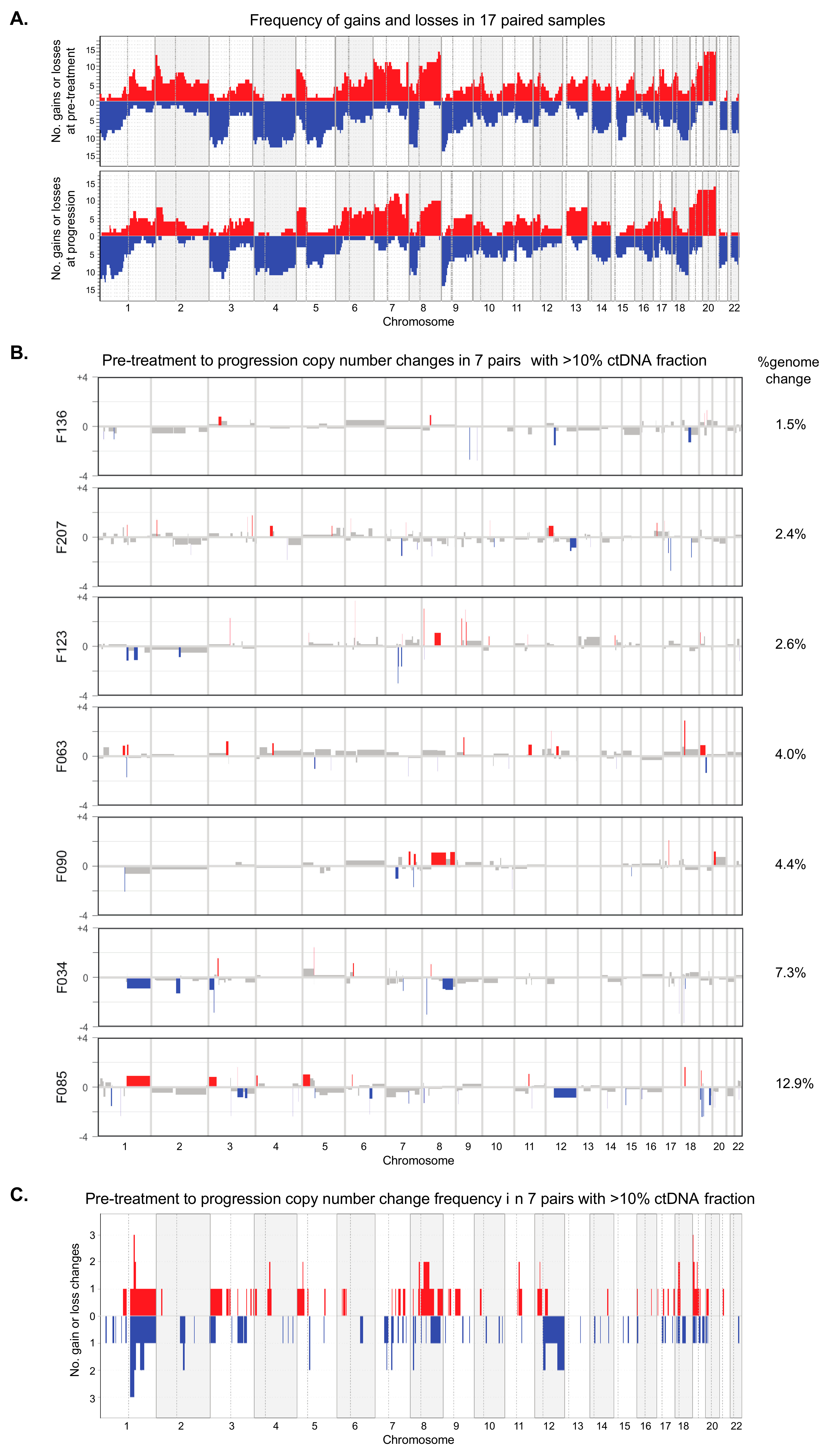
2. Results
3. Discussion
4. Methods
4.1. Trial Design and Sample Collection
4.2. Circulating Free (cf)DNA Extraction and Quantification
4.3. Low-Coverage Whole Genome Sequencing (lcWGS)
4.4. Somatic Copy Number Aberration (SCNA) Analysis
4.5. Survival Analyses by Pre-Treatment Circulating DNA Metrics
4.6. Data Availability
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| CSMD1 |
| LOC100287015 |
| MCPH1 |
| ANGPT2 |
| CLDN23 |
| MFHAS1 |
| ERI1 |
| MIR4660 |
| PPP1R3B |
| LOC157273 |
| TNKS |
| MIR597 |
| LINC00599 |
| MIR124-1 |
| MSRA |
| PRSS55 |
| RP1L1 |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.D.; Unverzagt, S.; Grothe, W.; Kleber, G.; Grothey, A.; Haerting, J.; Fleig, W.E. Chemotherapy for advanced gastric cancer. Cochrane database Syst. Rev. 2010, 3, CD004064. [Google Scholar] [CrossRef]
- Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; Shen, H.; et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar]
- Kim, J.; Bowlby, R.; Mungall, A.J.; Robertson, A.G.; Odze, R.D.; Cherniack, A.D.; Shih, J.; Pedamallu, C.S.; Cibulskis, C.; Dunford, A.; et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175. [Google Scholar]
- Liang, L.; Fang, J.-Y.; Xu, J. Gastric cancer and gene copy number variation: Emerging cancer drivers for targeted therapy. Oncogene 2016, 35, 1475–1482. [Google Scholar] [CrossRef]
- Labots, M.; Buffart, T.E.; Haan, J.C.; van Grieken, N.C.; Tijssen, M.; van de Velde, C.J.; Grabsch, H.I.; Ylstra, B.; Carvalho, B.; Fijneman, R.J.; et al. High-level copy number gains of established and potential drug target genes in gastric cancer as a lead for treatment development and selection. Cell. Oncol. 2014, 37, 41–52. [Google Scholar] [CrossRef]
- Zhang, Y. Epidemiology of esophageal cancer. World J. Gastroenterol. 2013, 19, 5598–5606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Ho, S.S.; Zhang, X.; Pattni, R.; Haraksingh, R.R.; Urban, A.E. Whole-genome sequencing analysis of CNV using low-coverage and paired-end strategies is efficient and outperforms array-based CNV analysis. J. Med. Genet. 2018, 55, 735–743. [Google Scholar] [CrossRef]
- Heitzer, E.; Auer, M.; Hoffmann, E.M.; Pichler, M.; Gasch, C.; Ulz, P.; Lax, S.; Waldispuehl-Geigl, J.; Mauermann, O.; Mohan, S.; et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int. J. Cancer 2013, 133, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Alsina, M.; Gullo, I.; Carneiro, F. Intratumoral heterogeneity in gastric cancer: A new challenge to face. Ann. Oncol. 2017, 28, 912–913. [Google Scholar] [CrossRef] [PubMed]
- Murugaesu, N.; Wilson, G.A.; Birkbak, N.J.; Watkins, T.; McGranahan, N.; Kumar, S.; Abbassi-Ghadi, N.; Salm, M.; Mitter, R.; Horswell, S.; et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 2015, 5, 821–831. [Google Scholar] [CrossRef]
- Lee, H.E.; Park, K.U.; Yoo, S.B.; Nam, S.K.; Park, D.J.; Kim, H.H.; Lee, H.S. Clinical significance of intratumoral HER2 heterogeneity in gastric cancer. Eur. J. Cancer 2013, 49, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Pearson, A.; Smyth, E.; Babina, I.S.; Herrera-Abreu, M.T.; Tarazona, N.; Peckitt, C.; Kilgour, E.; Smith, N.R.; Geh, C.; Rooney, C.; et al. High-Level Clonal FGFR Amplification and Response to FGFR Inhibition in a Translational Clinical Trial. Cancer Discov. 2016, 6, 838–851. [Google Scholar] [CrossRef]
- Petty, R.D.; Dahle-Smith, A.; Stevenson, D.A.J.; Osborne, A.; Massie, D.; Clark, C.; Murray, G.I.; Dutton, S.J.; Roberts, C.; Chong, I.Y.; et al. Gefitinib and EGFR Gene Copy Number Aberrations in Esophageal Cancer. J. Clin. Oncol. 2017, 35, 2279–2287. [Google Scholar] [CrossRef]
- Gao, J.; Wang, H.; Zang, W.; Li, B.; Rao, G.; Li, L.; Yu, Y.; Li, Z.; Dong, B.; Lu, Z.; et al. Circulating tumor DNA functions as an alternative for tissue to overcome tumor heterogeneity in advanced gastric cancer. Cancer Sci. 2017, 108, 1881–1887. [Google Scholar] [CrossRef]
- Pectasides, E.; Stachler, M.D.; Derks, S.; Liu, Y.; Maron, S.; Islam, M.; Alpert, L.; Kwak, H.; Kindler, H.; Polite, B.; et al. Genomic Heterogeneity as a Barrier to Precision Medicine in Gastroesophageal Adenocarcinoma. Cancer Discov. 2018, 8, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1324. [Google Scholar] [CrossRef]
- Chaplet, M.; Rai, R.; Jackson-Bernitsas, D.; Li, K.; Lin, S.Y. BRIT1/MCPH1: A guardian of genome and an enemy of tumors. Cell Cycle 2006, 5, 2579–2583. [Google Scholar] [CrossRef]
- Wei, Q.; Wang, X.; An, X.; Han, Q.; Meng, L.; Cao, W.L.Z. Effects of MCPH1 silencing on proliferation, apoptosis, and chemo-sensitivity of non-small cell lung cancer cells. Int. J. Clin. Exp. Med. 2018, 11, 6583–6595. [Google Scholar]
- Dulak, A.M.; Schumacher, S.E.; Van Lieshout, J.; Imamura, Y.; Fox, C.; Shim, B.; Ramos, A.H.; Saksena, G.; Baca, S.C.; Baselga, J.; et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012, 72, 4383–4393. [Google Scholar] [CrossRef]
- Dewhurst, S.M.; McGranahan, N.; Burrell, R.A.; Rowan, A.J.; Grönroos, E.; Endesfelder, D.; Joshi, T.; Mouradov, D.; Gibbs, P.; Ward, R.L.; et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 2014, 4, 175–185. [Google Scholar] [CrossRef]
- Vargas-Rondón, N.; Villegas, E.V.; Rondón-Lagos, M. The Role of Chromosomal Instability in Cancer and Therapeutic Responses. Cancers 2018, 10, 4. [Google Scholar] [CrossRef]
- Burrell, R.A.; Mcgranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef]
- Endesfelder, D.; Burrell, R.A.; Kanu, N.; McGranahan, N.; Howell, M.; Parker, P.J.; Downward, J.; Swanton, C.; Kschischo, M. Chromosomal instability selects gene copy-number variants encoding core regulators of proliferation in ER+ Breast cancer. Cancer Res. 2014, 74, 4853–4863. [Google Scholar] [CrossRef]
- Baslan, T.; Kendall, J.; Rodgers, L.; Cox, H.; Riggs, M.; Stepansky, A.; Troge, J.; Ravi, K.; Esposito, D.; Lakshmi, B.; et al. Genome-wide copy number analysis of single cells. Nat. Protoc. 2012, 7, 1024–1041. [Google Scholar] [CrossRef]
- Moorcraft, S.Y.; Gonzalez de Castro, D.; Cunningham, D.; Jones, T.; Walker, B.A.; Peckitt, C.; Yuan, L.C.; Frampton, M.; Begum, R.; Eltahir, Z.; et al. Investigating the feasibility of tumour molecular profiling in gastrointestinal malignancies in routine clinical practice. Ann. Oncol. 2018, 29, 230–236. [Google Scholar] [CrossRef]
- Janjigian, Y.Y.; Sanchez-Vega, F.; Jonsson, P.; Chatila, W.K.; Hechtman, J.F.; Ku, G.Y.; Riches, J.C.; Tuvy, Y.; Kundra, R.; Bouvier, N.; et al. Genetic Predictors of Response to Systemic Therapy in Esophagogastric Cancer. Cancer Discov. 2018, 8, 49–58. [Google Scholar] [CrossRef]
- Fang, W.-L.; Lan, Y.-T.; Huang, K.-H.; Liu, C.-A.; Hung, Y.-P.; Lin, C.-H.; Jhang, F.-Y.; Chang, S.-C.; Chen, M.-H.; Chao, Y.; et al. Clinical significance of circulating plasma DNA in gastric cancer. Int. J. Cancer 2016, 138, 2974–2983. [Google Scholar] [CrossRef]
- Saluja, H.; Karapetis, C.S.; Pedersen, S.K.; Young, G.P.; Symonds, E.L. The Use of Circulating Tumor DNA for Prognosis of Gastrointestinal Cancers. Front. Oncol. 2018, 8, 275. [Google Scholar] [CrossRef]
- Lee, A.J.X.; Endesfelder, D.; Rowan, A.J.; Walther, A.; Birkbak, N.J.; Futreal, P.A.; Downward, J.; Szallasi, Z.; Tomlinson, I.P.; Howell, M.; et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011, 71, 1858–1870. [Google Scholar] [CrossRef]
- Pietrantonio, F.; Fuca, G.; Morano, F.; Gloghini, A.; Corso, S.; Aprile, G.; Perrone, F.; De Vita, F.; Tamborini, E.; Tomasello, G.; et al. Biomarkers of primary resistance to trastuzumab in HER2-positive metastatic gastric cancer patients: The AMNESIA case-control study. Clin. Cancer Res. 2018, 24, 1082–1089. [Google Scholar] [CrossRef]
- Menyhart, O.; Santarpia, L.; Gyorffy, B. A Comprehensive Outline of Trastuzumab Resistance Biomarkers in HER2 Overexpressing Breast Cancer. Curr. Cancer Drug Targets 2015, 15, 665–683. [Google Scholar] [CrossRef]
- Berger, A.W.; Schwerdel, D.; Welz, H.; Marienfeld, R.; Schmidt, S.A.; Kleger, A.; Ettrich, T.J.; Seufferlein, T. Treatment monitoring in metastatic colorectal cancer patients by quantification and KRAS genotyping of circulating cell-free DNA. PLoS ONE 2017, 12, e0174308. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-S.; Liu, Z.-X.; Lu, Y.-X.; Bao, H.; Wu, X.; Zeng, Z.-L.; Liu, Z.; Zhao, Q.; He, C.-Y.; Lu, J.-H.; et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut 2018. [Google Scholar] [CrossRef]
- Zheng, X.; Fan, L.; Zhou, P.; Ma, H.; Huang, S.; Yu, D.; Zhao, L.; Yang, S.; Liu, J.; Huang, A.; et al. Detection of Circulating Tumor Cells and Circulating Tumor Microemboli in Gastric Cancer. Transl. Oncol. 2017, 10, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Matsusaka, S.; Chin, K.; Mikuniya, M.; Minowa, S.; Takayama, T.; Shibata, H.; Kuniyoshi, R.; Ogura, M.; Terui, Y.; et al. Detection of HER2 Amplification in Circulating Tumor Cells of HER2-Negative Gastric Cancer Patients. Target. Oncol. 2017, 12, 341–351. [Google Scholar] [CrossRef]
- Tagawa, S.T.; Antonarakis, E.S.; Gjyrezi, A.; Galletti, G.; Kim, S.; Worroll, D.; Stewart, J.; Zaher, A.; Szatrowski, T.P.; Ballman, K.V.; et al. Expression of AR-V7 and ARV 567Es in circulating tumor cells correlates with outcomes to taxane therapy in men with metastatic prostate cancer treated in taxynergy. Clin. Cancer Res. 2019, 25, 1880–1888. [Google Scholar] [CrossRef] [PubMed]
- Lallo, A.; Frese, K.K.; Morrow, C.J.; Sloane, R.; Gulati, S.; Schenk, M.W.; Trapani, F.; Simms, N.; Galvin, M.; Brown, S.; et al. The combination of the PARP inhibitor olaparib and the WEE1 Inhibitor AZD1775 as a new therapeutic option for small cell lung cancer. Clin. Cancer Res. 2018, 24, 5153–5164. [Google Scholar] [CrossRef] [PubMed]
- Lallo, A.; Gulati, S.; Schenk, M.W.; Khandelwal, G.; Berglund, U.W.; Pateras, I.S.; Chester, C.P.E.; Pham, T.M.; Kalderen, C.; Frese, K.K.; et al. Ex vivo culture of cells derived from circulating tumour cell xenograft to support small cell lung cancer research and experimental therapeutics. Br. J. Pharmacol. 2019, 176, 436–450. [Google Scholar] [CrossRef]
- Fu, M.; Gu, J.; Jiang, P.; Qian, H.; Xu, W.; Zhang, X. Exosomes in gastric cancer: Roles, mechanisms, and applications. Mol. Cancer 2019, 18, 41. [Google Scholar] [CrossRef]
- Mansukhani, S.; Barber, L.J.; Kleftogiannis, D.; Moorcraft, S.Y.; Davidson, M.; Woolston, A.; Proszek, P.Z.; Griffiths, B.; Fenwick, K.; Herman, B.; et al. Ultra-Sensitive mutation detection and genome-wide DNA copy number reconstruction by error- corrected circulating tumor DNA sequencing. Clin. Chem. 2018, 64, 1626–1635. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Belic, J.; Gutschi, S.; Quehenberger, F.; Fischereder, K.; Benezeder, T.; Auer, M.; Pischler, C.; Mannweiler, S.; et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013, 5, 30. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Ha, G.; Roth, A.; Lai, D.; Bashashati, A.; Ding, J.; Goya, R.; Giuliany, R.; Rosner, J.; Oloumi, A.; Shumansky, K.; et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 2012, 22, 1995–2007. [Google Scholar] [CrossRef]
- Nilsen, G.; Liestøl, K.; Van Loo, P.; Moen Vollan, H.K.; Eide, M.B.; Rueda, O.M.; Chin, S.F.; Russell, R.; Baumbusch, L.O.; Caldas, C.; et al. Copynumber: Efficient algorithms for single- and multi-track copy number segmentation. BMC Genom. 2012, 13, 591. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24. [Google Scholar] [CrossRef]

| Histopathological Variable | ||
|---|---|---|
| Number of Cases: | 30 | |
| Anatomic site of primary: | Gastric | 6 (20%) |
| OGJ/oesophageal | 24 (80%) | |
| Histological subtype: | Intestinal | 28 (93%) |
| Diffuse | 2 (7%) | |
| Clinical stage at presentation: | Locally advanced | 3 (10%) |
| Metastatic | 27 (90%) | |
| HER2 status *: | Positive | 6 (20%) |
| Negative | 24 (80%) | |
| First line chemotherapy: | Platinum/fluoropyrimidine doublet | 9 (30%) |
| Doublet+ anthracycline | 15 (50%) | |
| Doublet+ trastuzumab | 6 (20%) | |
| Metastatic sites: Liver | Yes | 16 (53%) |
| No | 14 (47%) | |
| Peritoneal | Yes | 6 (20%) |
| No | 24 (80%) | |
| Lung | Yes | 8 (27%) |
| No | 22 (73%) | |
| Number of metastatic organ sites: | 0–1 | 22 (73%) |
| ≥2 | 8 (27%) | |
| Primary tumour in situ: | Yes | 23 (77%) |
| No | 7 (23%) | |
| CA19-9 secretor: | Yes | 15 (50%) |
| No | 15 (50%) | |
| Histopathological Variable | n | Median cfDNA Concentration (ng/mL Plasma) | p-Value | Median ctDNA Fraction (%) | p-Value | Median ctDNA Concentration (ng/mL Plasma) | p-Value | |
|---|---|---|---|---|---|---|---|---|
| Primary tumour in situ | Yes | 23 | 9.66 | 0.0027 | 9.10 | 0.0046 | 2.14 | <0.0001 |
| No | 7 | 4.81 | 0.00 | 0.00 | ||||
| Liver metastases present | Yes | 16 | 10.09 | 0.1306 | 18.01 | 0.0043 | 2.18 | 0.0099 |
| No | 14 | 6.80 | 7.23 | 0.35 | ||||
| Primary tumour anatomic site | Gastric | 6 | 8.65 | 0.8996 | 3.33 | 0.0103 | 0.24 | 0.1401 |
| Nongastric | 24 | 9.05 | 9.31 | 0.84 | ||||
| No. of metastatic organ sites | 0–1 | 22 | 8.31 | 0.5042 | 7.77 | 0.1528 | 0.47 | 0.9814 |
| ≥2 | 8 | 1.22 | 14.47 | 0.58 | ||||
| HER2 status | Positive | 6 | 11.22 | 0.3739 | 8.81 | 0.4595 | 2.25 | 0.1713 |
| Negative | 24 | 8.32 | 8.22 | 0.47 | ||||
| CA19-9 secretion | Yes | 15 | 9.21 | 0.9999 | 8.10 | 0.5640 | 0.61 | 0.7733 |
| No | 15 | 8.54 | 9.02 | 0.78 | ||||
| Paired Pretreatment and Progression Cases | n | Median ctDNA Fraction (%) | p-Value | |
|---|---|---|---|---|
| All paired cases | Pretreatment | 20 | 15.18 | 0.1567 |
| Progression | 20 | 8.72 | ||
| Initial radiological response followed by progression to chemotherapy: ‘primary responders’ | Pretreatment | 12 | 17.00 | 0.0200 |
| Progression | 12 | 7.59 | ||
| Stable disease or primary radiological progression to chemotherapy: ‘primary nonresponders’ | Pretreatment | 8 | 11.27 | 0.7984 |
| Progression | 8 | 13.58 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davidson, M.; Barber, L.J.; Woolston, A.; Cafferkey, C.; Mansukhani, S.; Griffiths, B.; Moorcraft, S.-Y.; Rana, I.; Begum, R.; Assiotis, I.; et al. Detecting and Tracking Circulating Tumour DNA Copy Number Profiles during First Line Chemotherapy in Oesophagogastric Adenocarcinoma. Cancers 2019, 11, 736. https://doi.org/10.3390/cancers11050736
Davidson M, Barber LJ, Woolston A, Cafferkey C, Mansukhani S, Griffiths B, Moorcraft S-Y, Rana I, Begum R, Assiotis I, et al. Detecting and Tracking Circulating Tumour DNA Copy Number Profiles during First Line Chemotherapy in Oesophagogastric Adenocarcinoma. Cancers. 2019; 11(5):736. https://doi.org/10.3390/cancers11050736
Chicago/Turabian StyleDavidson, Michael, Louise J. Barber, Andrew Woolston, Catherine Cafferkey, Sonia Mansukhani, Beatrice Griffiths, Sing-Yu Moorcraft, Isma Rana, Ruwaida Begum, Ioannis Assiotis, and et al. 2019. "Detecting and Tracking Circulating Tumour DNA Copy Number Profiles during First Line Chemotherapy in Oesophagogastric Adenocarcinoma" Cancers 11, no. 5: 736. https://doi.org/10.3390/cancers11050736
APA StyleDavidson, M., Barber, L. J., Woolston, A., Cafferkey, C., Mansukhani, S., Griffiths, B., Moorcraft, S.-Y., Rana, I., Begum, R., Assiotis, I., Matthews, N., Rao, S., Watkins, D., Chau, I., Cunningham, D., Starling, N., & Gerlinger, M. (2019). Detecting and Tracking Circulating Tumour DNA Copy Number Profiles during First Line Chemotherapy in Oesophagogastric Adenocarcinoma. Cancers, 11(5), 736. https://doi.org/10.3390/cancers11050736





