Cul4A Modulates Invasion and Metastasis of Lung Cancer through Regulation of ANXA10
Abstract
1. Introduction
2. Material and Methods
2.1. Cell Lines and Cell Culture
2.2. Tissues
2.3. Immunohistochemistry (IHC)
2.4. Cell Migration Assay
2.5. Cell Invasion Assay
2.6. Western Bot Analysis
2.7. Semi-Quantitative Reverse Transcription-PCR (RT-PCR) Analysis
2.8. Transfection with Small Interfering RNA (siRNA) and Vectors
2.9. Protein Degradation Assay
2.10. Co-Immunoprecipitation Assay
2.11. In Vivo Ubiquitination Assay
2.12. Tail Vein Injection Mouse Xenograft Models
2.13. Statistical Analysis
3. Results
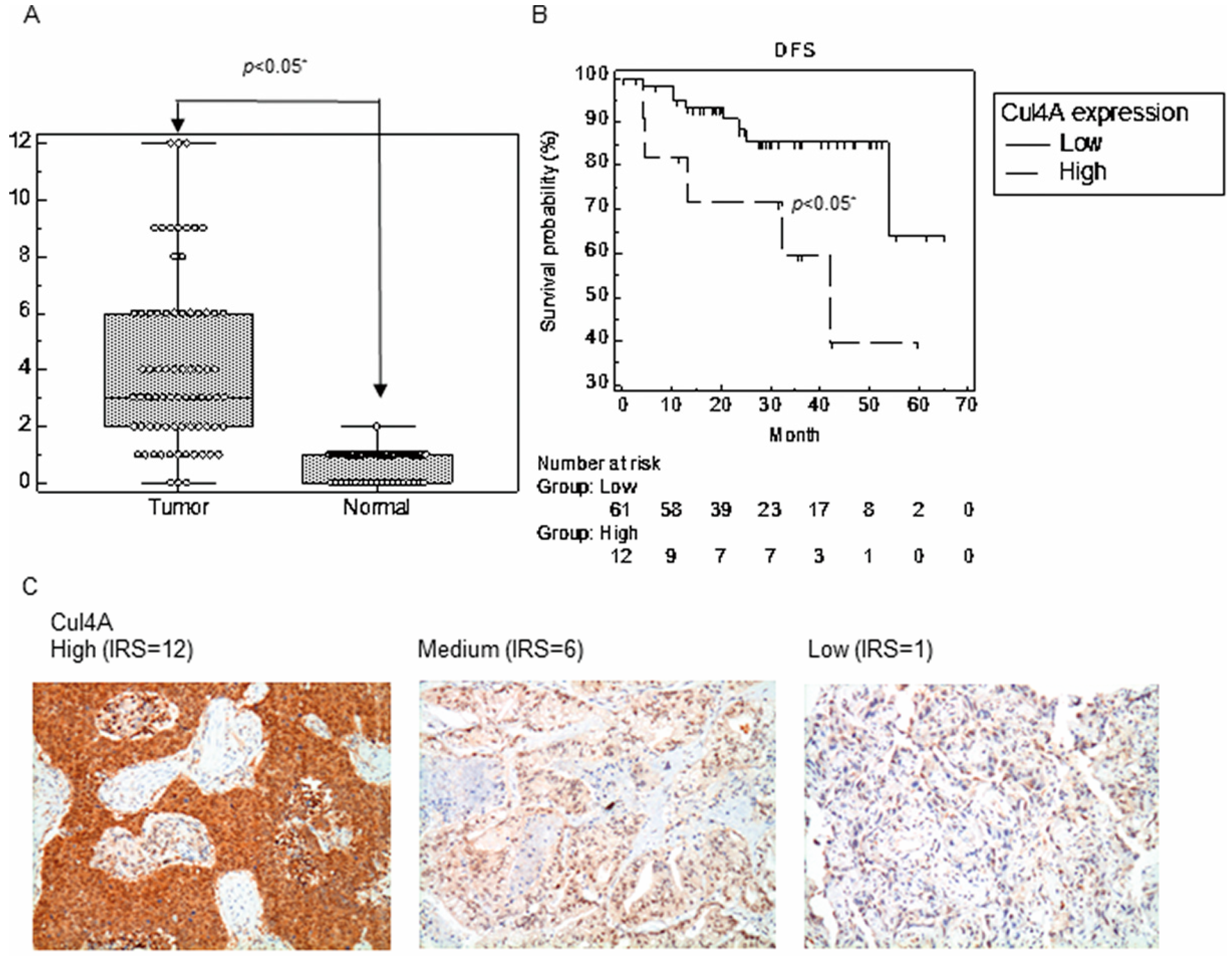
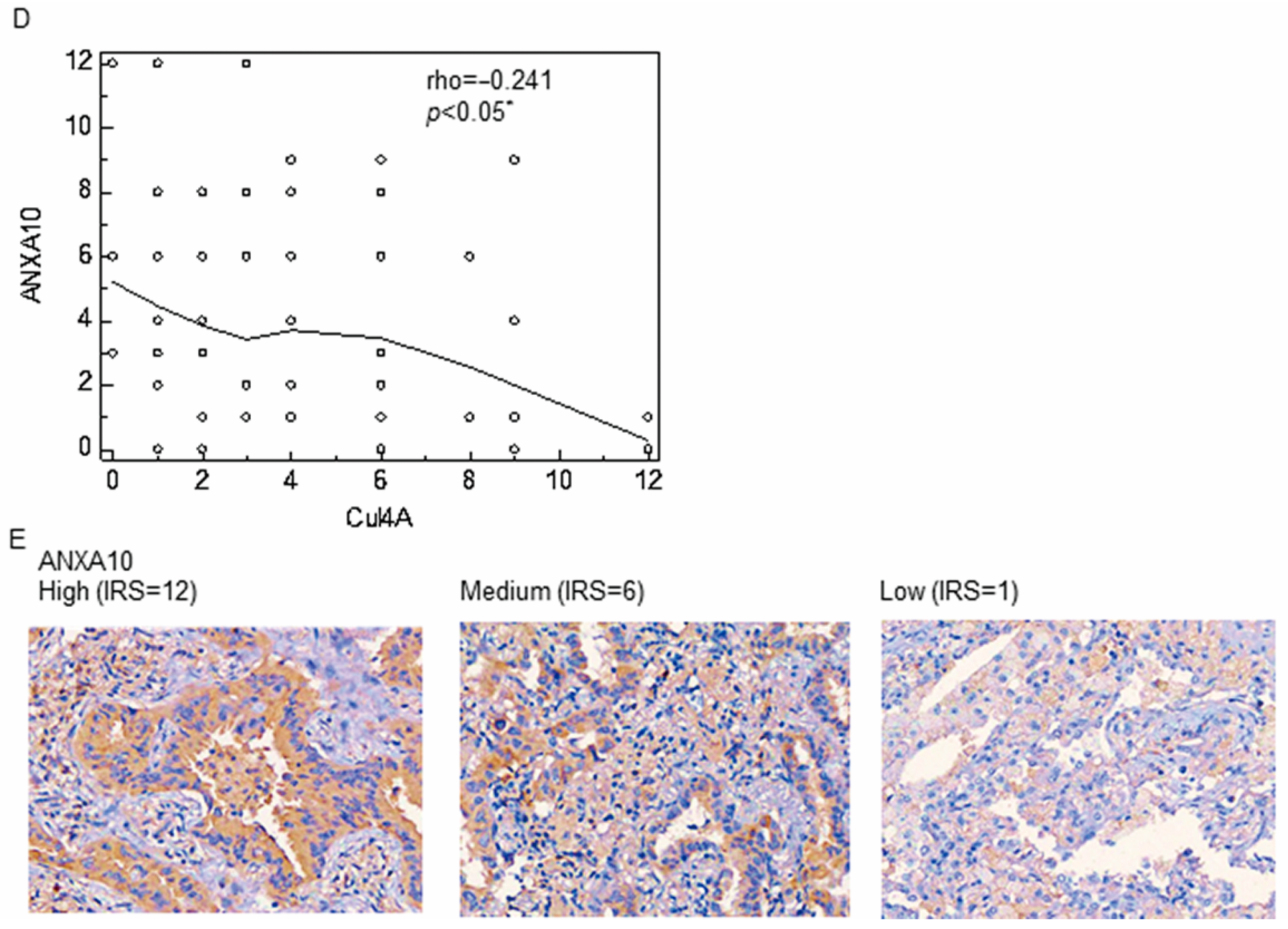
3.1. Cul4A Is Upregulated in the NSCLC Tissues
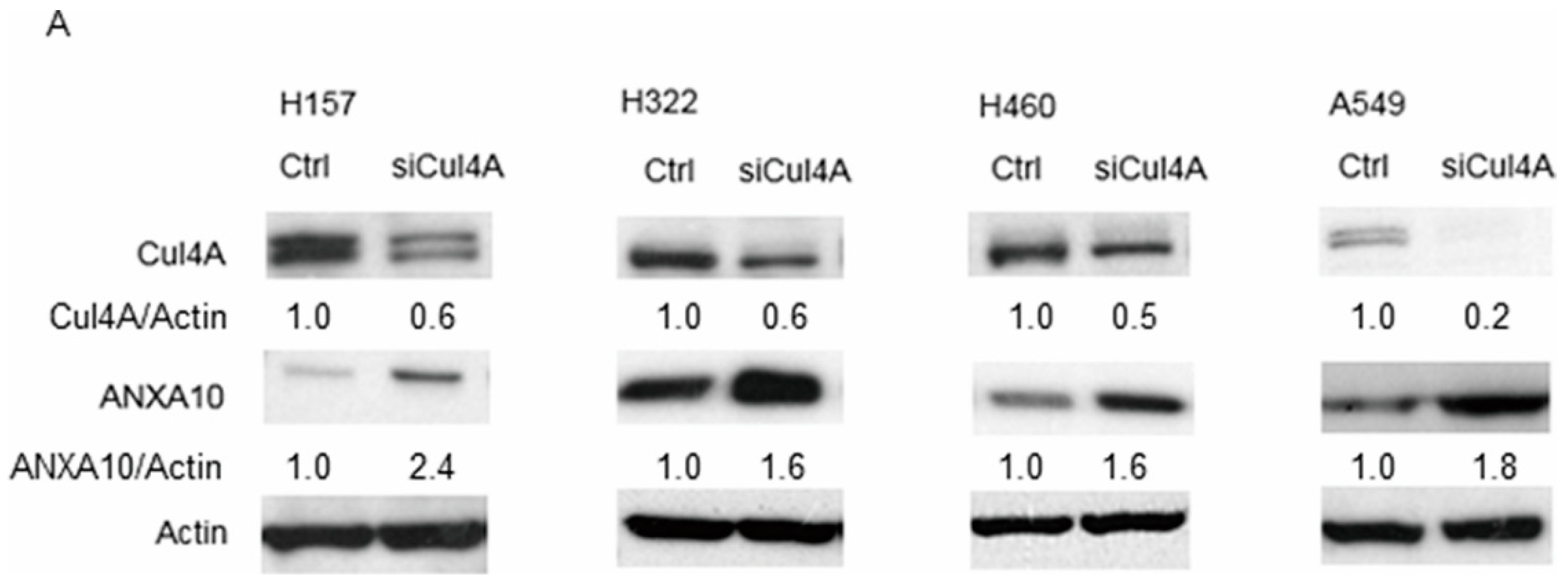
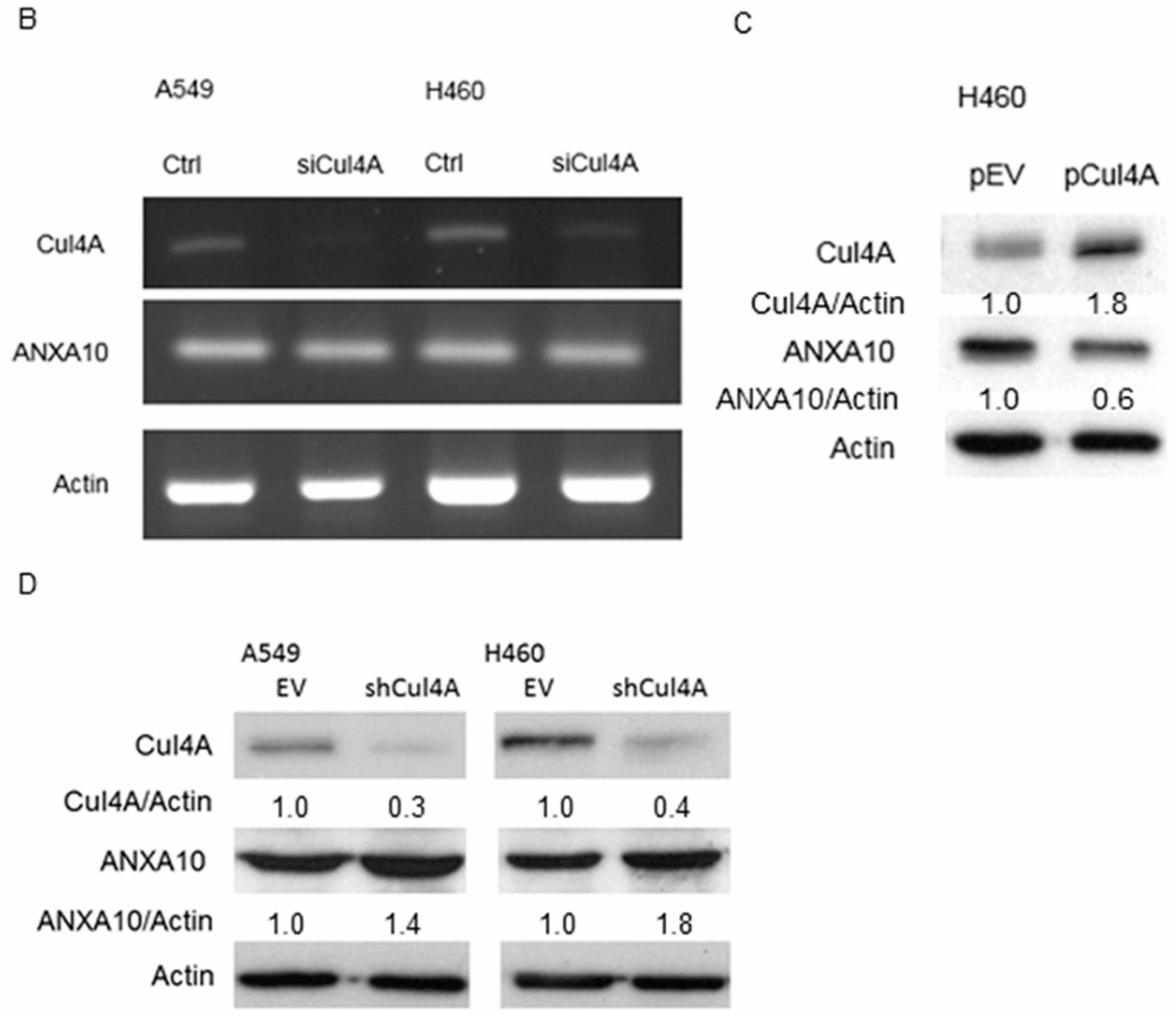
3.2. Knockdown of Cul4A Is Associated with the Upregulation of ANXA10 in Lung Cancer Cells
3.3. Knockdown of Cul4A Represses Metastasis and Invasion in Lung Cancer Cells
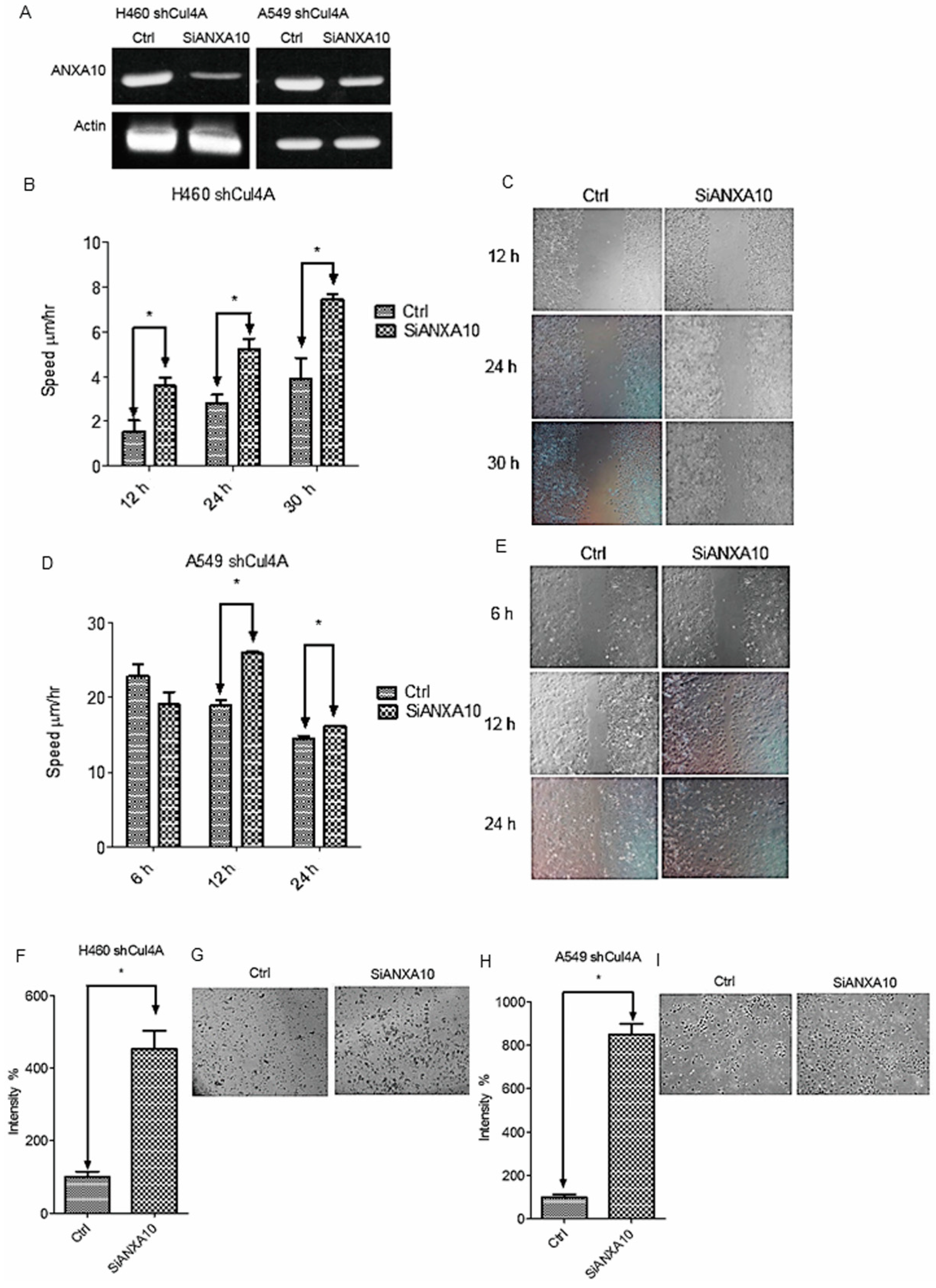
3.4. Cell Migration and Invasion Are Restored by Knockdown of ANXA10 in Cul4A Knockdown Lung Cancer Cells
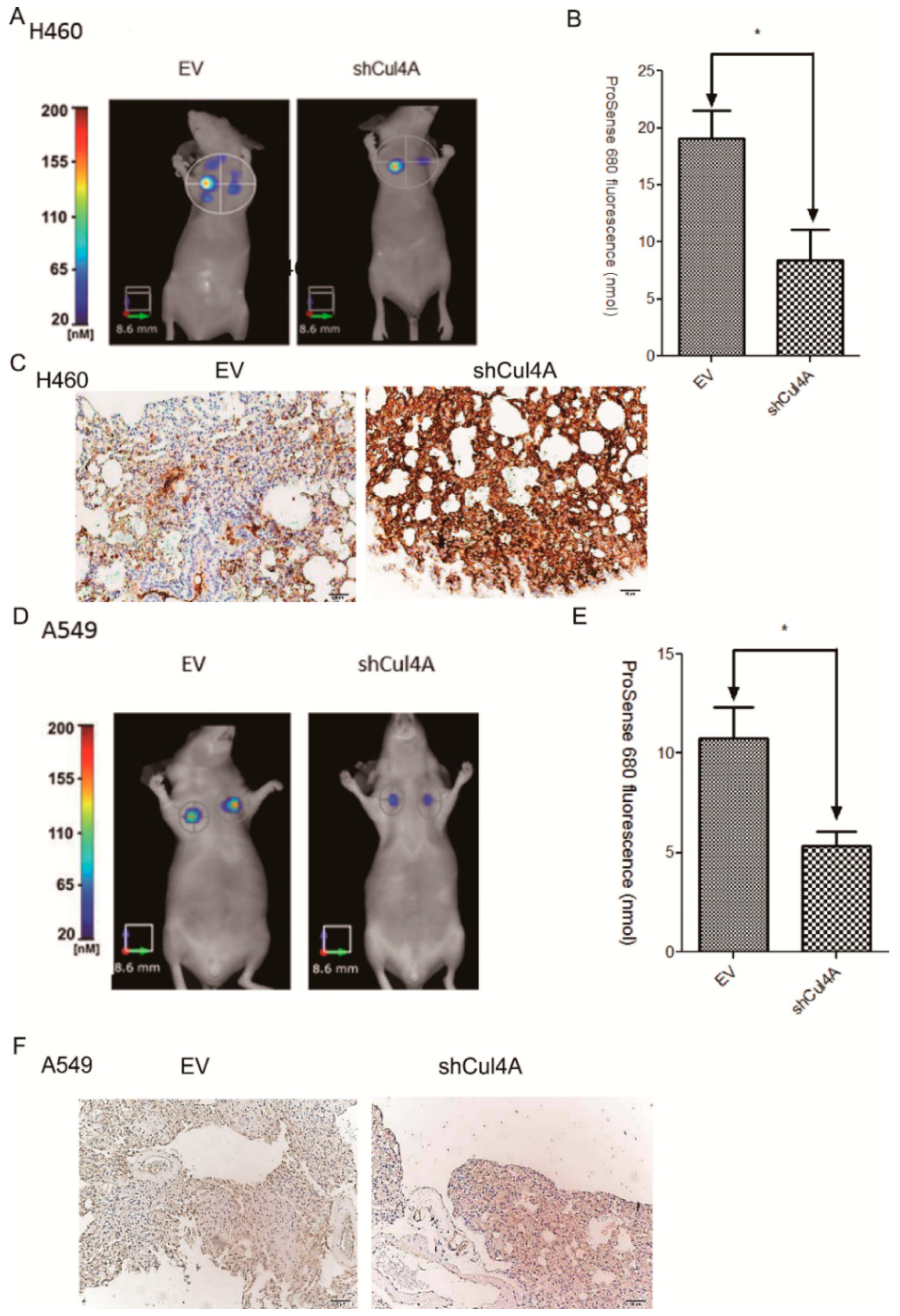
3.5. Knockdown of Cul4A Represses Metastasis of Lung Cancer Tumors in Tail Vein Injection Mouse Models
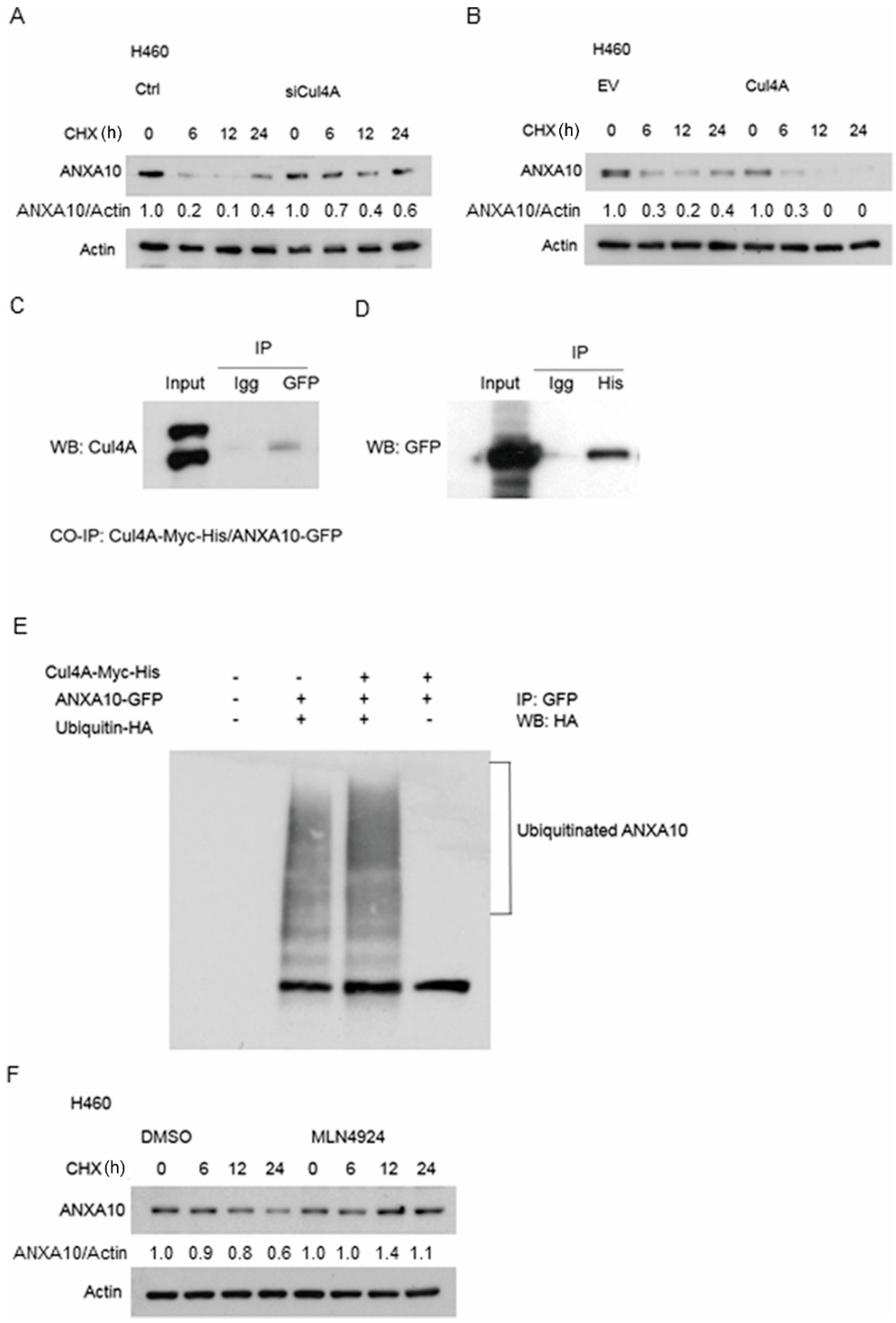
3.6. ANXA10 Expression Is Regulated by Protein Degradation
3.7. ANXA10 Is Ubiquitinated by Cul4A Through a Protein–Protein Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef]
- Waning, D.L.; Li, B.; Jia, N.; Naaldijk, Y.; Goebel, W.S.; HogenEsch, H.; Chun, K.T. Cul4A is required for hematopoietic cell viability and its deficiency leads to apoptosis. Blood 2008, 112, 320–329. [Google Scholar] [CrossRef][Green Version]
- Li, B.; Jia, N.; Kapur, R.; Chun, K.T. Cul4A targets p27 for degradation and regulates proliferation, cell cycle exit, and differentiation during erythropoiesis. Blood 2006, 107, 4291–4299. [Google Scholar] [CrossRef]
- Kopanja, D.; Stoyanova, T.; Okur, M.N.; Huang, E.; Bagchi, S.; Raychaudhuri, P. Proliferation defects and genome instability in cells lacking Cul4A. Oncogene 2009, 28, 2456–2465. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lee, S.; Zhang, J.; Peters, S.B.; Hannah, J.; Zhang, Y.; Yin, Y.; Koff, A.; Ma, L.; Zhou, P. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell 2009, 34, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhai, L.; Xu, J.; Joo, H.Y.; Jackson, S.; Erdjument-Bromage, H.; Tempst, P.; Xiong, Y.; Zhang, Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 2006, 22, 383–394. [Google Scholar] [CrossRef]
- Melchor, L.; Saucedo-Cuevas, L.P.; Munoz-Repeto, I.; Rodriguez-Pinilla, S.M.; Honrado, E.; Campoverde, A.; Palacios, J.; Nathanson, K.L.; Garcia, M.J.; Benitez, J. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res. 2009, 11, R86. [Google Scholar] [CrossRef]
- Hung, M.S.; Mao, J.H.; Xu, Z.; Yang, C.T.; Yu, J.S.; Harvard, C.; Lin, Y.C.; Bravo, D.T.; Jablons, D.M.; You, L. Cul4A is an oncogene in malignant pleural mesothelioma. J. Cell. Mol. Med. 2011, 15, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Yan, F.; Cao, W.; Chen, Z.; Zheng, H.; Li, H.; Pan, Y.; Narula, N.; Ren, X.; Li, H.; et al. Dysregulation of CUL4A and CUL4B Ubiquitin Ligases in Lung Cancer. J. Biol. Chem. 2017, 292, 2966–2978. [Google Scholar] [CrossRef]
- Chen, G.; Zhao, X.; Tan, Z.; Wang, D.; Luo, D.; Zhang, P.; Cao, J.; Wang, F.; Liu, Q.; Li, L. Investigation of the role of cullin 4A overexpression in human liver cancer. Mol. Med. Rep. 2018, 18, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Nag, A.; Bagchi, S.; Raychaudhuri, P. Cul4A physically associates with MDM2 and participates in the proteolysis of p53. Cancer Res. 2004, 64, 8152–8155. [Google Scholar] [CrossRef]
- Huang, J.; Chen, J. VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene 2008, 27, 4056–4064. [Google Scholar] [CrossRef]
- Jiang, L.; Rong, R.; Sheikh, M.S.; Huang, Y. Cullin-4A.DNA damage-binding protein 1 E3 ligase complex targets tumor suppressor RASSF1A for degradation during mitosis. J. Biol. Chem. 2011, 286, 6971–6978. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.S.; Chen, I.C.; You, L.; Jablons, D.M.; Li, Y.C.; Mao, J.H.; Xu, Z.; Hsieh, M.J.; Lin, Y.C.; Yang, C.T.; et al. Knockdown of Cul4A increases chemosensitivity to gemcitabine through upregulation of TGFBI in lung cancer cells. Oncol. Rep. 2015, 34, 3187–3195. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, H.; Shiomi, Y.; Iida, H.; Michishita, M.; Takami, T.; Tsurimoto, T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 2008, 283, 29045–29052. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Z.; Mao, J.H.; Hung, M.S.; Hsieh, D.; Au, A.; Jablons, D.M.; You, L. Analysis of lung tumor initiation and progression in transgenic mice for Cre-inducible overexpression of Cul4A gene. Thorac. Cancer 2015, 6, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hung, M.S.; Wang, Y.; Mao, J.H.; Tan, J.L.; Jahan, K.; Roos, H.; Xu, Z.; Jablons, D.M.; You, L. Transgenic mice for cre-inducible overexpression of the Cul4A gene. Genesis 2011, 49, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Uramoto, H.; Tanaka, F. Recurrence after surgery in patients with NSCLC. Transl. Lung Cancer Res. 2014, 3, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, I.; Yohena, T.; Kitajima, M.; Ushijima, C.; Nishioka, K.; Ichinose, Y.; Sugimachi, K. Survival of non-small cell lung cancer patients with postoperative recurrence at distant organs. Ann. Thorac. Cardiovasc. Surg. 2001, 7, 204–209. [Google Scholar]
- Zhong, J.; Shaik, S.; Wan, L.; Tron, A.E.; Wang, Z.; Sun, L.; Inuzuka, H.; Wei, W. SCF beta-TRCP targets MTSS1 for ubiquitination-mediated destruction to regulate cancer cell proliferation and migration. Oncotarget 2013, 4, 2339–2353. [Google Scholar] [CrossRef]
- Hung, M.S.; Chen, I.C.; You, L.; Jablons, D.M.; Li, Y.C.; Mao, J.H.; Xu, Z.; Lung, J.H.; Yang, C.T.; Liu, S.T. Knockdown of cullin 4A inhibits growth and increases chemosensitivity in lung cancer cells. J. Cell. Mol. Med. 2016, 20, 1295–1306. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, P.; Liu, Z.; Wang, Q.; Wen, M.; Wang, Y.; Yuan, H.; Mao, J.H.; Wei, G. CUL4A overexpression enhances lung tumor growth and sensitizes lung cancer cells to erlotinib via transcriptional regulation of EGFR. Mol. Cancer 2014, 13, 252. [Google Scholar] [CrossRef]
- Kim, J.K.; Kim, P.J.; Jung, K.H.; Noh, J.H.; Eun, J.W.; Bae, H.J.; Xie, H.J.; Shan, J.M.; Ping, W.Y.; Park, W.S.; et al. Decreased expression of annexin A10 in gastric cancer and its overexpression in tumor cell growth suppression. Oncol. Rep. 2010, 24, 607–612. [Google Scholar]
- Miyazawa, Y.; Sekine, Y.; Kato, H.; Furuya, Y.; Koike, H.; Suzuki, K. Simvastatin Up-Regulates Annexin A10 That Can Inhibit the Proliferation, Migration, and Invasion in Androgen-Independent Human Prostate Cancer Cells. Prostate 2017, 77, 337–349. [Google Scholar] [CrossRef]
- Munksgaard, P.P.; Mansilla, F.; Brems Eskildsen, A.S.; Fristrup, N.; Birkenkamp-Demtroder, K.; Ulhoi, B.P.; Borre, M.; Agerbaek, M.; Hermann, G.G.; Orntoft, T.F.; et al. Low ANXA10 expression is associated with disease aggressiveness in bladder cancer. Br. J. Cancer 2011, 105, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Shimoji, T.; Murakami, K.; Sugiyama, Y.; Matsuda, M.; Inubushi, S.; Nasu, J.; Shirakura, M.; Suzuki, T.; Wakita, T.; Kishino, T.; et al. Identification of annexin A1 as a novel substrate for E6AP-mediated ubiquitylation. J. Cell. Biochem. 2009, 106, 1123–1135. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Jing, B.; Xing, T.; Hou, L.; Yang, Z. Overexpression of annexin A2 is associated with abnormal ubiquitination in breast cancer. Genom. Proteom. Bioinform. 2012, 10, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.J.; Zhan, S.K.; Ji, W.Z.; Pan, Y.X.; Liu, W.; Li, D.Y.; Huang, P.; Zhang, X.X.; Cao, C.Y.; Zhang, J.; et al. Ubiquitin-protein ligase E3C promotes glioma progression by mediating the ubiquitination and degrading of Annexin A7. Sci. Rep. 2015, 5, 11066. [Google Scholar] [CrossRef]
- Remmele, W.; Stegner, H.E. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Der. Pathol. 1987, 8, 138–140. [Google Scholar]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef]
- Song, J.; Zhang, J.; Shao, J. Knockdown of CUL4A inhibits invasion and induces apoptosis in osteosarcoma cells. Int. J. Immunopathol. Pharmacol. 2015, 28, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wen, M.; Kwon, Y.; Xu, Y.; Liu, Y.; Zhang, P.; He, X.; Wang, Q.; Huang, Y.; Jen, K.Y.; et al. CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res. 2014, 74, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Zhou, H.; Zhu, L.; Wang, D.; Fan, S.; Zhao, W. CUL4A promotes proliferation and metastasis of colorectal cancer cells by regulating H3K4 trimethylation in epithelial-mesenchymal transition. Onco Targets Ther. 2017, 10, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Lei, W.; Xiang, X.; Zhang, L.; Lei, J.; Gong, Y.; Song, M.; Wang, Y.; Fang, Z.; Yu, F.; et al. Cullin 4A (CUL4A), a direct target of miR-9 and miR-137, promotes gastric cancer proliferation and invasion by regulating the Hippo signaling pathway. Oncotarget 2016, 7, 10037–10050. [Google Scholar] [CrossRef] [PubMed]
- Mussunoor, S.; Murray, G.I. The role of annexins in tumour development and progression. J. Pathol. 2008, 216, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.H.; Lin, C.Y.; Peng, S.Y.; Jeng, Y.M.; Pan, H.W.; Lai, P.L.; Liu, C.L.; Hsu, H.C. Down-regulation of annexin A10 in hepatocellular carcinoma is associated with vascular invasion, early recurrence, and poor prognosis in synergy with p53 mutation. Am. J. Pathol. 2002, 160, 1831–1837. [Google Scholar] [CrossRef]
- Kim, J.; Kim, M.A.; Jee, C.D.; Jung, E.J.; Kim, W.H. Reduced expression and homozygous deletion of annexin A10 in gastric carcinoma. Int. J. Cancer 2009, 125, 1842–1850. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, D.; Niki, T.; Ishikawa, S.; Goto, A.; Ohara, E.; Yokomizo, T.; Heizmann, C.W.; Aburatani, H.; Moriyama, S.; Moriyama, H.; et al. Differential expression of S100A2 and S100A4 in lung adenocarcinomas: Clinicopathological significance, relationship to p53 and identification of their target genes. Cancer Sci. 2005, 96, 844–857. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hung, M.-S.; Chen, Y.-C.; Lin, P.-Y.; Li, Y.-C.; Hsu, C.-C.; Lung, J.-H.; You, L.; Xu, Z.; Mao, J.-H.; Jablons, D.M.; et al. Cul4A Modulates Invasion and Metastasis of Lung Cancer through Regulation of ANXA10. Cancers 2019, 11, 618. https://doi.org/10.3390/cancers11050618
Hung M-S, Chen Y-C, Lin P-Y, Li Y-C, Hsu C-C, Lung J-H, You L, Xu Z, Mao J-H, Jablons DM, et al. Cul4A Modulates Invasion and Metastasis of Lung Cancer through Regulation of ANXA10. Cancers. 2019; 11(5):618. https://doi.org/10.3390/cancers11050618
Chicago/Turabian StyleHung, Ming-Szu, Yi-Chuan Chen, Paul-Yann Lin, Ya-Chin Li, Chia-Chen Hsu, Jr-Hau Lung, Liang You, Zhidong Xu, Jian-Hua Mao, David M. Jablons, and et al. 2019. "Cul4A Modulates Invasion and Metastasis of Lung Cancer through Regulation of ANXA10" Cancers 11, no. 5: 618. https://doi.org/10.3390/cancers11050618
APA StyleHung, M.-S., Chen, Y.-C., Lin, P.-Y., Li, Y.-C., Hsu, C.-C., Lung, J.-H., You, L., Xu, Z., Mao, J.-H., Jablons, D. M., & Yang, C.-T. (2019). Cul4A Modulates Invasion and Metastasis of Lung Cancer through Regulation of ANXA10. Cancers, 11(5), 618. https://doi.org/10.3390/cancers11050618






