Immunohistochemical and Molecular Features of Melanomas Exhibiting Intratumor and Intertumor Histomorphologic Heterogeneity
Abstract
:1. Introduction
2. Results
2.1. Clinicopathological Characteristics of Patients and Tumors Evaluated by NGS
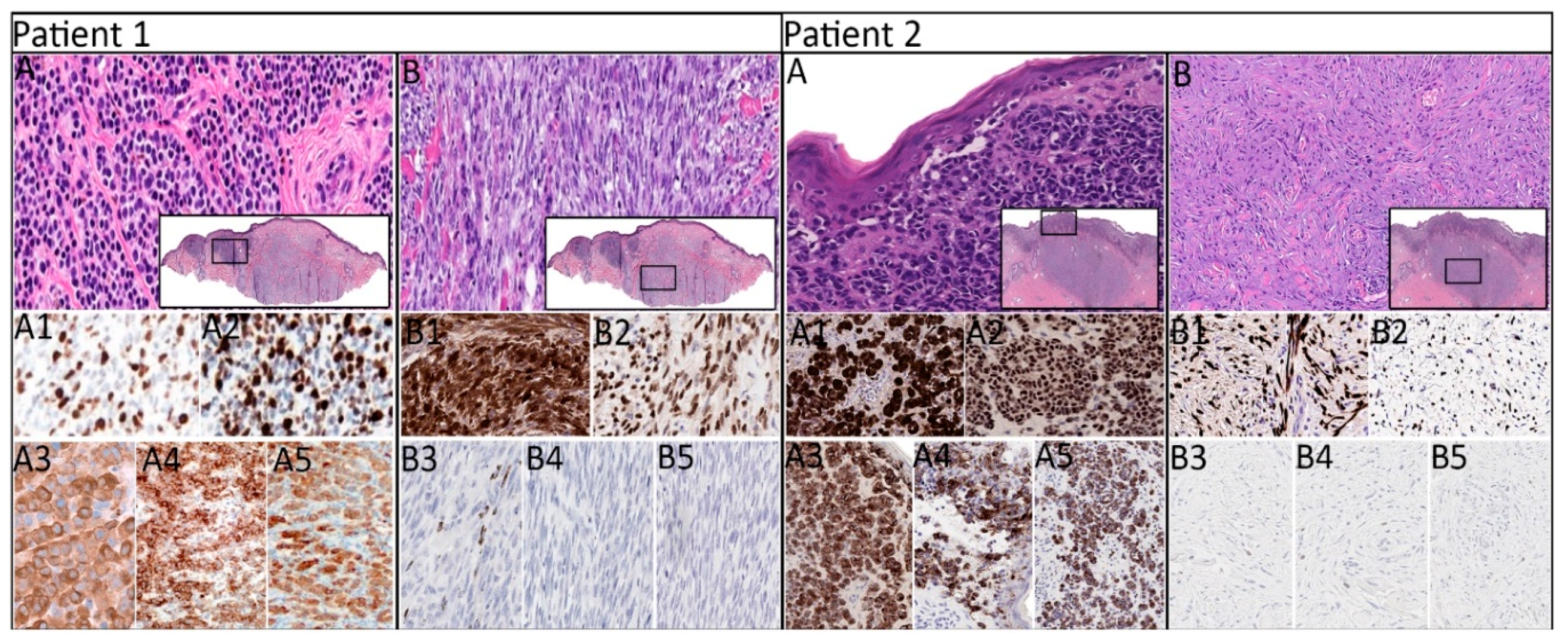
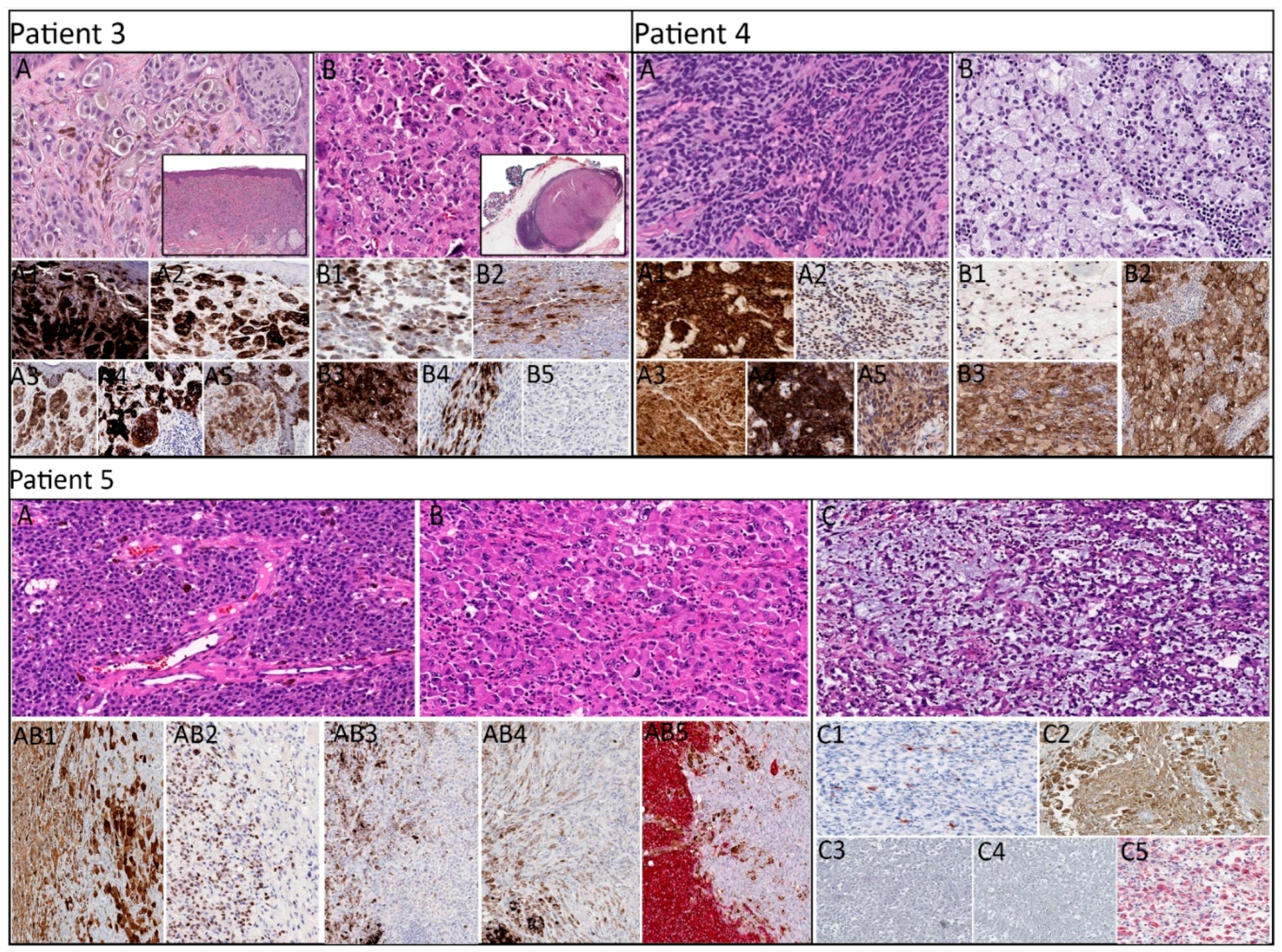
2.2. Histopathologic, Immunohistochemical, and Molecular Characteristics by Patient
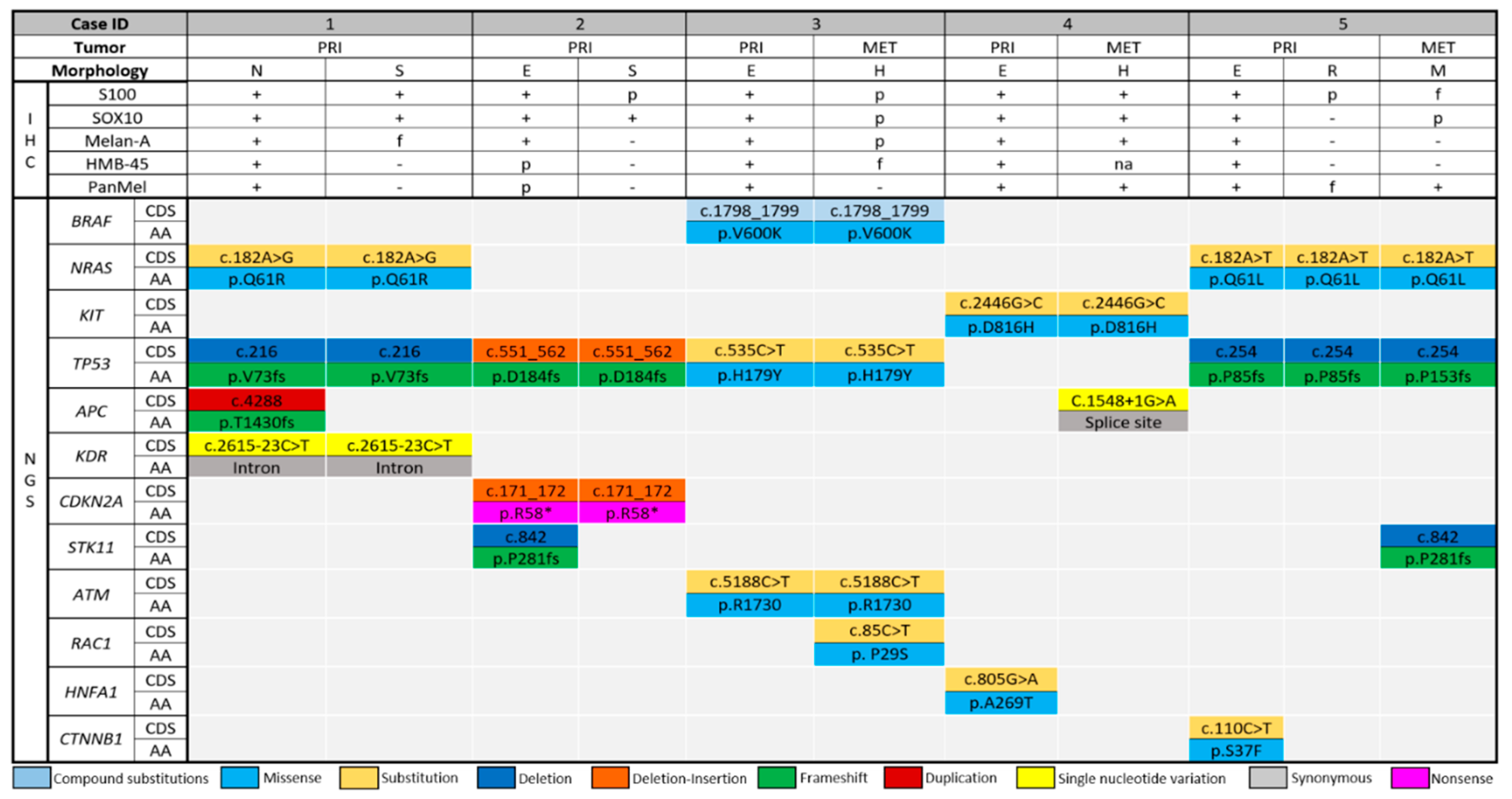
2.3. Distribution of the Detected Driver Mutations
3. Discussion
4. Materials and Methods
4.1. Laser Capture Microdissection and DNA Extraction
4.2. Test Platform
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Andor, N.; Graham, T.A.; Jansen, M.; Xia, L.C.; Aktipis, C.A.; Petritsch, C.; Ji, H.P.; Maley, C.C. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 2016, 22, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, B.; Bai, X.; Cohen, C.; Zhong, H.; Kilroy, S.; Louis, G.; Moses, M.; Arbiser, J.L. Malignant Transformation of Melanocytes to Melanoma by Constitutive Activation of Mitogen-activated Protein Kinase Kinase (MAPKK) Signaling. J. Boil. Chem. 2003, 278, 9790–9795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Choi, J.W.; Kim, Y.S. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: A meta-analysis. Br. J. Dermatol. 2011, 164, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Boursault, L.; Haddad, V.; Vergier, B.; Cappellen, D.; Verdon, S.; Bellocq, J.P.; Jouary, T.; Merlio, J.P. Tumor Homogeneity between Primary and Metastatic Sites for BRAF Status in Metastatic Melanoma Determined by Immunohistochemical and Molecular Testing. PLoS ONE 2013, 8, e70826. [Google Scholar] [CrossRef]
- Colombino, M.; Capone, M.; Lissia, A.; Cossu, A.G.M.; Rubino, C.; De Giorgi, V.; Massi, D.; Fonsatti, E.; Staibano, S.; Nappi, O.; et al. BRAF/NRAS Mutation Frequencies Among Primary Tumors and Metastases in Patients with Melanoma. J. Clin. Oncol. 2012, 30, 2522–2529. [Google Scholar] [CrossRef]
- Dong, J.; Phelps, R.G.; Qiao, R.; Yao, S.; Benard, O.; Ronai, Z.; Aaronson, S.A. BRAF oncogenic mutations correlate with progression rather than initiation of human melanoma. Cancer Res. 2003, 63, 3883–3885. [Google Scholar]
- Hélias-Rodzewicz, Z.; Funck-Brentano, E.; Baudoux, L.; Jung, C.K.; Zimmermann, U.; Marin, C.; Clérici, T.; Le Gall, C.; Peschaud, F.; Taly, V.; et al. Variations of BRAF mutant allele percentage in melanomas. BMC Cancer 2015, 15, 497. [Google Scholar] [CrossRef]
- Chiappetta, C.; Proietti, I.; Soccodato, V.; Puggioni, C.; Zaralli, R.; Pacini, L.; Porta, N.; Skroza, N.; Petrozza, V.; Potenza, C.; et al. BRAF and NRAS Mutations are Heterogeneous and Not Mutually Exclusive in Nodular Melanoma. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Omholt, K.; Platz, A.; Kanter, L.; Ringborg, U.; Hansson, J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin. Cancer Res. 2003, 9, 6483–6488. [Google Scholar]
- Sensi, M.; Nicolini, G.; Petti, C.; Bersani, I.; Lozupone, F.; Molla, A.; Vegetti, C.; Nonaka, D.; Mortarini, R.; Parmiani, G.; et al. Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene 2006, 25, 3357–3364. [Google Scholar] [CrossRef] [Green Version]
- Akbani, R.; Akdemir, K.C.; Aksoy, B.A.; Albert, M.; Ally, A.; Amin, S.B.; Arachchi, H.; Arora, A.; Auman, J.T.; Ayala, B.; et al. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef] [PubMed]
- Curtin, J.A.; Busam, K.; Pinkel, D.; Bastian, B.C. Somatic Activation of KIT in Distinct Subtypes of Melanoma. J. Clin. Oncol. 2006, 24, 4340–4346. [Google Scholar] [CrossRef] [PubMed]
- Bekers, E.M.; Grunsven, A.C.H.V.E.V.; Groenen, P.J.T.A.; Westdorp, H.; Koornstra, R.H.T.; Bonenkamp, J.J.; Flucke, U.; Blokx, W.A.M. Metastatic melanoma mimicking solitary fibrous tumor: Report of two cases. Virchows Arch. 2014, 464, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Huang, L.; Xu, A.; Zhang, A.; Kong, X.; Ding, M.; Hu, W.; Guo, Z.; Zhong, W.; Sun, S.; et al. Undifferentiated sinonasal malignant melanoma: A case report. Oncol. Lett. 2018, 16, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Crowson, A.N.; Magro, C.; Mihm, M.C. Unusual histologic and clinical variants of melanoma: Implications for therapy. Curr. Oncol. Rep. 2007, 9, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.; Kumarapeli, A.R.; Gokden, N.; Cox, R.M.; Hutchins, L.; Gardner, J.M. Metastatic melanoma with dedifferentiation and extensive rhabdomyosarcomatous heterologous component. J. Cutan. Pathol. 2018, 45, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, A.; Asai, J.; Wakabayashi, Y.; Komori, S.; Hanada, K.; Takenaka, H.; Konishi, E.; Katoh, N. Malignant melanoma with probable smooth muscle differentiation. Case Rep. Dermatol. 2014, 6, 16–19. [Google Scholar] [CrossRef]
- Wasserman, J.K.; Sekhon, H.S.; Ayroud, Y. Malignant Melanoma with Osteoclast-Like Differentiation. Int. J. Surg. Pathol. 2015, 23, 478–482. [Google Scholar] [CrossRef]
- Vivancos, A.; Caratú, G.; Matito, J.; Muñoz, E.; Ferrer, B.; Hernández-Losa, J.; Bodet, D.; Pérez-Alea, M.; Cortés, J.; Garcia-Patos, V.; et al. Genetic evolution of nevus of Ota reveals clonal heterogeneity acquiring BAP1 and TP53 mutations. Pigment. Cell Melanoma Res. 2016, 29, 247–253. [Google Scholar] [CrossRef]
- Landau, D.A.; Carter, S.L.; Stojanov, P.; McKenna, A.; Stevenson, K.; Lawrence, M.S.; Sougnez, C.; Stewart, C.; Sivachenko, A.; Wang, L.; et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013, 152, 714–726. [Google Scholar] [CrossRef]
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta 2010, 1805, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shain, A.H.; Yeh, I.; Kovalyshyn, I.; Sriharan, A.; Talevich, E.; Gagnon, A.; Dummer, R.; North, J.; Pincus, L.; Ruben, B.; et al. The Genetic Evolution of Melanoma from Precursor Lesions. N. Engl. J. Med. 2015, 373, 1926–1936. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Harbst, K.; Lauss, M.; Cirenajwis, H.; Isaksson, K.; Rosengren, F.; Torngren, T.; Kvist, A.; Johansson, M.C.; Vallon-Christersson, J.; Baldetorp, B.; et al. Multiregion Whole-Exome Sequencing Uncovers the Genetic Evolution and Mutational Heterogeneity of Early-Stage Metastatic Melanoma. Cancer Res. 2016, 76, 4765–4774. [Google Scholar] [CrossRef]
- Satzger, I.; Marks, L.; Kerick, M.; Klages, S.; Berking, C.; Herbst, R.; Völker, B.; Schacht, V.; Timmermann, B.; Gutzmer, R. Allele frequencies of BRAFV600 mutations in primary melanomas and matched metastases and their relevance for BRAF inhibitor therapy in metastatic melanoma. Oncotarget 2015, 6, 37895–37905. [Google Scholar] [CrossRef] [Green Version]
- Yancovitz, M.; Litterman, A.; Yoon, J.; Ng, E.; Shapiro, R.L.; Berman, R.S.; Pavlick, A.C.; Darvishian, F.; Christos, P.; Mazumdar, M. Intra- and inter-tumor heterogeneity of BRAF(V600E))mutations in primary and metastatic melanoma. PLoS ONE 2012, 7, e29336. [Google Scholar] [CrossRef]
- Mesbah Ardakani, N.; Leslie, C.; Grieu-Iacopetta, F.; Lam, W.S.; Budgeon, C.; Millward, M.; Amanuel, B. Clinical and therapeutic implications of BRAF mutation heterogeneity in metastatic melanoma. Pigment Cell Melanoma Res. 2017, 30, 233–242. [Google Scholar] [CrossRef]
- Lamy, P.J.; Castan, F.; Lozano, N.; Montélion, C.; Audran, P.; Bibeau, F.; Roques, S.; Montels, F.; Laberenne, A.C. Next-Generation Genotyping by Digital PCR to Detect and Quantify the BRAF V600E Mutation in Melanoma Biopsies. J. Mol. Diagn. 2015, 17, 366–373. [Google Scholar] [CrossRef] [Green Version]
- Sakaizawa, K.; Goto, Y.; Kiniwa, Y.; Uchiyama, A.; Harada, K.; Shimada, S.; Saida, T.; Ferrone, S.; Takata, M.; Uhara, H.; et al. Mutation analysis of BRAF and KIT in circulating melanoma cells at the single cell level. Br. J. Cancer 2012, 106, 939–946. [Google Scholar] [CrossRef]
- Lin, J.; Goto, Y.; Murata, H.; Sakaizawa, K.; Uchiyama, A.; Saida, T.; Takata, M. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. Br. J. Cancer 2011, 104, 464–468. [Google Scholar] [CrossRef]
- Casula, M.; Colombino, M.; Manca, A.; Caracò, C.; Botti, G.; Ascierto, P.A.; Lissia, A.; Cossu, A.G.M.; Palmieri, G.; Mozzillo, N.; et al. Low Levels of Genetic Heterogeneity in Matched Lymph Node Metastases from Patients with Melanoma. J. Investig. Dermatol. 2016, 136, 1917–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goswami, R.S.; Patel, K.P.; Singh, R.R.; Meric-Bernstam, F.; Kopetz, E.S.; Subbiah, V.; Alvarez, R.H.; Davies, M.A.; Jabbar, K.J.; Roy-Chowdhuri, S.; et al. Hotspot mutation panel testing reveals clonal evolution in a study of 265 paired primary and metastatic tumors. Clin. Cancer Res. 2015, 21, 2644–2651. [Google Scholar] [CrossRef] [PubMed]
- Turajlic, S.; Furney, S.J.; Lambros, M.B.; Mitsopoulos, C.; Kozarewa, I.; Geyer, F.C.; Mackay, A.; Hakas, J.; Zvelebil, M.; Lord, C.J. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 2012, 22, 196–207. [Google Scholar] [CrossRef]
- Reuben, A.; Spencer, C.N.; Prieto, P.A.; Gopalakrishnan, V.; Reddy, S.M.; Miller, J.P.; Mao, X.; De Macedo, M.P.; Chen, J.; Song, X.; et al. Genomic and immune heterogeneity are associated with differential responses to therapy in melanoma. NPJ Genom. Med. 2017, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Watson, I.R.; Li, L.; Cabeceiras, P.K.; Mahdavi, M.; Fang, Z.; Stemke-Hale, K.; Mills, G.B.; Chin, L. Abstract LB-214: The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Exp. Mol. Ther. 2014, 74, 4845–4852. [Google Scholar]
- Anaka, M.; Hudson, C.; Lo, P.H.; Do, H.; Caballero, O.L.; Davis, I.D.; Dobrovic, A.; Cebon, J.; Behren, A. Intratumoral genetic heterogeneity in metastatic melanoma is accompanied by variation in malignant behaviors. BMC Med. Genom. 2013, 6, 40. [Google Scholar] [CrossRef]
- Horak, P.; Fröhling, S.; Glimm, H. Integrating next-generation sequencing into clinical oncology: Strategies, promises and pitfalls. ESMO Open 2016, 1, e000094. [Google Scholar] [CrossRef]
- de Unamuno Bustos, B.; Estal, R.M.; Simó, G.P.; de Juan Jimenez, I.; Muñoz, B.E.; Serna, M.R.; de Miquel, V.A.; Ros, M.L.; Sánchez, R.B.; Enguídanos, E.N.; et al. Towards Personalized Medicine in Melanoma: Implementation of a Clinical Next-Generation Sequencing Panel. Sci. Rep. 2017, 7, 495. [Google Scholar] [CrossRef]
| Patient | Age/Sex | Tumor Site(s) | SLN Metastasis | Treatment and Disease Outcome |
|---|---|---|---|---|
| Primary lesion only | ||||
| 1 | 79/M | Left occipital scalp | No | WLE with negative margins. Multiple metastases in the lung detected 5 months after the initial diagnosis. Pembrolizumab 200 mg/mm2 + albumin-bound paclitaxel (Abraxane) 200 mg/mm2 for 6 one-month cycles. Hepatic and ischial bone metastases detected 11 months after the initial diagnosis. At last follow-up, 21 months after the initial diagnosis, the patient was alive with stable metastatic disease. |
| 2 | 70/M | Left clavicular area | No | WLE with negative margins. At last follow-up, 20 months after the initial diagnosis, the patient was disease-free. |
| Paired primary tumor and metastatic lesion | ||||
| 3 | 72/M | Primary: right forehead Metastasis: right intraparotid lymph node | No | WLE with negative margins. Nivolumab therapy was initiated, but the patient was lost to follow-up and died of unknown cause 12 months after the initial diagnosis. |
| 4 | 81/F | Primary: plantar surface of right foot Metastasis: right groin lymph node | Yes | WLE with negative margins. Nivolumab × 6 cycles. Metastasis to right pelvic soft tissue was detected 21 months after the initial diagnosis. |
| 5 | 75/M | Primary: Left lateral arm Metastasis: Left lateral forearm (in-transit) | Yes | Treatment and follow-up not available. |
| Case | Histologic Subtype | Immunohistochemical Profile | Breslow Thickness (PRI) or Largest Dimension (MET) in mm | Mitoses Per mm2 | Ulceration Width in mm | LVI/PNI (PRI Only) | Detected Mutations | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Morphology | S100 | SOX10 | PanMel | HMB-45 | Melan-A | |||||||
| Primary lesion only | ||||||||||||
| 1 | LMM | Nevoid | + | + | + | + | + | 7.1 | 5 | - | +/+ | NRAS, TP53, APC, KDR |
| Spindled | + | + | + (focal) | - | - | NRAS, TP53, KDR | ||||||
| 2 | SSM | Epithelioid | + | + | + | + (patchy) | + (patchy) | 2.3 | 2 | - | -/+ | TP53, CDKN2A, STK11 |
| Spindled | + (patchy) | + | - | - | - | TP53, CDKN2A | ||||||
| Paired primary and metastatic lesions | ||||||||||||
| 3 | SSM | Primary: Epithelioid | + | + | + | + | + | 1.2 | 3 | - | -/- | BRAF, TP53, ATM |
| Metastasis: Rhabdoid | + (patchy) | + (patchy) | + (patchy) | + (focal) | - | 1.3 | BRAF, TP53, ATM, RAC1 | |||||
| 4 | ALM | Primary: Epithelioid | + | + | + | + | + | 3.1 | 6 | 0.1 | +/- | KIT, HNFA1 |
| Metastasis: Foamy/balloon-like | + | + | + | NA | + | 60 | KIT, APC | |||||
| 5 | NM | Primary: Epithelioid | + | + | + | + | + | 12 | 14 | 18 | +/+ | NRAS, TP53, CTNNB1 |
| Primary: Rhabdoid | + (patchy) | - | - | - | + (focal) | NRAS, TP53 | ||||||
| Metastasis: Myxoid stroma | + (focal) | + (patchy) | - | - | + (weak) | 1.6 | NRAS, TP53, STK11 | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mejbel, H.A.; Arudra, S.K.C.; Pradhan, D.; Torres-Cabala, C.A.; Nagarajan, P.; Tetzlaff, M.T.; Curry, J.L.; Ivan, D.; Duose, D.Y.; Luthra, R.; et al. Immunohistochemical and Molecular Features of Melanomas Exhibiting Intratumor and Intertumor Histomorphologic Heterogeneity. Cancers 2019, 11, 1714. https://doi.org/10.3390/cancers11111714
Mejbel HA, Arudra SKC, Pradhan D, Torres-Cabala CA, Nagarajan P, Tetzlaff MT, Curry JL, Ivan D, Duose DY, Luthra R, et al. Immunohistochemical and Molecular Features of Melanomas Exhibiting Intratumor and Intertumor Histomorphologic Heterogeneity. Cancers. 2019; 11(11):1714. https://doi.org/10.3390/cancers11111714
Chicago/Turabian StyleMejbel, Haider A., Sri Krishna C. Arudra, Dinesh Pradhan, Carlos A. Torres-Cabala, Priyadharsini Nagarajan, Michael T. Tetzlaff, Jonathan L. Curry, Doina Ivan, Dzifa Y. Duose, Raja Luthra, and et al. 2019. "Immunohistochemical and Molecular Features of Melanomas Exhibiting Intratumor and Intertumor Histomorphologic Heterogeneity" Cancers 11, no. 11: 1714. https://doi.org/10.3390/cancers11111714
APA StyleMejbel, H. A., Arudra, S. K. C., Pradhan, D., Torres-Cabala, C. A., Nagarajan, P., Tetzlaff, M. T., Curry, J. L., Ivan, D., Duose, D. Y., Luthra, R., Prieto, V. G., Ballester, L. Y., & Aung, P. P. (2019). Immunohistochemical and Molecular Features of Melanomas Exhibiting Intratumor and Intertumor Histomorphologic Heterogeneity. Cancers, 11(11), 1714. https://doi.org/10.3390/cancers11111714








