Transcriptome Profiling Reveals Matrisome Alteration as a Key Feature of Ovarian Cancer Progression
Abstract
1. Introduction
2. Results
2.1. Study Design
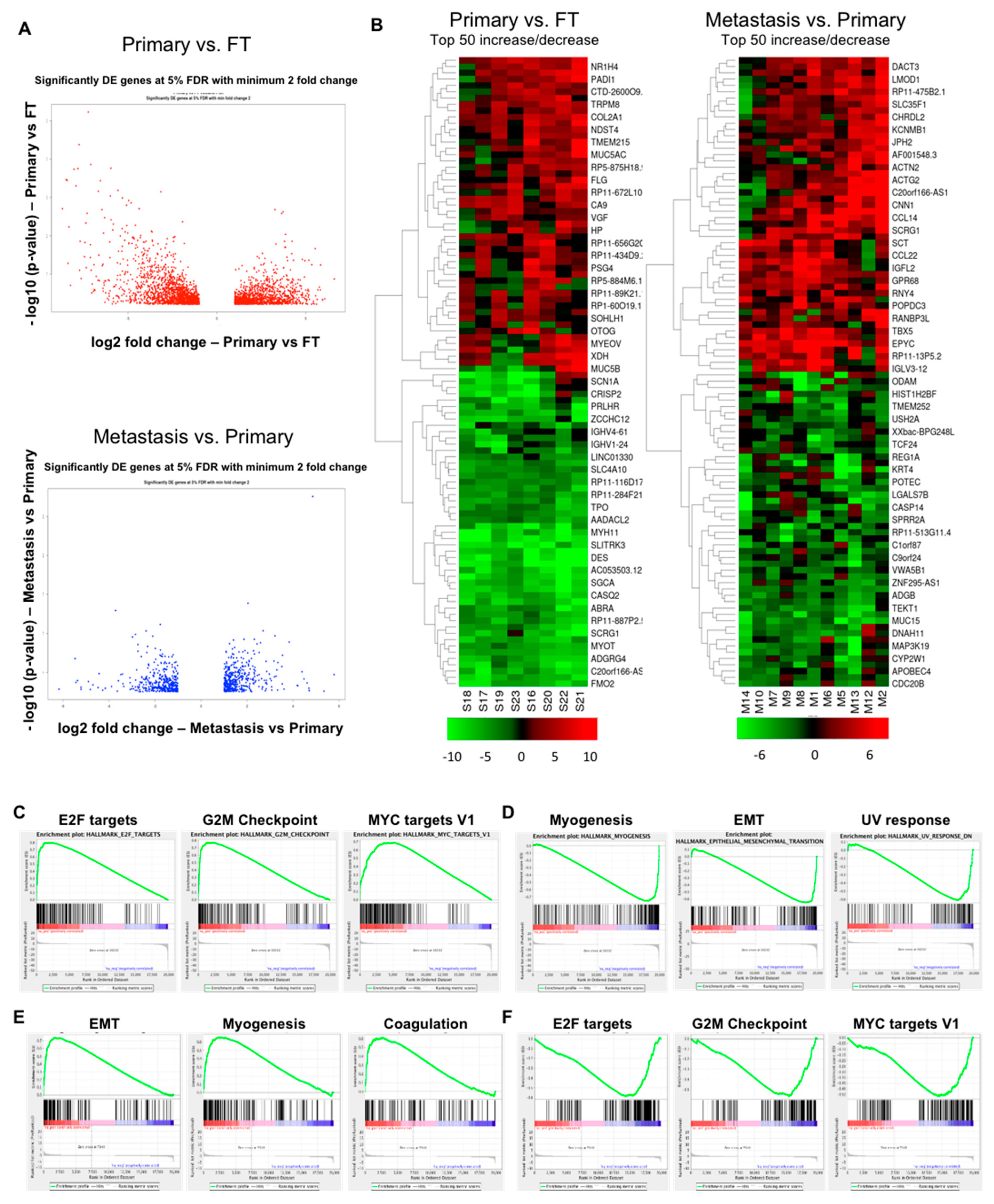
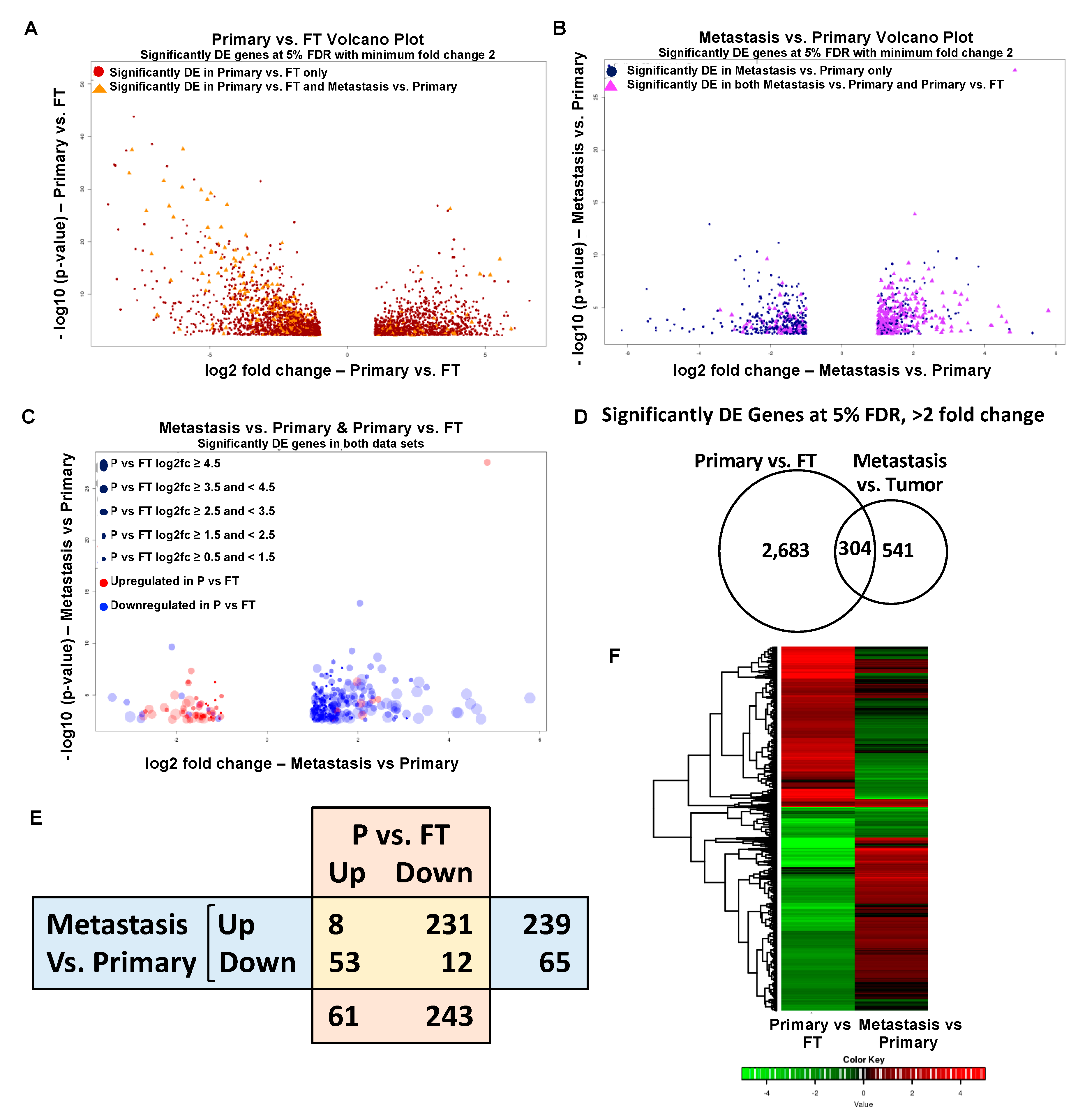
2.2. Gene Expression Changes in Normal FT versus Primary Tumors and Primary Tumors versus Metastasis
2.3. Overlapping Genes in Primary Tumors versus FT and Metastasis versus Primary Tumors
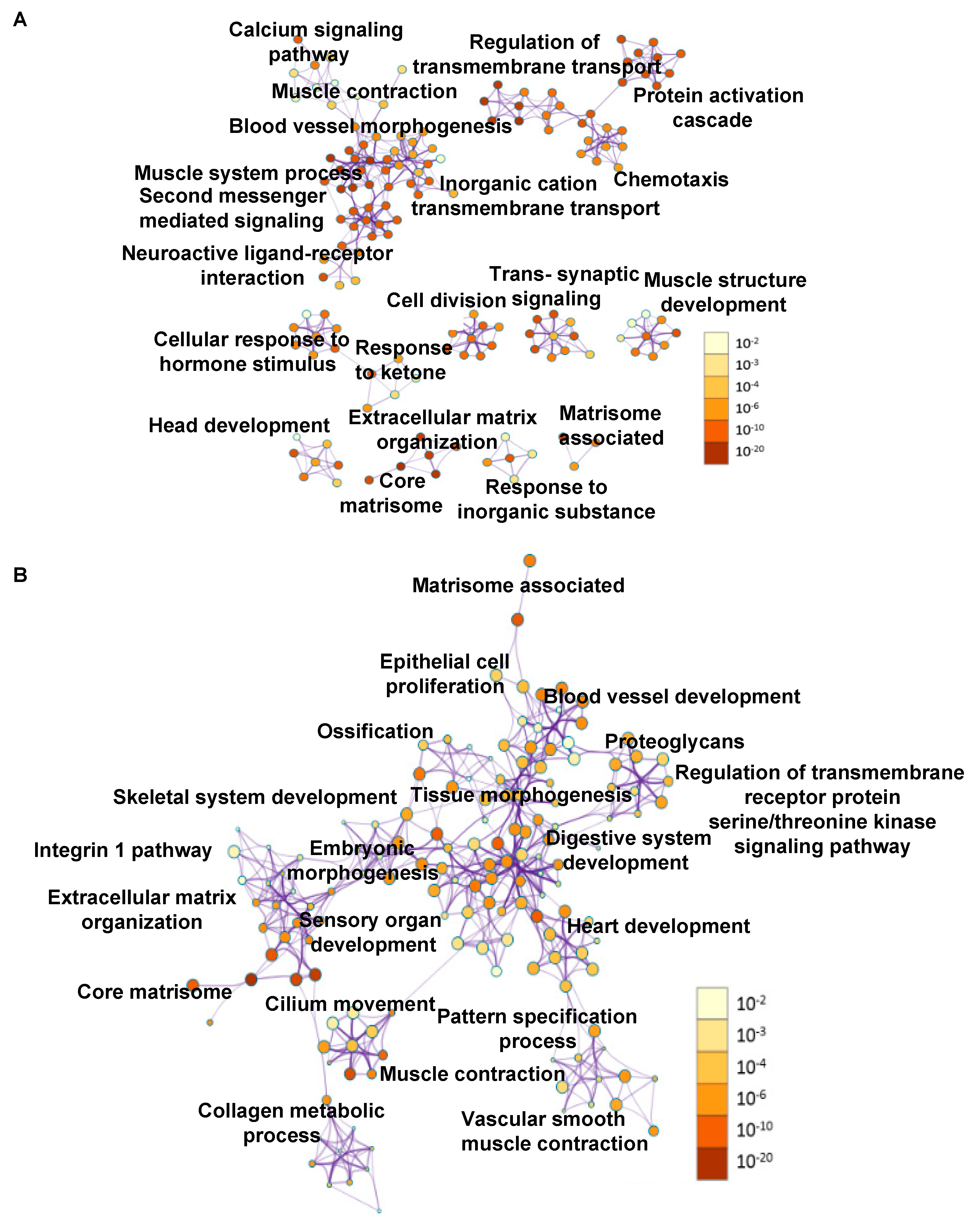
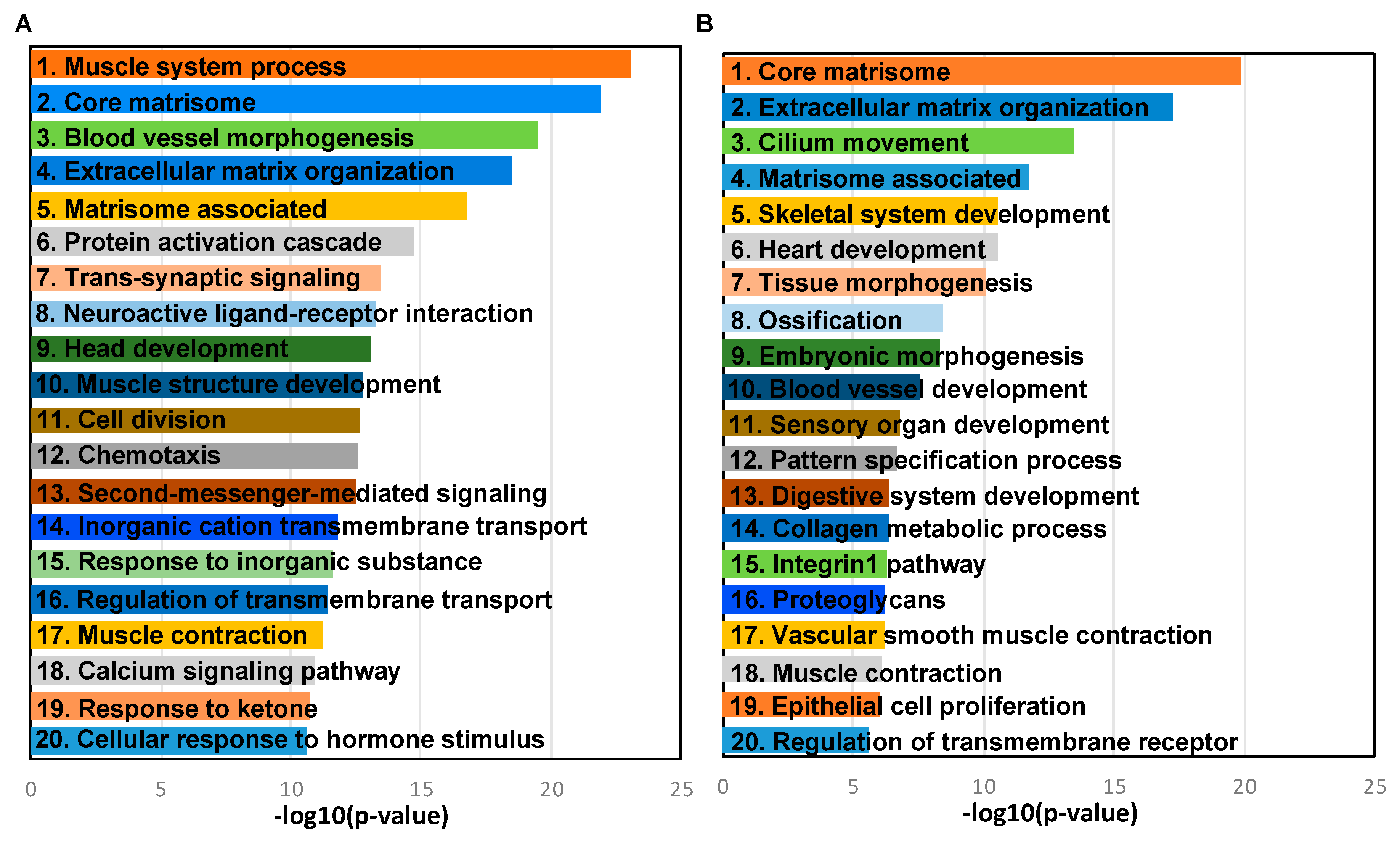
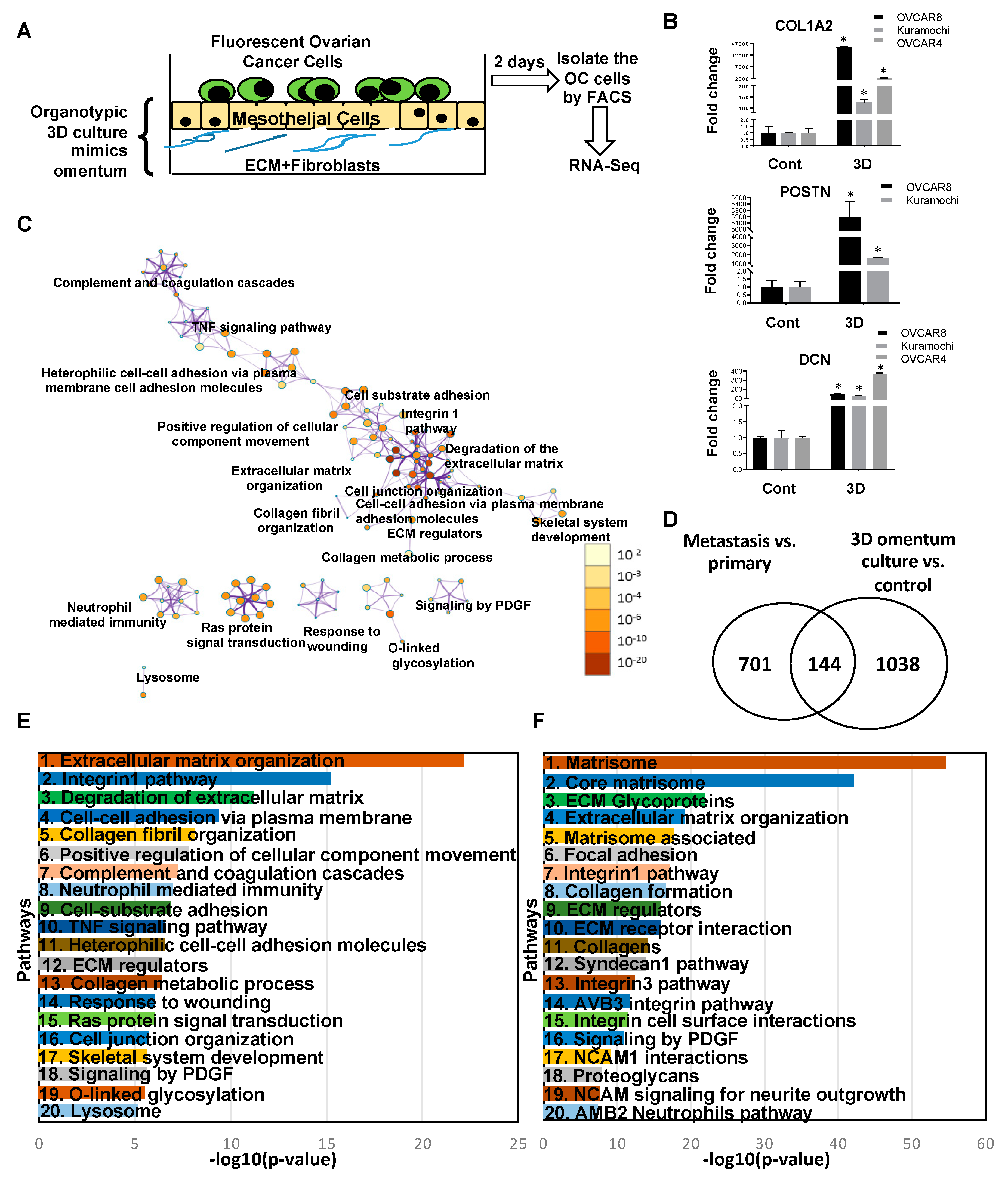
2.4. Transcriptome Changes During Early Metastatic Colonization
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Patient Samples
4.3. Cell Lines
4.4. D Omental Culture and RNA Isolation
4.5. RNA Sequencing
4.6. Analysis of Sequencing Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian Cancer Statistics, 2018. CA A Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Integrated Genomic Analyses of Ovarian Carcinoma. Nature 2011, 474, 609–615. [CrossRef] [PubMed]
- Vaughan, S.; Coward, J.I.; Bast, R.C., Jr.; Berchuck, A.; Berek, J.S.; Brenton, J.D.; Coukos, G.; Crum, C.C.; Drapkin, R.; Etemadmoghadam, D.; et al. Rethinking Ovarian Cancer: Recommendations for Improving Outcomes. Nat. Rev. Cancer 2011, 11, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Koonings, P.P.; Campbell, K.; Mishell, D.R., Jr.; Grimes, D.A. Relative Frequency of Primary Ovarian Neoplasms: A 10-Year Review. Obstet. Gynecol. 1989, 74, 921–926. [Google Scholar] [CrossRef]
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel Molecular Subtypes of Serous and Endometrioid Ovarian Cancer Linked to Clinical Outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Budczies, J.; Darb-Esfahani, S.; Gyorffy, B.; Sehouli, J.; Konsgen, D.; Zeillinger, R.; Weichert, W.; Noske, A.; Buckendahl, A.C.; et al. A Prognostic Gene Expression Index in Ovarian Cancer—Validation across Different Independent Data Sets. J. Pathol. 2009, 218, 273–280. [Google Scholar] [CrossRef]
- Piek, J.M.; van Diest, P.J.; Zweemer, R.P.; Jansen, J.W.; Poort–Keesom, R.J.; Menko, F.H.; Gille, J.J.; Jongsma, A.P.; Pals, G.; Kenemans, P.; et al. Dysplastic Changes in Prophylactically Removed Fallopian Tubes of Women Predisposed to Developing Ovarian Cancer. J. Pathol. 2001, 195, 451–456. [Google Scholar] [CrossRef]
- Callahan, M.J.; Crum, C.P.; Medeiros, F.; Kindelberger, D.W.; Elvin, J.A.; Garber, J.E.; Feltmate, C.M.; Berkowitz, R.S.; Muto, M.G. Primary Fallopian Tube Malignancies in BRCA-Positive Women Undergoing Surgery for Ovarian Cancer Risk Reduction. J. Clin. Oncol. 2007, 25, 3985–3990. [Google Scholar] [CrossRef]
- Kindelberger, D.W.; Lee, Y.; Miron, A.; Hirsch, M.S.; Feltmate, C.; Medeiros, F.; Callahan, M.J.; Garner, E.O.; Gordon, R.W.; Birch, C.; et al. Intraepithelial Carcinoma of the Fimbria and Pelvic Serous Carcinoma: Evidence for a Causal Relationship. Am. J. Surg. Pathol. 2007, 31, 161–169. [Google Scholar] [CrossRef]
- Ducie, J.; Dao, F.; Considine, M.; Olvera, N.; Shaw, P.A.; Kurman, R.J.; Shih, I.M.; Soslow, R.A.; Cope, L.; Levine, D.A. Molecular Analysis of High-Grade Serous Ovarian Carcinoma With and Without Associated Serous Tubal Intra-Epithelial Carcinoma. Nat. Commun. 2017, 8, 990. [Google Scholar] [CrossRef]
- Li, J.; Abushahin, N.; Pang, S.; Xiang, L.; Chambers, S.K.; Fadare, O.; Kong, B.; Zheng, W. Tubal Origin of ‘Ovarian’ Low-Grade Serous Carcinoma. Mod. Pathol. 2011, 24, 1488–1499. [Google Scholar]
- Grellety, T.; Lucchesi, C.; Hostein, I.; Auzanneau, C.; Khalifa, E.; Soubeyran, I.; Italiano, A. High-depth Sequencing of Paired Primary and Metastatic Tumours: Implications for Personalised Medicine. Eur. J. Cancer (Oxford, England: 1990) 2017, 84, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Marchion, D.C.; Xiong, Y.; Chon, H.S.; Al Sawah, E.; Bou Zgheib, N.; Ramirez, I.J.; Abbasi, F.; Stickles, X.B.; Judson, P.L.; Hakam, A.; et al. Gene Expression Data Reveal Common Pathways that Characterize the Unifocal Nature of Ovarian Cancer. Am. J. Obstet. Gynecol. 2013, 209, 576.e1–576.e16. [Google Scholar] [CrossRef] [PubMed]
- Chien, J.; Neums, L.; Powell, A.; Torres, M.; Kalli, K.R.; Multinu, F.; Shridhar, V.; Mariani, A. Genetic Evidence for Early Peritoneal Spreading in Pelvic High-Grade Serous Cancer. Front. Oncol. 2018, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, A.S.; Fischer, A.; Miller, D.H.; Vang, S.; MacLaughlan, S.; Wu, H.T.; Yu, J.; Steinhoff, M.; Collins, C.; Smith, P.J.; et al. Expression Profiling of Primary and Metastatic Ovarian Tumors Reveals Differences Indicative of Aggressive Disease. PLoS ONE 2014, 9, e94476. [Google Scholar] [CrossRef] [PubMed]
- Glasgow, M.A.; Argenta, P.; Abrahante, J.E.; Shetty, M.; Talukdar, S.; Croonquist, P.A.; Khalifa, M.A.; Starr, T.K. Biological Insights into Chemotherapy Resistance in Ovarian Cancer. Int. J. Mol. Sci. 2019, 20, 2131. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Yoon, J.K.; Kim, B.; Kim, S.; Kim, M.A.; Lim, H.; Bang, D.; Song, Y.S. Tumor Evolution and Intratumor Heterogeneity of An Epithelial Ovarian Cancer Investigated Using Next-Generation Sequencing. BMC Cancer 2015, 15, 85. [Google Scholar] [CrossRef]
- Eckert, M.A.; Pan, S.; Hernandez, K.M.; Loth, R.M.; Andrade, J.; Volchenboum, S.L.; Faber, P.; Montag, A.; Lastra, R.; Peter, M.E.; et al. Genomics of Ovarian Cancer Progression Reveals Diverse Metastatic Trajectories Including Intraepithelial Metastasis to the Fallopian Tube. Cancer Discov. 2016, 6, 1342–1351. [Google Scholar] [CrossRef]
- Bowtell, D.D.; Bohm, S.; Ahmed, A.A.; Aspuria, P.J.; Bast, R.C., Jr.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking Ovarian Cancer II: Reducing Mortality from High-Grade Serous Ovarian Cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef]
- Tomar, S.; Plotnik, J.P.; Haley, J.; Scantland, J.; Dasari, S.; Sheikh, Z.; Emerson, R.; Lenz, D.; Hollenhorst, P.C.; Mitra, A.K. ETS1 Induction by the Microenvironment Promotes Ovarian Cancer Metastasis Through Focal Adhesion Kinase. Cancer Lett. 2018, 414, 190–204. [Google Scholar] [CrossRef]
- Mitra, A.K.; Chiang, C.Y.; Tiwari, P.; Tomar, S.; Watters, K.M.; Peter, M.E.; Lengyel, E. Microenvironment-Induced Downregulation of miR-193b Drives Ovarian Cancer Metastasis. Oncogene 2015, 34, 5923–5932. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.K. Ovarian Cancer Metastasis: A Unique Mechanism of Dissemination; Tumor Metastasis: London, UK, 2016; pp. 43–58. ISBN 978-953-51-2631-7. [Google Scholar] [CrossRef]
- Watters, K.M.; Bajwa, P.; Kenny, H.A. Organotypic 3D Models of the Ovarian Cancer Tumor Microenvironment. Cancers 2018, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Kenny, H.A.; Chiang, C.Y.; White, E.A.; Schryver, E.M.; Habis, M.; Romero, I.L.; Ladanyi, A.; Penicka, C.V.; George, J.; Matlin, K.; et al. Mesothelial Cells Promote Early Ovarian Cancer Metastasis Through Fibronectin Secretion. J. Clin. Investig. 2014, 124, 4614–4628. [Google Scholar] [CrossRef] [PubMed]
- Iwanicki, M.P.; Davidowitz, R.A.; Ng, M.R.; Besser, A.; Muranen, T.; Merritt, M.; Danuser, G.; Ince, T.A.; Brugge, J.S. Ovarian Cancer Spheroids Use Myosin-Generated Force to Clear the Mesothelium. Cancer Discov. 2011, 1, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, C.L.; Weinberg, R.A. A Perspective on Cancer Cell Metastasis. Science (New York, N.Y.) 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and Growth of Cancer Cells in Metastatic Sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Peters, P.N.; Schryver, E.M.; Lengyel, E.; Kenny, H. Modeling the Early Steps of Ovarian Cancer Dissemination in an Organotypic Culture of the Human Peritoneal Cavity. J. Vis. Exp. 2015, e53541. [Google Scholar] [CrossRef]
- Klymenko, Y.; Kim, O.; Loughran, E.; Yang, J.; Lombard, R.; Alber, M.; Stack, M.S. Cadherin Composition and Multicellular Aggregate Invasion in Organotypic Models of Epithelial Ovarian Cancer Intraperitoneal Metastasis. Oncogene 2017, 36, 5840–5851. [Google Scholar] [CrossRef]
- Yin, J.G.; Liu, X.Y.; Wang, B.; Wang, D.Y.; Wei, M.; Fang, H.; Xiang, M. Gene Expression Profiling Analysis of Ovarian Cancer. Oncol. Lett. 2016, 12, 405–412. [Google Scholar] [CrossRef]
- Cretu, A.; Brooks, P.C. Impact of the Non-Cellular Tumor Microenvironment on Metastasis: Potential Therapeutic and Imaging Opportunities. J. Cell. Physiol. 2007, 213, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Van Kempen, L.C.; Ruiter, D.J.; van Muijen, G.N.; Coussens, L.M. The Tumor Microenvironment: A Critical Determinant of Neoplastic Evolution. Eur. J. Cell Biol. 2003, 82, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, S.; Ross, K.N.; Lander, E.S.; Golub, T.R. A Molecular Signature of Metastasis in Primary Solid Tumors. Nat. Genet. 2003, 33, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Begum, S.; Hearn, J.D.; Hynes, R.O. GPR56, an Atypical G Protein-Coupled Receptor, Binds Tissue Transglutaminase, TG2, and Inhibits Melanoma Tumor Growth and Metastasis. Proc. Natl. Acad. Sci. USA 2006, 103, 9023–9028. [Google Scholar] [CrossRef] [PubMed]
- Oudin, M.J.; Jonas, O.; Kosciuk, T.; Broye, L.C.; Guido, B.C.; Wyckoff, J.; Riquelme, D.; Lamar, J.M.; Asokan, S.B.; Whittaker, C.; et al. Tumor Cell-Driven Extracellular Matrix Remodeling Drives Haptotaxis during Metastatic Progression. Cancer Discov. 2016, 6, 516–531. [Google Scholar] [CrossRef]
- Gehler, S.; Ponik, S.M.; Riching, K.M.; Keely, P.J. Bi-directional Signaling: Extracellular Matrix and Integrin Regulation of Breast Tumor Progression. Crit. Rev. Eukaryot. Gene Expr. 2013, 23, 139–157. [Google Scholar] [CrossRef]
- Naba, A.; Clauser, K.R.; Hoersch, S.; Liu, H.; Carr, S.A.; Hynes, R.O. The Matrisome: In Silico Definition and in vivo Characterization by Proteomics of Normal and Tumor Extracellular Matrices. Mol. Cell. Proteom. 2012, 11, M111.014647. [Google Scholar] [CrossRef]
- Cho, A.; Howell, V.M.; Colvin, E.K. The Extracellular Matrix in Epithelial Ovarian Cancer—A Piece of a Puzzle. Front. Oncol. 2015, 5, 245. [Google Scholar] [CrossRef]
- Paszek, M.J.; Zahir, N.; Johnson, K.R.; Lakins, J.N.; Rozenberg, G.I.; Gefen, A.; Reinhart-King, C.A.; Margulies, S.S.; Dembo, M.; Boettiger, D.; et al. Tensional Homeostasis and the Malignant Phenotype. Cancer Cell 2005, 8, 241–254. [Google Scholar] [CrossRef]
- Butcher, D.T.; Alliston, T.; Weaver, V.M. A Tense Situation: Forcing Tumour Progression. Nat. Rev. Cancer 2009, 9, 108–122. [Google Scholar] [CrossRef]
- Zhu, G.G.; Risteli, L.; Makinen, M.; Risteli, J.; Kauppila, A.; Stenback, F. Immunohistochemical Study of Type I Collagen and Type I pN-Collagen in Benign and Malignant Ovarian Neoplasms. Cancer 1995, 75, 1010–1017. [Google Scholar] [CrossRef]
- Huijbers, I.J.; Iravani, M.; Popov, S.; Robertson, D.; Al-Sarraj, S.; Jones, C.; Isacke, C.M. A Role for Fibrillar Collagen Deposition and the Collagen Internalization Receptor endo180 in Glioma Invasion. PLoS ONE 2010, 5, e9808. [Google Scholar] [CrossRef] [PubMed]
- Kehlet, S.N.; Sanz-Pamplona, R.; Brix, S.; Leeming, D.J.; Karsdal, M.A.; Moreno, V. Excessive Collagen Turnover Products are Released During Colorectal Cancer Progression and Elevated in Serum from Metastatic Colorectal Cancer Patients. Sci. Rep. 2016, 6, 30599. [Google Scholar] [CrossRef] [PubMed]
- Sherman-Baust, C.A.; Weeraratna, A.T.; Rangel, L.B.; Pizer, E.S.; Cho, K.R.; Schwartz, D.R.; Shock, T.; Morin, P.J. Remodeling of the Extracellular Matrix through Overexpression of Collagen VI Contributes to Cisplatin Resistance in Ovarian Cancer Cells. Cancer Cell 2003, 3, 377–386. [Google Scholar] [CrossRef]
- Wu, Y.H.; Chang, T.H.; Huang, Y.F.; Huang, H.D.; Chou, C.Y. COL11A1 Promotes Tumor Progression and Predicts Poor Clinical Outcome in Ovarian Cancer. Oncogene 2014, 33, 3432–3440. [Google Scholar] [CrossRef] [PubMed]
- Raglow, Z.; Thomas, S.M. Tumor Matrix Protein Collagen XIalpha1 in Cancer. Cancer Lett. 2015, 357, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Teng, P.N.; Wang, G.; Hood, B.L.; Conrads, K.A.; Hamilton, C.A.; Maxwell, G.L.; Darcy, K.M.; Conrads, T.P. Identification of Candidate Circulating Cisplatin-Resistant Biomarkers from Epithelial Ovarian Carcinoma Cell Secretomes. Br. J. Cancer 2014, 110, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Pearce, O.M.T.; Delaine-Smith, R.M.; Maniati, E.; Nichols, S.; Wang, J.; Bohm, S.; Rajeeve, V.; Ullah, D.; Chakravarty, P.; Jones, R.R.; et al. Deconstruction of a Metastatic Tumor Microenvironment Reveals a Common Matrix Response in Human Cancers. Cancer Discov. 2018, 8, 304–319. [Google Scholar] [CrossRef]
- De Pascalis, C.; Etienne-Manneville, S. Single and Collective Cell Migration: The Mechanics of Adhesions. Mol. Biol. Cell 2017, 28, 1833–1846. [Google Scholar] [CrossRef]
- Maziveyi, M.; Alahari, S.K. Cell Matrix Adhesions in Cancer: The Proteins that Form the Glue. Oncotarget 2017, 8, 48471–48487. [Google Scholar] [CrossRef]
- Burridge, K.; Guilluy, C. Focal Adhesions, Stress Fibers and Mechanical Tension. Exp. Cell Res. 2016, 343, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Tilghman, R.W.; Parsons, J.T. Focal Adhesion Kinase as a Regulator of Cell Tension in the Progression of Cancer. Semin. Cancer Biol. 2008, 18, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.K.; Sawada, K.; Tiwari, P.; Mui, K.; Gwin, K.; Lengyel, E. Ligand-Independent Activation of c-Met by Fibronectin and alpha(5)beta(1)-Integrin Regulates Ovarian Cancer Invasion and Metastasis. Oncogene 2011, 30, 1566–1576. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Kenny, H.A.; Jagadeeswaran, S.; Zillhardt, M.R.; Montag, A.G.; Kistner, E.; Yamada, S.D.; Mitra, A.K.; Lengyel, E. β3-Integrin Expression on Tumor Cells Inhibits Tumor Progression, Reduces Metastasis, and is Associated with a Favorable Prognosis in Patients with Ovarian Cancer. Am. J. Pathol. 2009, 175, 2184–2196. [Google Scholar]
- Kobayashi, M.; Sawada, K.; Kimura, T. Potential of Integrin Inhibitors for Treating Ovarian Cancer: A Literature Review. Cancers 2017, 9, 83. [Google Scholar] [CrossRef]
- Noh, K.; Mangala, L.S.; Han, H.D.; Zhang, N.; Pradeep, S.; Wu, S.Y.; Ma, S.; Mora, E.; Rupaimoole, R.; Jiang, D.; et al. Differential Effects of EGFL6 on Tumor versus Wound Angiogenesis. Cell Rep. 2017, 21, 2785–2795. [Google Scholar] [CrossRef]
- Choi, H.J.; Armaiz Pena, G.N.; Pradeep, S.; Cho, M.S.; Coleman, R.L.; Sood, A.K. Anti-vascular Therapies in Ovarian Cancer: Moving beyond Anti-VEGF Approaches. Cancer Metastasis Rev. 2015, 34, 19–40. [Google Scholar] [CrossRef]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef]
- Nakamura, M.; Ono, Y.J.; Kanemura, M.; Tanaka, T.; Hayashi, M.; Terai, Y.; Ohmichi, M. Hepatocyte Growth Factor Secreted by Ovarian Cancer Cells Stimulates Peritoneal Implantation via the Mesothelial-Mesenchymal Transition of the Peritoneum. Gynecol. Oncol. 2015, 139, 345–354. [Google Scholar] [CrossRef]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating Cell Lines as Tumour Models by Comparison of Genomic Profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef]
- Haley, J.; Tomar, S.; Pulliam, N.; Xiong, S.; Perkins, S.M.; Karpf, A.R.; Mitra, S.; Nephew, K.P.; Mitra, A.K. Functional Characterization of a Panel of High-Grade Serous Ovarian Cancer Cell Lines As Representative Experimental Models of the Disease. Oncotarget 2016, 7, 32810–32820. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.K.; Davis, D.A.; Tomar, S.; Roy, L.; Gurler, H.; Xie, J.; Lantvit, D.D.; Cardenas, H.; Fang, F.; Liu, Y.; et al. In vivo Tumor Growth of high-grade Serous Ovarian Cancer Cell Lines. Gynecol. Oncol. 2015, 138, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics (Oxford, England) 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics (Oxford, England) 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Dorum, G.; Snipen, L.; Solheim, M.; Saebo, S. Rotation Testing in Gene Set Enrichment Analysis for Small Direct Comparison Experiments. Stat. Appl. Genet. Mol. Biol. 2009, 8, 1–24. [Google Scholar] [CrossRef]
| A | Ingenuity Canonical Pathways: FT vs. Primary Tumors | −log(p-Value) | D | Ingenuity Canonical Pathways: Metastasis vs. Primary Tumors | −log(p-Value) | ||
| 1 | Hepatic Fibrosis / Hepatic Stellate Cell Activation | 10.2 | 1 | Hepatic Fibrosis / Hepatic Stellate Cell Activation | 6.58 | ||
| 2 | cAMP-mediated signaling | 10.1 | 2 | Osteoarthritis Pathway | 5.15 | ||
| 3 | G-Protein Coupled Receptor Signaling | 8.16 | 3 | Agranulocyte Adhesion and Diapedesis | 4.12 | ||
| 4 | Calcium Signaling | 6.94 | 4 | Inhibition of Matrix Metalloproteases | 2.98 | ||
| 5 | Amyotrophic Lateral Sclerosis Signaling | 6.08 | 5 | Bladder Cancer Signaling | 2.86 | ||
| 6 | eNOS Signaling | 5.8 | 6 | Atherosclerosis Signaling | 2.27 | ||
| 7 | MSP-RON Signaling Pathway | 5.77 | 7 | Granulocyte Adhesion and Diapedesis | 2.07 | ||
| 8 | Cellular Effects of Sildenafil (Viagra) | 5.34 | 8 | Neuroprotective Role of THOP1 in Alzheimer’s Disease | 1.99 | ||
| 9 | Intrinsic Prothrombin Activation Pathway | 5.31 | 9 | Eicosanoid Signaling | 1.8 | ||
| 10 | Breast Cancer Regulation by Stathmin1 | 5.27 | 10 | ILK Signaling | 1.76 | ||
| B | Top Diseases and Functions: FT vs. Primary Tumors | E | Top Diseases and Functions: Metastasis vs. Primary Tumors | ||||
| 1 | Cancer, Cellular Development, Organismal Injury and Abnormalities | 1 | Digestive System Development and Function, Embryonic Development, Organismal Development | ||||
| 2 | Nervous System Development and Function, Cell Death and Survival, Tissue Morphology | 2 | Cell-mediated Immune Response, Cellular Movement, Hematological System Development and Function | ||||
| 3 | Cell Morphology, Cellular Assembly and Organization, Cellular Function and Maintenance | 3 | Cardiovascular Disease, Hereditary Disorder, Organismal Injury and Abnormalities | ||||
| 4 | Cell Signaling, Neurological Disease, Organismal Injury and Abnormalities | 4 | Cardiac Arrythmia, Cardiovascular Disease, Hereditary Disorder | ||||
| 5 | Cellular Development, Cellular Growth and Proliferation, Hematological System Development and Function | 5 | Skeletal and Muscular System Development and Function, Gastrointestinal Disease, Organismal Injury and Abnormalities | ||||
| 6 | Cancer, Organismal Injury and Abnormalities, Carbohydrate Metabolism | 6 | Cardiovascular Disease, Cell-To-Cell Signaling and Interaction, Drug Metabolism | ||||
| 7 | Cell Signaling, Carbohydrate Metabolism, Small Molecule Biochemistry | 7 | Digestive System Development and Function, Connective Tissue Development and Function, Connective Tissue Disorders | ||||
| 8 | Molecular Transport, Connective Tissue Disorders, Developmental Disorder | 8 | Developmental Disorder, Hereditary Disorder, Ophthalmic Disease | ||||
| 9 | Skeletal and Muscular System Development and Function, Skeletal and Muscular Disorders, Hereditary Disorder | 9 | Dermatological Diseases and Conditions, Inflammatory Disease, Organismal Injury and Abnormalities | ||||
| 10 | Cell-To-Cell Signaling and Interaction, Cellular Assembly and Organization, Nervous System Development and Function | 10 | Endocrine System Disorders, Organ Morphology, Organismal Injury and Abnormalities | ||||
| C | Upstream Regulator | Molecule Type | p-Value of Overlap | F | Upstream Regulator | Molecule Type | p-Value of Overlap |
| 1 | TGFB1 | growth factor | 1.06 × 10−38 | 1 | MYOCD | transcription regulator | 1.08 × 10−13 |
| 2 | Vegf | Group | 1.48 × 10−33 | 2 | BMP4 | growth factor | 2.57 × 10−10 |
| 3 | beta-estradiol | chemical—endogenous mammalian | 3.47 × 10−29 | 3 | RUNX2 | transcription regulator | 6.84 × 10−10 |
| 4 | ERBB2 | Kinase | 2.48 × 10−28 | 4 | BMP2 | growth factor | 1.43 × 10−8 |
| 5 | dexamethasone | chemical drug | 3.2 × 10−26 | 5 | miR-199a-5p (and other miRNAs w/seed CCAGUGU) | mature microrna | 2.25 × 10−8 |
| 6 | HGF | growth factor | 7.39 × 10−25 | 6 | U0126 | chemical—kinase inhibitor | 2.42 × 10−8 |
| 7 | FGF2 | growth factor | 6.31 × 10−24 | 7 | TGFB3 | growth factor | 2.73 × 10−8 |
| 8 | progesterone | chemical—endogenous mammalian | 1.43 × 10−23 | 8 | TGFB1 | growth factor | 3.72 × 10−8 |
| 9 | TNF | cytokine | 3.83 × 10−22 | 9 | GNA13 | enzyme | 1.29 × 10−7 |
| 10 | IL6 | cytokine | 1.87 × 10−21 | 10 | HAND2 | transcription regulator | 2.1 × 10−7 |
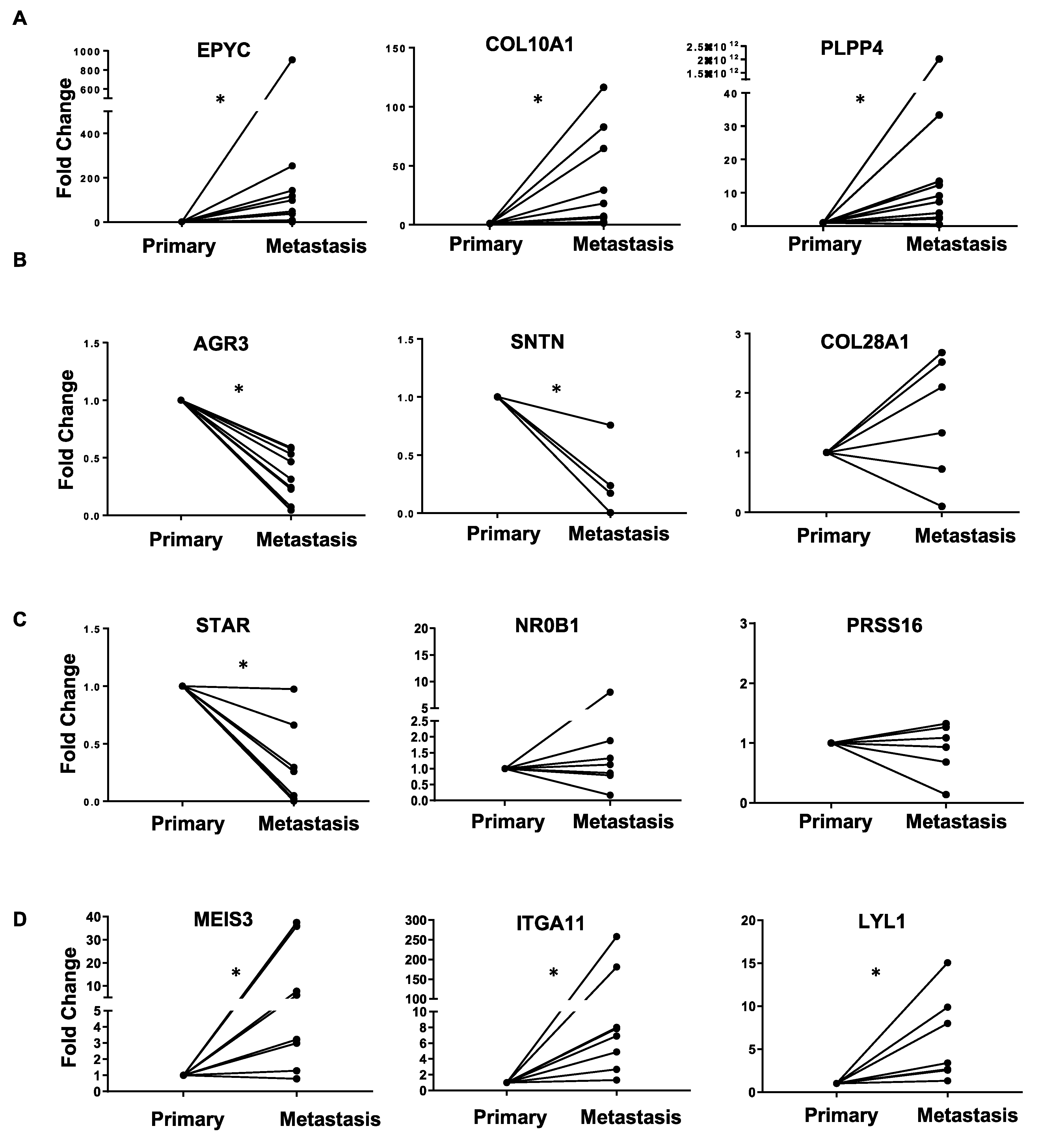
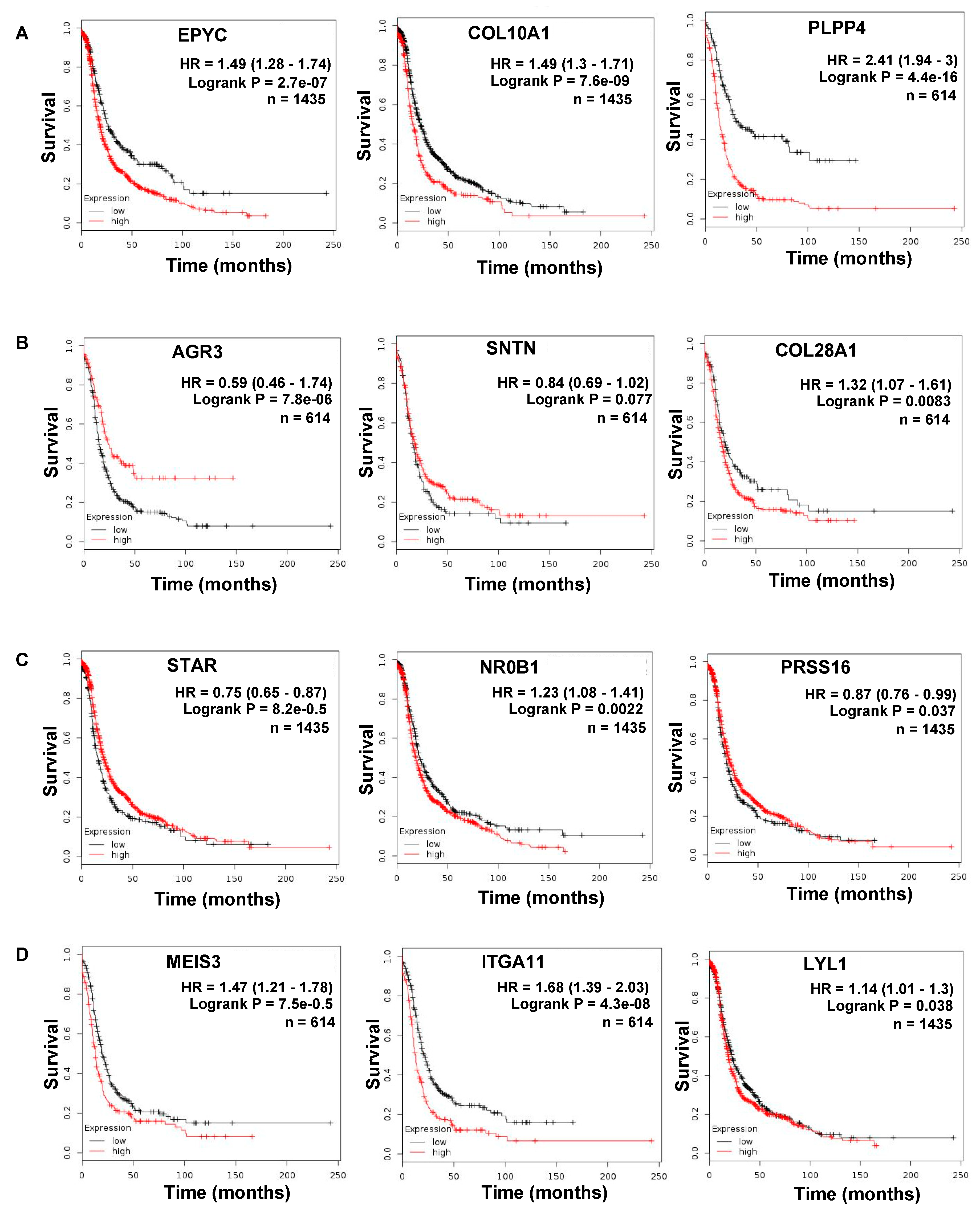
| (A). Common Upregulated Genes in Ovarian Cancer Metastasis versus Primary and Primary versus FT | (B). Common Downregulated Genes in Ovarian Cancer Metastasis versus Primary and Primary versus FT | ||||
| Gene Name | log2 Fold Change | p-Value | Gene Name | log2 Fold Change | p-Value |
| EPYC | 4.84 | 2.78 × 10−28 | AGR3 | −2.09 | 2.25 × 10−10 |
| COL10A1 | 1.98 | 5.19 × 10−7 | SNTN | −3.09 | 4.80 × 10−5 |
| PLPP4 | 2.06 | 5.95 × 10−5 | ANKUB1 | −3.41 | 1.64 × 10−5 |
| RP11-13P5.2 | 2.44 | 2.46 × 10−5 | COL28A1 | −1.29 | 0.000236 |
| PODNL1 | 1.44 | 0.002166 | DCDC2B | −3.01 | 0.001219 |
| CILP2 | 1.54 | 0.000325 | ADGB | −2.75 | 0.000667 |
| FNDC1 | 2.32 | 3.26 × 10−5 | CFAP52 | −1.88 | 1.19 × 10−5 |
| SLC35D3 | 2.09 | 0.000901 | RP11-356K23.1 | −2.77 | 0.001993 |
| WDR38 | −1.49 | 8.60 × 10−5 | |||
| CFAP221 | −1.19 | 0.000604 | |||
| NWD1 | −1.72 | 0.000766 | |||
| FAM166B | −1.07 | 0.001892 | |||
| (C). Top 10 Opposite: Upregulated Genes in Metastasis versus Primary and Downregulated in Primary versus FT | (D). Top 10 Opposite: Downregulated Genes in Metastasis versus Primary and Upregulated in Primary versus FT | ||||
| Gene Name | log2 Fold Change | p-Value | Gene Name | log2 Fold Change | p-Value |
| MEIS3 | 2.04 | 1.29 × 10−14 | ATP6V1C2 | −1.73 | 6.83 × 10−7 |
| ITGA11 | 2.19 | 1.58 × 10−7 | STAR | −1.67 | 4.38 × 10−8 |
| RP1-79C4.4 | 1.10 | 2.54 × 10−8 | NR0B1 | −1.84 | 5.01 × 10−4 |
| LYL1 | 1.42 | 9.63 × 10−7 | KCNG3 | −1.58 | 9.16 × 10−4 |
| ASPA | 1.49 | 3.70 × 10−5 | PRSS16 | −1.14 | 3.02 × 10−5 |
| AOC3 | 1.49 | 9.69 × 10−6 | FAM167A | −1.45 | 6.31 × 10−5 |
| GIMAP8 | 1.41 | 4.79 × 10−8 | C3orf67 | −1.30 | 1.00 × 10−4 |
| LRRN4CL | 1.23 | 5.73 × 10−5 | JPH1 | −1.62 | 1.85 × 10−4 |
| IGHV1-24 | 1.99 | 5.70 × 10−4 | HOOK1 | −1.56 | 1.93 × 10−5 |
| CCL14 | 2.85 | 7.45 × 10−6 | ESM1 | −1.78 | 5.89 × 10−5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitra, S.; Tiwari, K.; Podicheti, R.; Pandhiri, T.; Rusch, D.B.; Bonetto, A.; Zhang, C.; Mitra, A.K. Transcriptome Profiling Reveals Matrisome Alteration as a Key Feature of Ovarian Cancer Progression. Cancers 2019, 11, 1513. https://doi.org/10.3390/cancers11101513
Mitra S, Tiwari K, Podicheti R, Pandhiri T, Rusch DB, Bonetto A, Zhang C, Mitra AK. Transcriptome Profiling Reveals Matrisome Alteration as a Key Feature of Ovarian Cancer Progression. Cancers. 2019; 11(10):1513. https://doi.org/10.3390/cancers11101513
Chicago/Turabian StyleMitra, Sumegha, Kartikeya Tiwari, Ram Podicheti, Taruni Pandhiri, Douglas B. Rusch, Andrea Bonetto, Chi Zhang, and Anirban K. Mitra. 2019. "Transcriptome Profiling Reveals Matrisome Alteration as a Key Feature of Ovarian Cancer Progression" Cancers 11, no. 10: 1513. https://doi.org/10.3390/cancers11101513
APA StyleMitra, S., Tiwari, K., Podicheti, R., Pandhiri, T., Rusch, D. B., Bonetto, A., Zhang, C., & Mitra, A. K. (2019). Transcriptome Profiling Reveals Matrisome Alteration as a Key Feature of Ovarian Cancer Progression. Cancers, 11(10), 1513. https://doi.org/10.3390/cancers11101513






