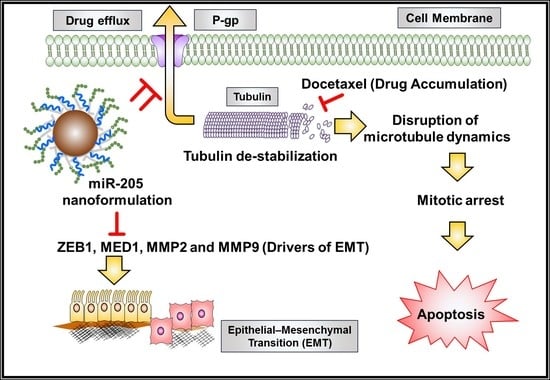
miRNA-205 Nanoformulation Sensitizes Prostate Cancer Cells to Chemotherapy
Abstract
1. Introduction
2. Results
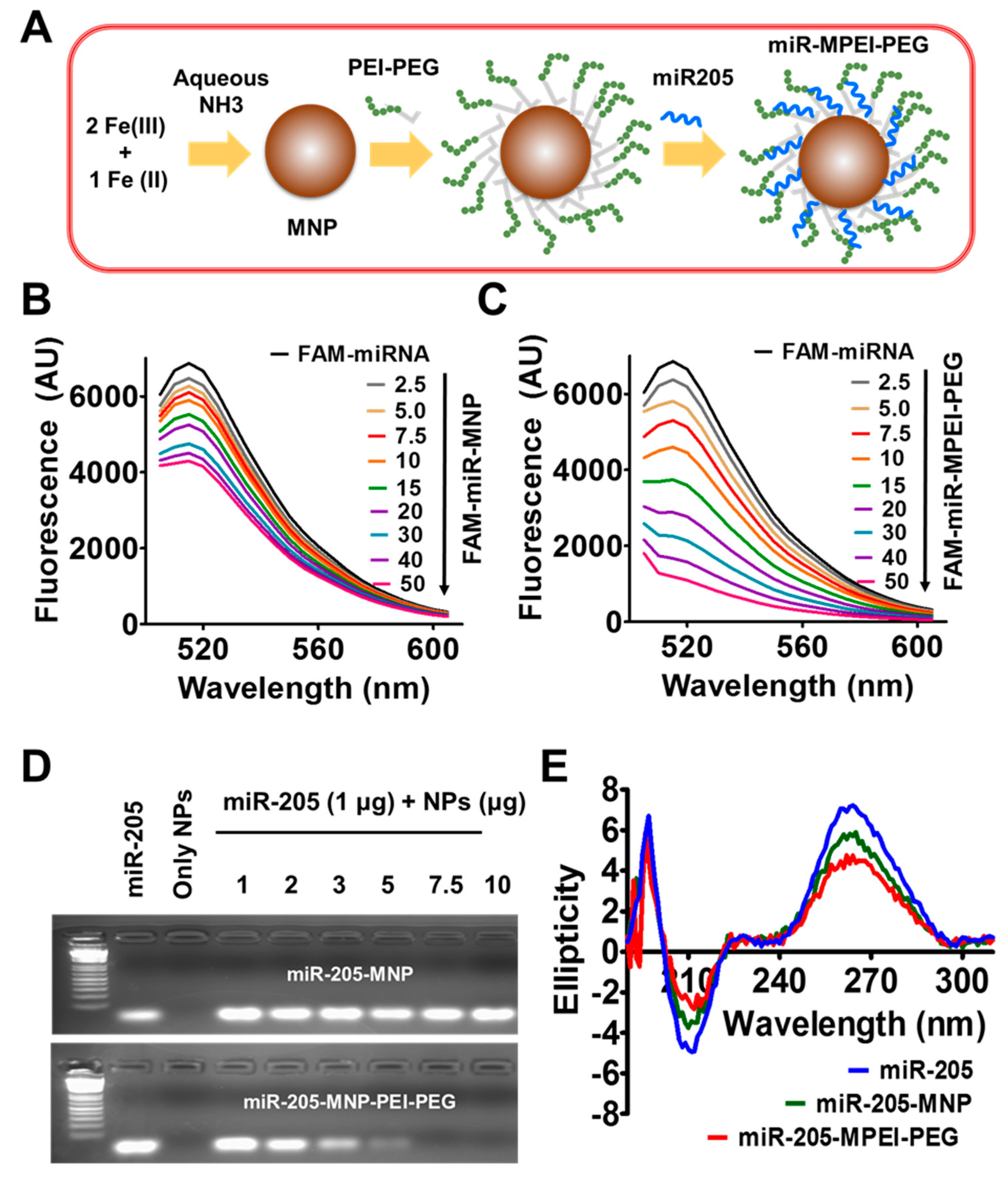
2.1. Preparation and Characterization of miR-205-MPEI-PEG (NPs) Formulations
2.2. miR-205 Efficiently Complexes with MPEI-PEG Formulation
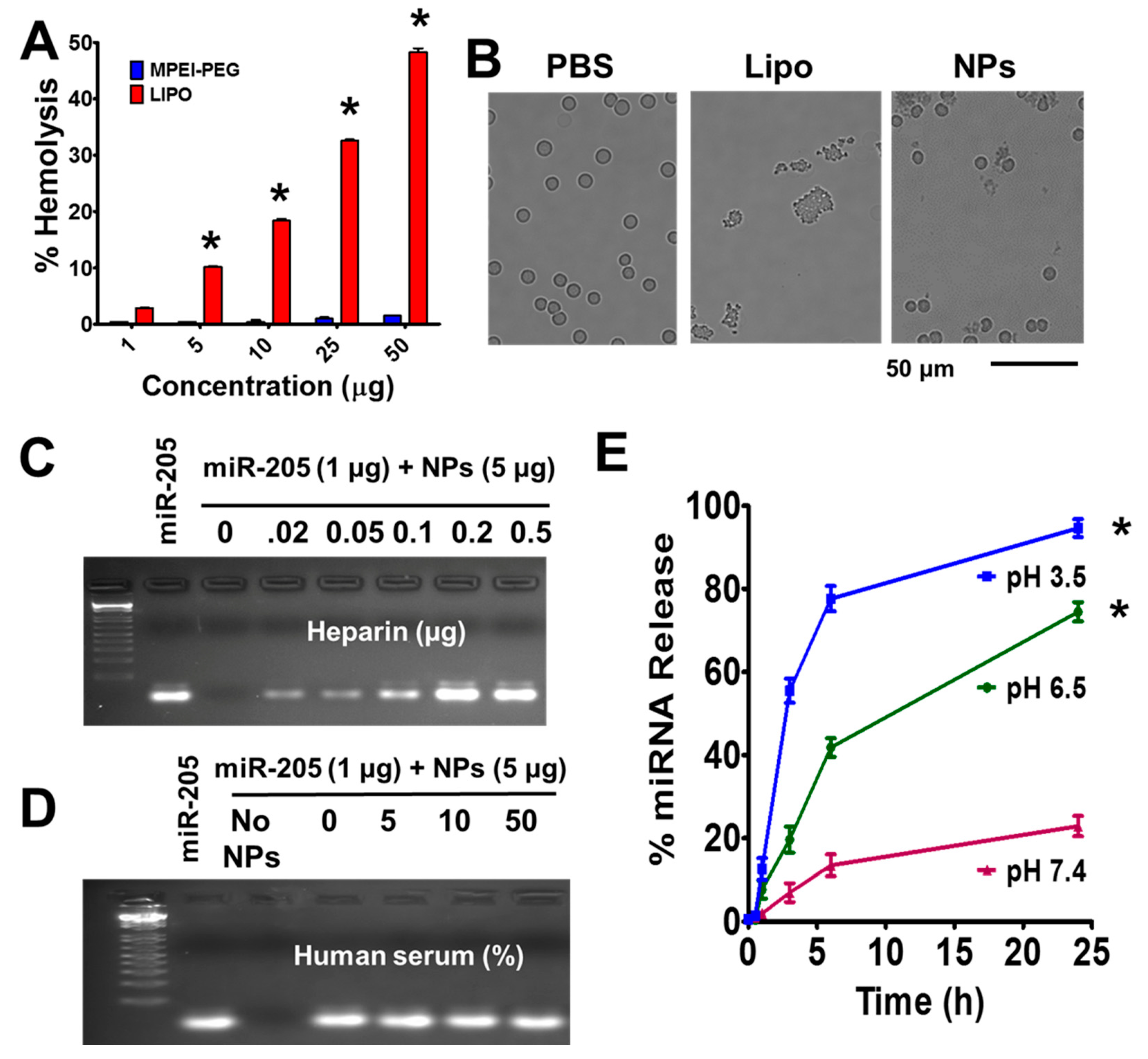
2.3. MPEI-PEG NPs Showed Superior Hemocompatibility
2.4. Fate of miR-205 in MPEI-PEG NPs Formulation
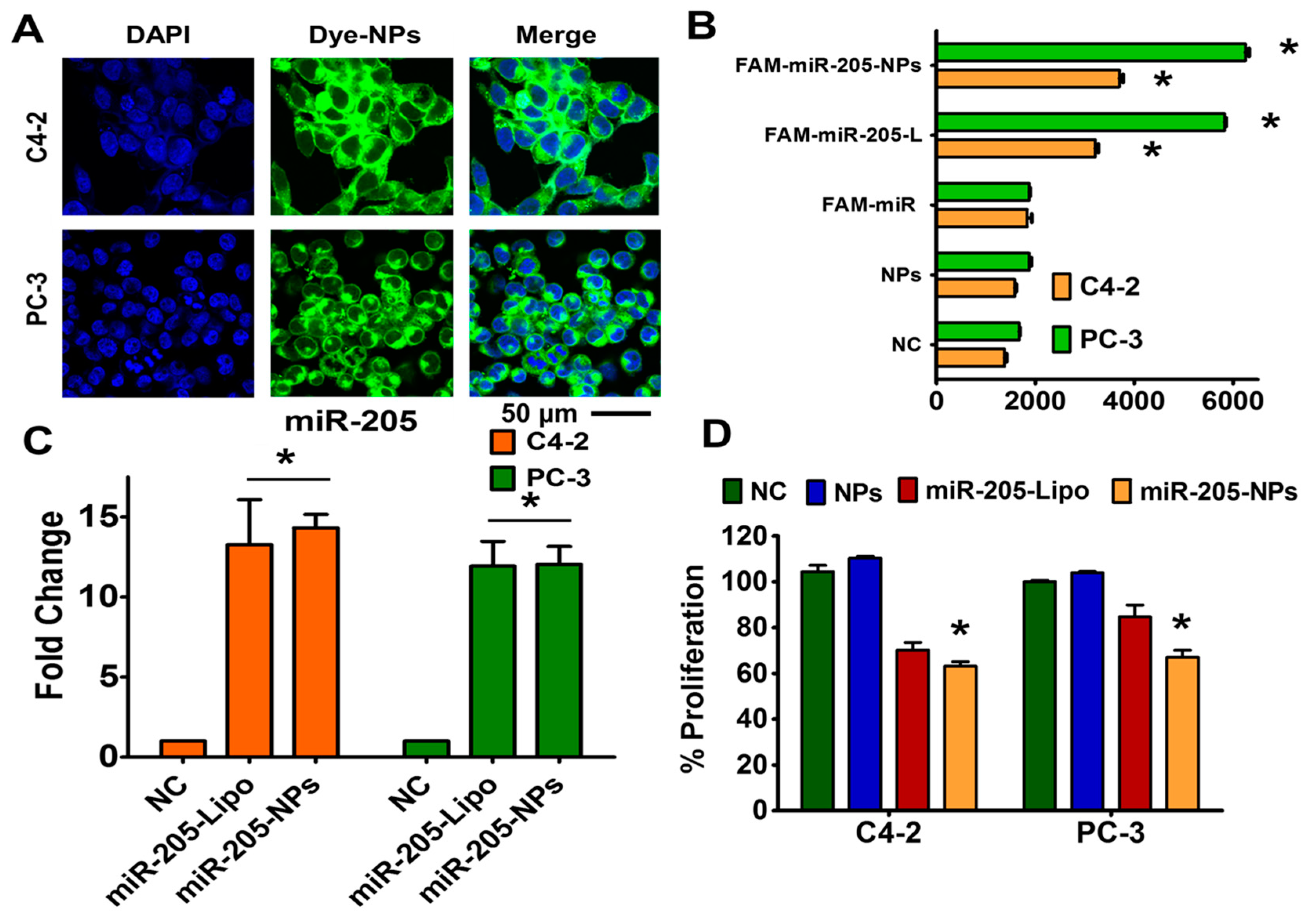
2.5. miR-205 is Efficiently Delivered and Preserved its Activity through NPs Formulation
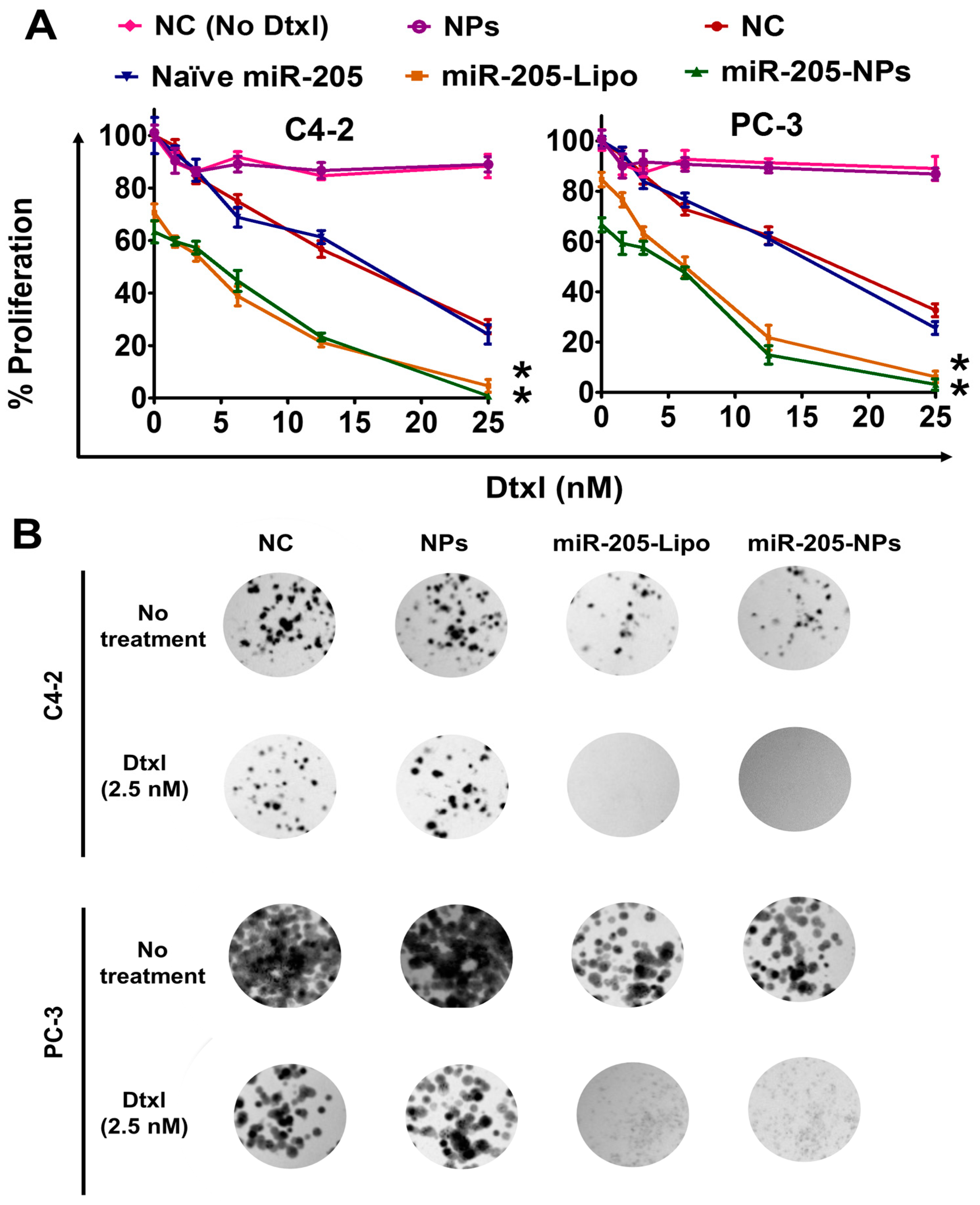
2.6. miR-205-NPs Formulation Induces Chemosensitization in Prostate Cancer Cells
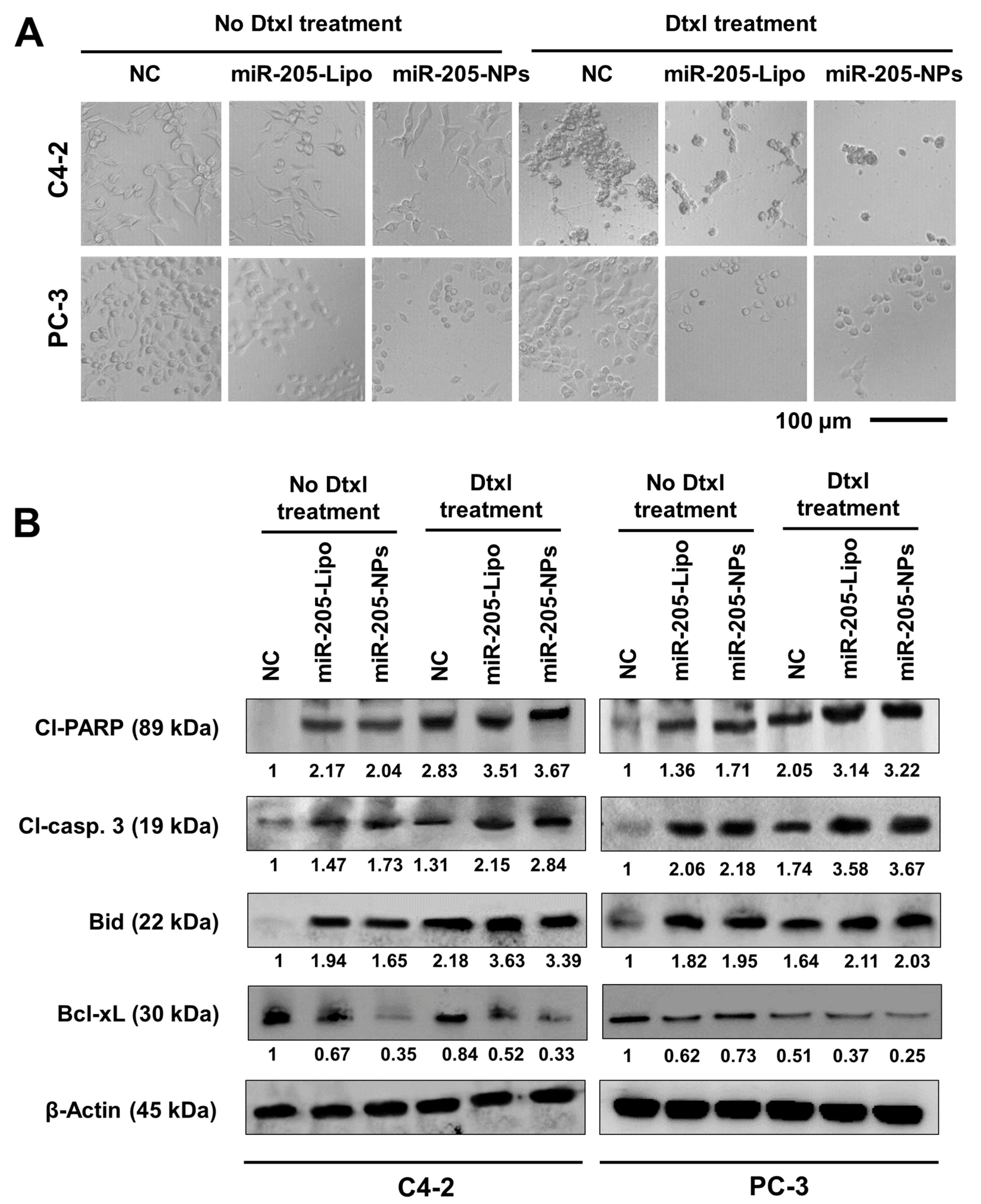
2.7. miR-205-NPs Induce Apoptotic Signaling during Docetaxel Treatment
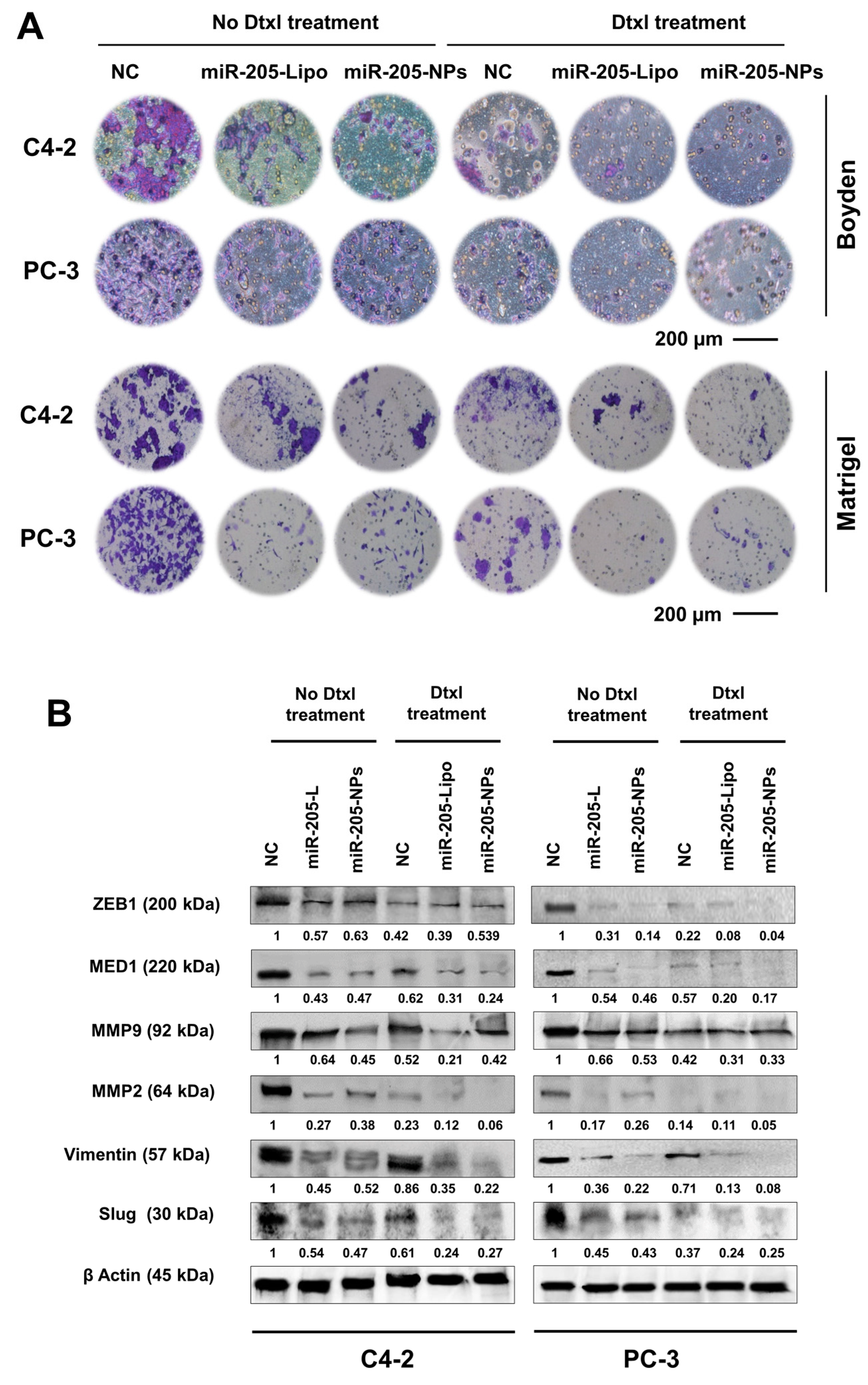
2.8. miR-205-NPs Formulation with Docetaxel Treatment Inhibits Metastatic Phenotype of PrCa Cells
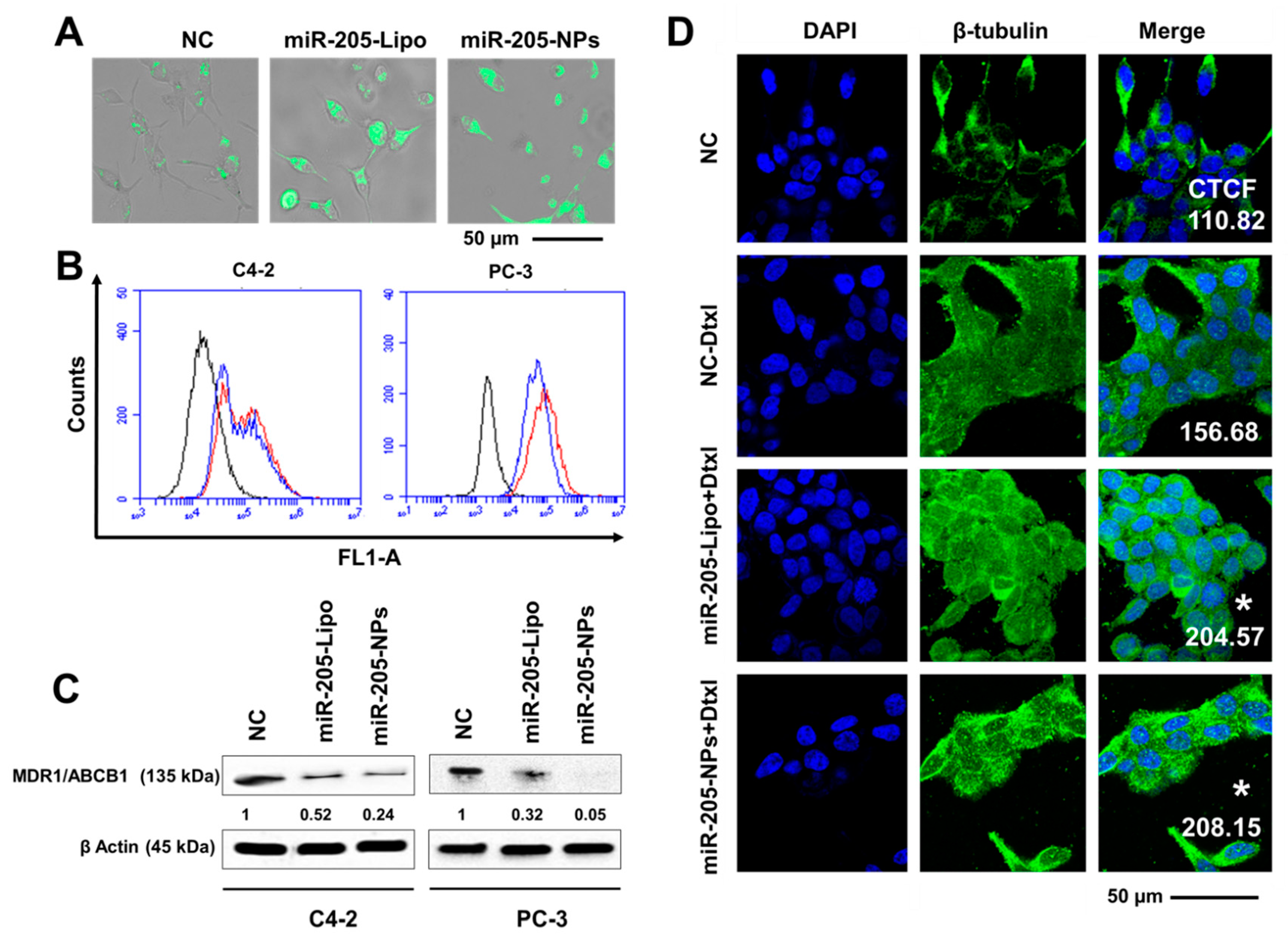
2.9. miR-205-NPs Exhibit Enhanced Intracellular Uptake of Rh 123 in PrCa Cells
2.10. miR-205-NPs Show Pronounced Tubulin Stabilization
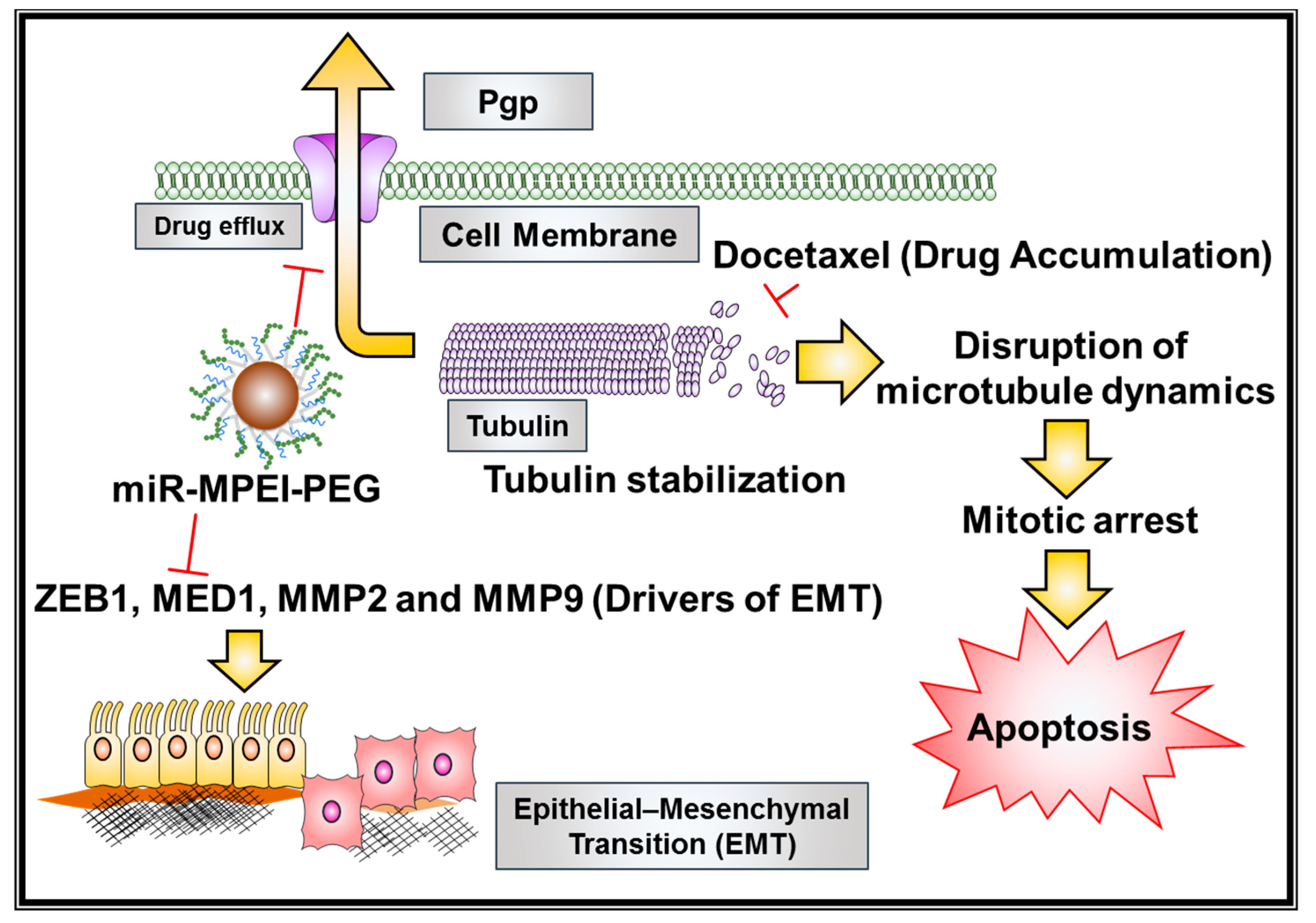
3. Discussion
4. Materials and Methods
4.1. Materials and Cell Culture
4.2. Synthesis of Magnetic Nanoparticles
4.3. Particle Characterization
4.4. miR-205 Binding with Nanoparticles
4.5. Hemolytic Assay
4.6. Stability of microRNA
4.7. miRNA Stability in Serum
4.8. miRNA Release Assay
4.9. Cellular Uptake and Transfection Efficiency
4.10. Transfection and Treatments
4.11. Anticancer Efficacy of miR-205
4.11.1. MTS Assay
4.11.2. Colony Formation Assay
4.11.3. Cell Migration and Invasion Assays
4.12. Western Blotting
4.13. Rhodamine 123 Uptake/Retention Assays
4.14. Tubulin Stabilization Assay
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Rani, S.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; Crown, J.; et al. Docetaxel-resistance in prostate cancer: Evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PloS ONE 2012, 7, e50999. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C. Overcoming docetaxel resistance in prostate cancer: A perspective review. Ther. Adv. Med. Oncol. 2012, 4, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Cengiz, E.; Karaca, B.; Kucukzeybek, Y.; Gorumlu, G.; Gul, M.K.; Erten, C.; Atmaca, H.; Uzunoglu, S.; Karabulut, B.; Sanli, U.A.; et al. Overcoming drug resistance in hormone- and drug-refractory prostate cancer cell line, pc-3 by docetaxel and gossypol combination. Mol. Biol. Rep. 2010, 37, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Pekarik, V.; Gumulec, J.; Masarik, M.; Kizek, R.; Adam, V. Prostate cancer, miRNAs, metallothioneins and resistance to cytostatic drugs. Curr. Med. Chem. 2013, 20, 534–544. [Google Scholar] [PubMed]
- Casanova-Salas, I.; Rubio-Briones, J.; Fernandez-Serra, A.; Lopez-Guerrero, J.A. MiRNAs as biomarkers in prostate cancer. Clin. Transl. Oncol. 2012, 14, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, Q.; Feng, N.H.; Cheng, G.; Guan, Z.L.; Wang, Y.; Qin, C.; Yin, C.J.; Hua, L.X. Mir-205 is frequently downregulated in prostate cancer and acts as a tumor suppressor by inhibiting tumor growth. Asian J. Androl. 2013, 6, 735. [Google Scholar] [CrossRef] [PubMed]
- Hagman, Z.; Haflidadottir, B.S.; Ceder, J.A.; Larne, O.; Bjartell, A.; Lilja, H.; Edsjo, A.; Ceder, Y. Mir-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br. J. Cancer 2013, 108, 1668–1676. [Google Scholar] [CrossRef] [PubMed]
- Gandellini, P.; Profumo, V.; Casamichele, A.; Fenderico, N.; Borrelli, S.; Petrovich, G.; Santilli, G.; Callari, M.; Colecchia, M.; Pozzi, S.; et al. Mir-205 regulates basement membrane deposition in human prostate: Implications for cancer development. Cell Death Differ. 2012, 19, 1750–1760. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, N.; Li, X.; Padi, S.K.; Zhang, Q.; Tang, M.S.; Guo, B. Downregulation of mir-205 and mir-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010, 1, e105. [Google Scholar] [CrossRef] [PubMed]
- Hulf, T.; Sibbritt, T.; Wiklund, E.D.; Patterson, K.; Song, J.Z.; Stirzaker, C.; Qu, W.; Nair, S.; Horvath, L.G.; Armstrong, N.J.; et al. Epigenetic-induced repression of microRNA-205 is associated with med1 activation and a poorer prognosis in localized prostate cancer. Oncogene 2013, 32, 2891–2899. [Google Scholar] [CrossRef] [PubMed]
- Puhr, M.; Hoefer, J.; Schafer, G.; Erb, H.H.; Oh, S.J.; Klocker, H.; Heidegger, I.; Neuwirt, H.; Culig, Z. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of mir-200c and mir-205. Am. J. Pathol. 2012, 181, 2188–2201. [Google Scholar] [CrossRef] [PubMed]
- Gardlik, R.; Palffy, R.; Hodosy, J.; Lukacs, J.; Turna, J.; Celec, P. Vectors and delivery systems in gene therapy. Med. Sci. Monit. 2005, 11, 110–121. [Google Scholar]
- Khatri, N.; Rathi, M.; Baradia, D.; Trehan, S.; Misra, A. In vivo delivery aspects of miRNA, shrna and siRNA. Crit. Rev. Ther. Drug Carrier Syst. 2012, 29, 487–527. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Schwind, S.; Yu, B.; Santhanam, R.; Wang, H.; Hoellerbauer, P.; Mims, A.; Klisovic, R.; Walker, A.R.; Chan, K.K.; et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: A novel therapeutic strategy in acute myeloid leukemia. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 2355–2367. [Google Scholar] [CrossRef] [PubMed]
- Hobel, S.; Koburger, I.; John, M.; Czubayko, F.; Hadwiger, P.; Vornlocher, H.P.; Aigner, A. Polyethylenimine/small interfering RNA-mediated knockdown of vascular endothelial growth factor in vivo exerts anti-tumor effects synergistically with bevacizumab. J. Gene. Med. 2010, 12, 287–300. [Google Scholar] [PubMed]
- Chen, Y.; Zhu, X.; Zhang, X.; Liu, B.; Huang, L. Nanoparticles modified with tumor-targeting scfv deliver siRNAand miRNA for cancer therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 1650–1656. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.F.; Weirauch, U.; Thomas, M.; Grunweller, A.; Hartmann, R.K.; Aigner, A. MicroRNA replacement therapy for mir-145 and mir-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011, 71, 5214–5224. [Google Scholar] [CrossRef] [PubMed]
- JP, B. The proton sponge: A trick to enter cells the viruses did not exploit. Chimia. 1997, 51, 34–36. [Google Scholar]
- Yallapu, M.M.; Ebeling, M.C.; Khan, S.; Sundram, V.; Chauhan, N.; Gupta, B.K.; Puumala, S.E.; Jaggi, M.; Chauhan, S.C. Novel curcumin-loaded magnetic nanoparticles for pancreatic cancer treatment. Mol. Cancer Ther. 2013, 12, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Yallapu, M.M.; Othman, S.F.; Curtis, E.T.; Gupta, B.K.; Jaggi, M.; Chauhan, S.C. Multi-functional magnetic nanoparticles for magnetic resonance imaging and cancer therapy. Biomater. 2011, 32, 1890–1905. [Google Scholar] [CrossRef] [PubMed]
- Setua, S.; Khan, S.; Yallapu, M.M.; Behrman, S.W.; Sikander, M.; Khan, S.S.; Jaggi, M.; Chauhan, S.C. Restitution of tumor suppressor microRNA-145 using magnetic nanoformulation for pancreatic cancer therapy. J. Gastrointest. Surg. 2017, 21, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Diener, Y.; Walenda, T.; Jost, E.; Brümmendorf, T.H.; Bosio, A.; Wagner, W.; Bissels, U. MicroRNA expression profiles of serum from patients before and after chemotherapy. Genom. Data 2015, 6, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Nordby, Y.; Richardsen, E.; Ness, N.; Donnem, T.; Patel, H.R.H.; Busund, L.-T.; Bremnes, R.M.; Andersen, S. High mir-205 expression in normal epithelium is associated with biochemical failure—An argument for epithelial crosstalk in prostate cancer? Sci. Rep. 2017, 7, 16308. [Google Scholar] [CrossRef] [PubMed]
- Stankevicins, L.; Barat, A.; Dessen, P.; Vassetzky, Y.; de Moura Gallo, C.V. The microRNA-205-5p is correlated to metastatic potential of 21t series: A breast cancer progression model. PLoS ONE 2017, 12, e0173756. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Vu, T.; Wang, J.; Mesmar, F.; Mukhopadhyay, S.; Saxena, A.; McCollum, C.W.; Gustafsson, J.A.; Bondesson, M.; Williams, C. Estrogen receptor beta reduces colon cancer metastasis through a novel mir-205-prox1 mechanism. Oncotarget 2016, 7, 42159–42171. [Google Scholar] [CrossRef] [PubMed]
- Dieli-Conwright, C.M.; Lee, K.; Kiwata, J.L. Reducing the risk of breast cancer recurrence: An evaluation of the effects and mechanisms of diet and exercise. Curr. Breast Cancer Rep. 2016, 8, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Sinibaldi, V.J. Docetaxel treatment in the elderly patient with hormone refractory prostate cancer. Clin. Interv. Aging 2007, 2, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Gandellini, P.; Giannoni, E.; Casamichele, A.; Taddei, M.L.; Callari, M.; Piovan, C.; Valdagni, R.; Pierotti, M.A.; Zaffaroni, N.; Chiarugi, P. Mir-205 hinders the malignant interplay between prostate cancer cells and associated fibroblasts. Antioxid. Redox. Signal 2014, 20, 1045–1059. [Google Scholar] [CrossRef] [PubMed]
- Gandellini, P.; Folini, M.; Longoni, N.; Pennati, M.; Binda, M.; Colecchia, M.; Salvioni, R.; Supino, R.; Moretti, R.; Limonta, P.; et al. Mir-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase cepsilon. Cancer Res. 2009, 69, 2287–2295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sun, Y.; Ma, L. Zeb1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle 2015, 14, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Forster, S.; Thumser, A.E.; Hood, S.R.; Plant, N. Characterization of rhodamine-123 as a tracer dye for use in in vitro drug transport assays. PLoS ONE 2012, 7, e33253. [Google Scholar] [CrossRef] [PubMed]
- Puente, J.; Grande, E.; Medina, A.; Maroto, P.; Lainez, N.; Arranz, J.A. Docetaxel in prostate cancer: A familiar face as the new standard in a hormone-sensitive setting. Ther. Adv. Med. Oncol. 2017, 9, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. MicroRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [PubMed]
- DeCola, A.; Volpe, S.; Budani, M.C.; Ferracin, M.; Lattanzio, R.; Turdo, A.; D’Agostino, D.; Capone, E.; Stassi, G.; Todaro, M.; et al. Mir-205-5p-mediated downregulation of erbb/her receptors in breast cancer stem cells results in targeted therapy resistance. Cell Death Dis. 2015, 6, e1823. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Qiu, Y.; Yague, E.; Ji, W.; Liu, J.; Zhang, J. MiRNA-205 targets vegfa and fgf2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. 2016, 7, e2291. [Google Scholar] [CrossRef] [PubMed]
- Nagesh, P.K.; Chowdhury, P.; Boya, V.N.; Kashyap, V.K.; Khan, S.; Hafeez, B.B.; Zafar, N.; Behrman, S.W.; Chauhan, S.C.; Jaggi, M.; et al. Abstract 1206: Mir-205 replenishment in prostate cancer cells: A novel nanoparticle approach. Cancer Res. 2017, 77, 1206. [Google Scholar] [CrossRef]
- Verdoodt, B.; Neid, M.; Vogt, M.; Kuhn, V.; Liffers, S.T.; Palisaar, R.J.; Noldus, J.; Tannapfel, A.; Mirmohammadsadegh, A. MicroRNA-205, a novel regulator of the anti-apoptotic protein bcl2, is downregulated in prostate cancer. Int. J. Oncol. 2013, 43, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Tucci, P.; Agostini, M.; Grespi, F.; Markert, E.K.; Terrinoni, A.; Vousden, K.H.; Muller, P.A.; Dotsch, V.; Kehrloesser, S.; Sayan, B.S.; et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 15312–15317. [Google Scholar] [CrossRef] [PubMed]
- Boll, K.; Reiche, K.; Kasack, K.; Morbt, N.; Kretzschmar, A.K.; Tomm, J.M.; Verhaegh, G.; Schalken, J.; von Bergen, M.; Horn, F.; et al. Mir-130a, mir-203 and mir-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene 2013, 32, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, D.Y.; Huang, L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 2015, 81, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Li, X.; Li, Y.; Che, J.; Li, X.; Zhao, X.; Chen, Y.; Zheng, X.; Yuan, W. Biodegradable and biocompatible cationic polymer delivering microRNA-221/222 promotes nerve regeneration after sciatic nerve crush. Int. J. Nanomed. 2017, 12, 4195–4208. [Google Scholar] [CrossRef] [PubMed]
- Nagesh, P.K.B.; Johnson, N.R.; Boya, V.K.N.; Chowdhury, P.; Othman, S.F.; Khalilzad-Sharghi, V.; Hafeez, B.B.; Ganju, A.; Khan, S.; Behrman, S.W.; et al. Psma targeted docetaxel-loaded superparamagnetic iron oxide nanoparticles for prostate cancer. Colloids Surf. B Biointerfaces 2016, 144, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Kalita, H.; Prashanth Kumar, B.N.; Konar, S.; Tantubay, S.; Kr. Mahto, M.; Mandal, M.; Pathak, A. Sonochemically synthesized biocompatible zirconium phosphate nanoparticles for ph sensitive drug delivery application. Mater. Sci. Eng. C 2016, 60, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Puvvada, N.; Rajput, S.; Kumar, B.N.P.; Mandal, M.; Pathak, A. Exploring the fluorescence switching phenomenon of curcumin encapsulated niosomes: In vitro real time monitoring of curcumin release to cancer cells. RSC Adv. 2013, 3, 2553–2557. [Google Scholar] [CrossRef]
- Boya, V.N.; Lovett, R.; Setua, S.; Gandhi, V.; Nagesh, P.K.B.; Khan, S.; Jaggi, M.; Yallapu, M.M.; Chauhan, S.C. Probing mucin interaction behavior of magnetic nanoparticles. J. Colloid Interface Sci. 2017, 488, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Yallapu, M.M.; Othman, S.F.; Curtis, E.T.; Bauer, N.A.; Chauhan, N.; Kumar, D.; Jaggi, M.; Chauhan, S.C. Curcumin-loaded magnetic nanoparticles for breast cancer therapeutics and imaging applications. Int. J. Nanomed. 2012, 7, 1761–1779. [Google Scholar]
- Liang, G.F.; Zhu, Y.L.; Sun, B.; Hu, F.H.; Tian, T.; Li, S.C.; Xiao, Z.D. Plga-based gene delivering nanoparticle enhance suppression effect of miRNA in hepg2 cells. Nanoscale Res. Lett. 2011, 6, 447. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Mondal, G.; Slavik, P.; Rachagani, S.; Batra, S.K.; Mahato, R.I. Codelivery of small molecule hedgehog inhibitor and miRNA for treating pancreatic cancer. Mol. Pharm. 2015, 12, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Bülbül, G.; Hayat, A.; Mustafa, F.; Andreescu, S. DNA assay based on nanoceria as fluorescence quenchers (nanoceracq DNA assay). Sci. Rep. 2018, 8, 2426. [Google Scholar] [CrossRef] [PubMed]
- Karthik, S.; Prashanth Kumar, B.N.; Gangopadhyay, M.; Mandal, M.; Singh, N.D.P. A targeted, image-guided and dually locked photoresponsive drug delivery system. J. Mater. Chem. B 2015, 3, 728–732. [Google Scholar] [CrossRef]
- Khan, S.; Ebeling, M.C.; Zaman, M.S.; Sikander, M.; Yallapu, M.M.; Chauhan, N.; Yacoubian, A.M.; Behrman, S.W.; Zafar, N.; Kumar, D.; et al. MicroRNA-145 targets muc13 and suppresses growth and invasion of pancreatic cancer. Oncotarget 2014, 5, 7599–7609. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, P.; Nagesh, P.K.B.; Khan, S.; Hafeez, B.B.; Chauhan, S.C.; Jaggi, M.; Yallapu, M.M. Development of polyvinylpyrrolidone/paclitaxel self-assemblies for breast cancer. Acta Pharm. Sin. B 2017, 8, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.N.; Rajput, S.; Dey, K.K.; Parekh, A.; Das, S.; Mazumdar, A.; Mandal, M. Celecoxib alleviates tamoxifen-instigated angiogenic effects by ros-dependent vegf/vegfr2 autocrine signaling. BMC Cancer 2013, 13, 273. [Google Scholar] [CrossRef] [PubMed]
- Hafeez, B.B.; Ganju, A.; Sikander, M.; Kashyap, V.K.; Hafeez, Z.B.; Chauhan, N.; Malik, S.; Massey, A.E.; Tripathi, M.K.; Halaweish, F.T.; et al. Ormeloxifene suppresses prostate tumor growth and metastatic phenotypes via inhibition of oncogenic beta-catenin signaling and emt progression. Mol. Cancer Ther. 2017, 16, 2267–2280. [Google Scholar] [CrossRef] [PubMed]
- Nagesh, P.; Hatami, E.; Chowdhury, P.; Kashyap, V.; Khan, S.; Hafeez, B.; Chauhan, S.; Jaggi, M.; Yallapu, M. Tannic acid induces endoplasmic reticulum stress-mediated apoptosis in prostate cancer. Cancers 2018, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Rajput, S.; Kumar, B.N.; Banik, P.; Parida, S.; Mandal, M. Thymoquinone restores radiation-induced tgf-beta expression and abrogates emt in chemoradiotherapy of breast cancer cells. J. Cell Physiol. 2015, 230, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Petriz, J.; Garcia-Lopez, J. Flow cytometric analysis of p-glycoprotein function using rhodamine 123. Leukemia 1997, 11, 1124–1130. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagesh, P.K.B.; Chowdhury, P.; Hatami, E.; Boya, V.K.N.; Kashyap, V.K.; Khan, S.; Hafeez, B.B.; Chauhan, S.C.; Jaggi, M.; Yallapu, M.M. miRNA-205 Nanoformulation Sensitizes Prostate Cancer Cells to Chemotherapy. Cancers 2018, 10, 289. https://doi.org/10.3390/cancers10090289
Nagesh PKB, Chowdhury P, Hatami E, Boya VKN, Kashyap VK, Khan S, Hafeez BB, Chauhan SC, Jaggi M, Yallapu MM. miRNA-205 Nanoformulation Sensitizes Prostate Cancer Cells to Chemotherapy. Cancers. 2018; 10(9):289. https://doi.org/10.3390/cancers10090289
Chicago/Turabian StyleNagesh, Prashanth K. B., Pallabita Chowdhury, Elham Hatami, Vijaya K. N. Boya, Vivek K. Kashyap, Sheema Khan, Bilal B. Hafeez, Subhash C. Chauhan, Meena Jaggi, and Murali M. Yallapu. 2018. "miRNA-205 Nanoformulation Sensitizes Prostate Cancer Cells to Chemotherapy" Cancers 10, no. 9: 289. https://doi.org/10.3390/cancers10090289
APA StyleNagesh, P. K. B., Chowdhury, P., Hatami, E., Boya, V. K. N., Kashyap, V. K., Khan, S., Hafeez, B. B., Chauhan, S. C., Jaggi, M., & Yallapu, M. M. (2018). miRNA-205 Nanoformulation Sensitizes Prostate Cancer Cells to Chemotherapy. Cancers, 10(9), 289. https://doi.org/10.3390/cancers10090289