Dysregulation of Nrf2 in Hepatocellular Carcinoma: Role in Cancer Progression and Chemoresistance
Abstract
1. Introduction
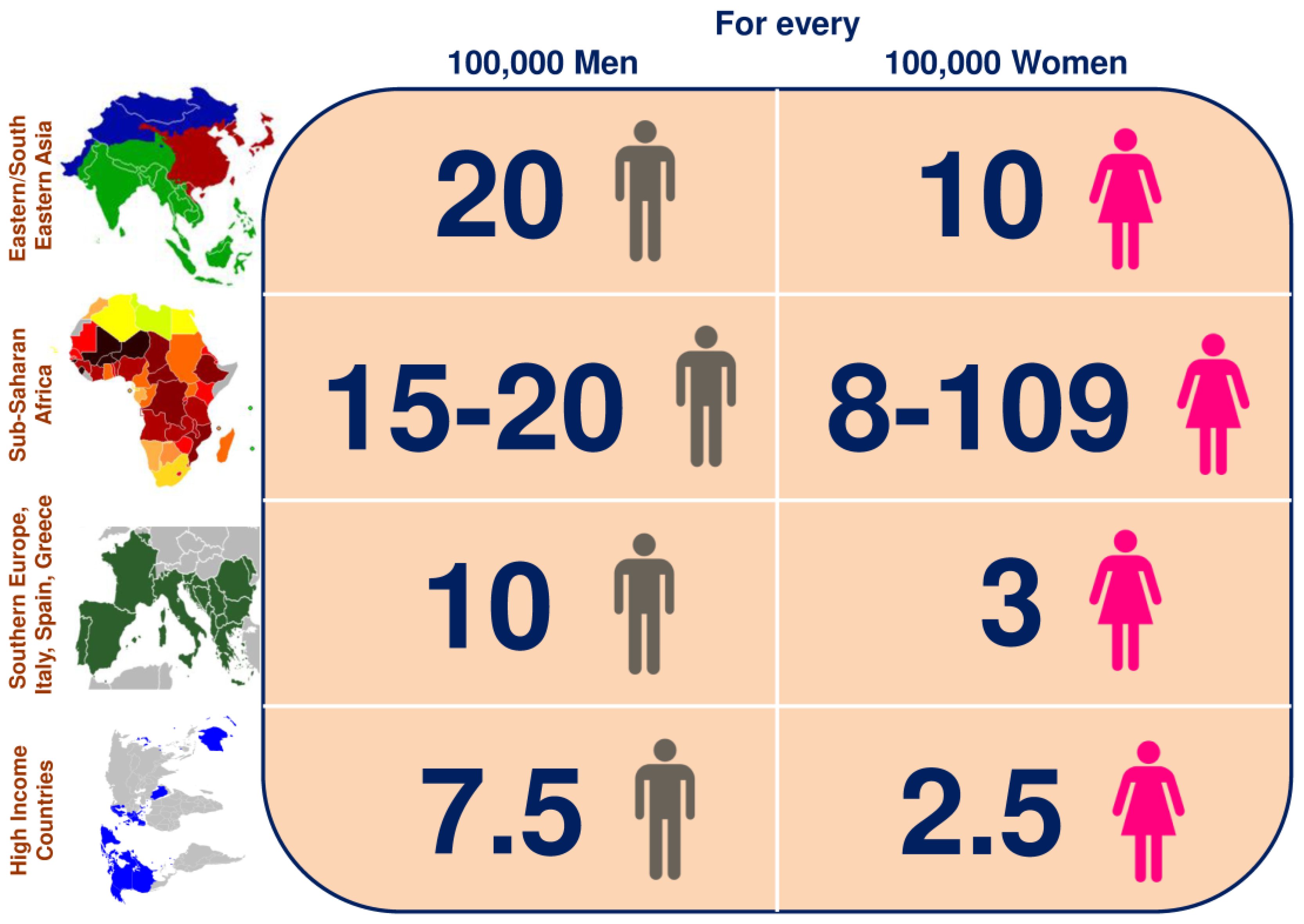
2. Epidemiology of HCC
3. Molecular Pathogenesis of HCC
4. Oxidative Stress and Inflammation in HCC
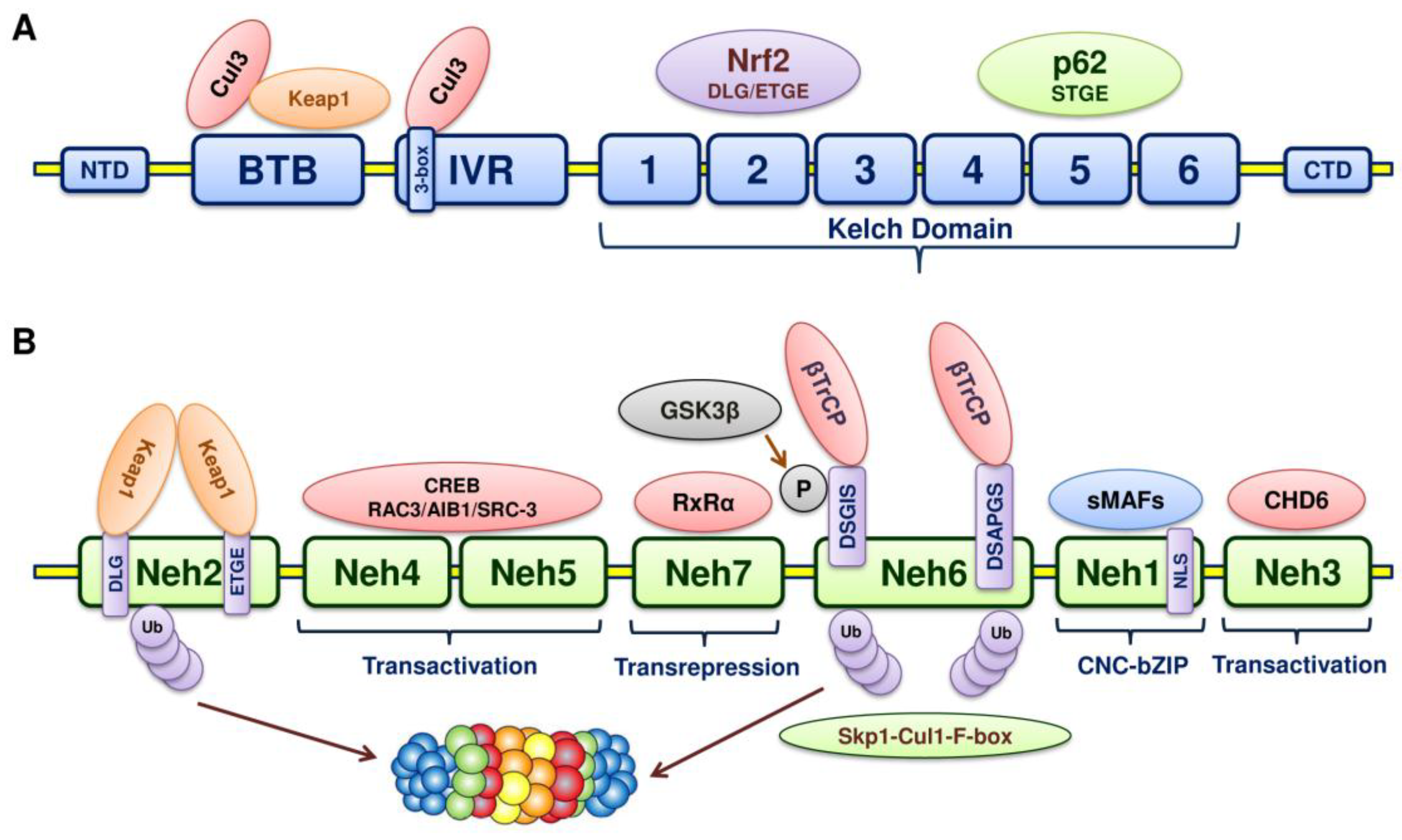
5. Structure of Nrf2
6. Structure of Keap1
7. Keap1-Nrf2-ARE Pathway
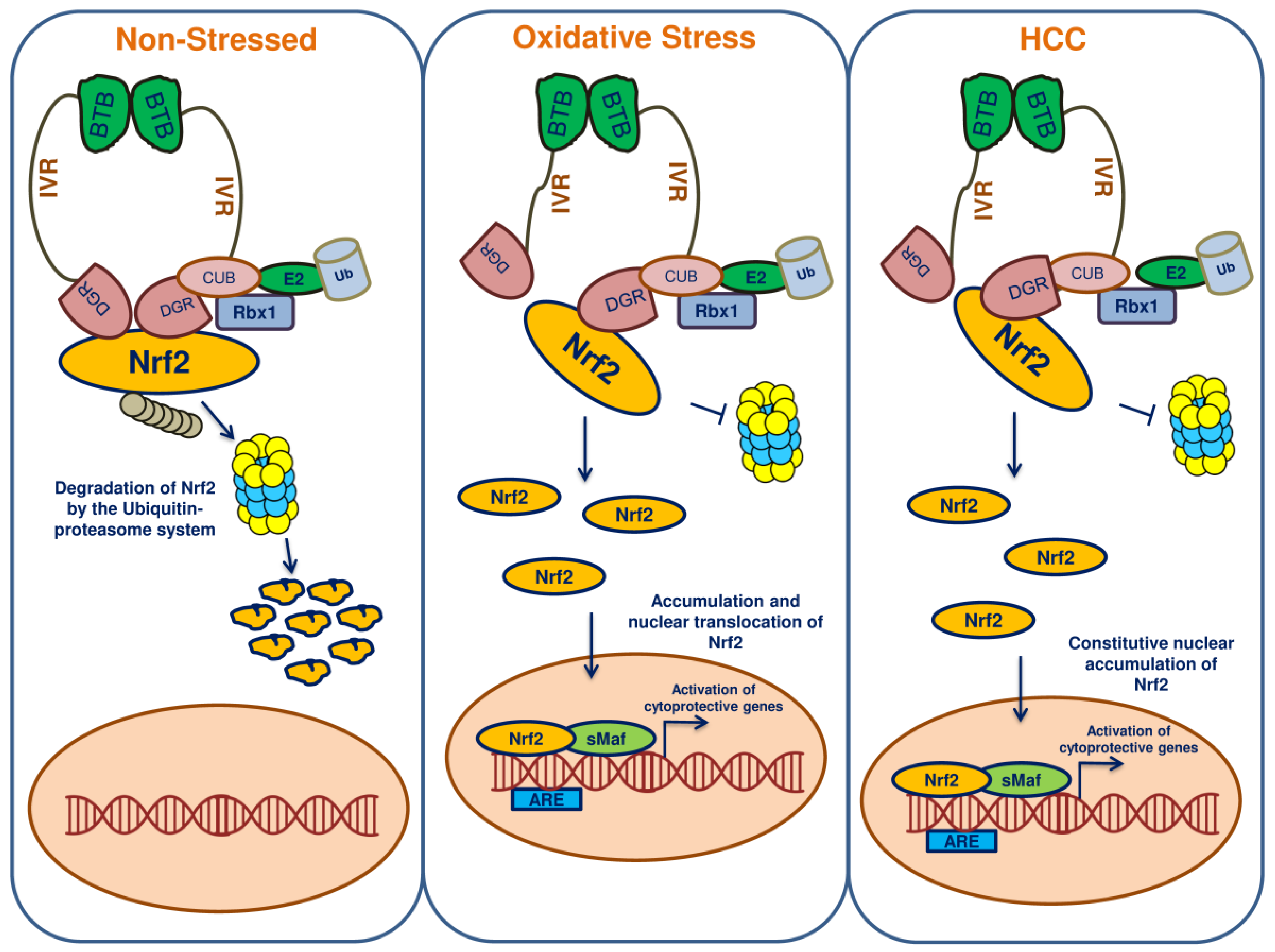
8. Nrf2 and Keap1 Mutations Lead to HCC
9. Nrf2 in HCC
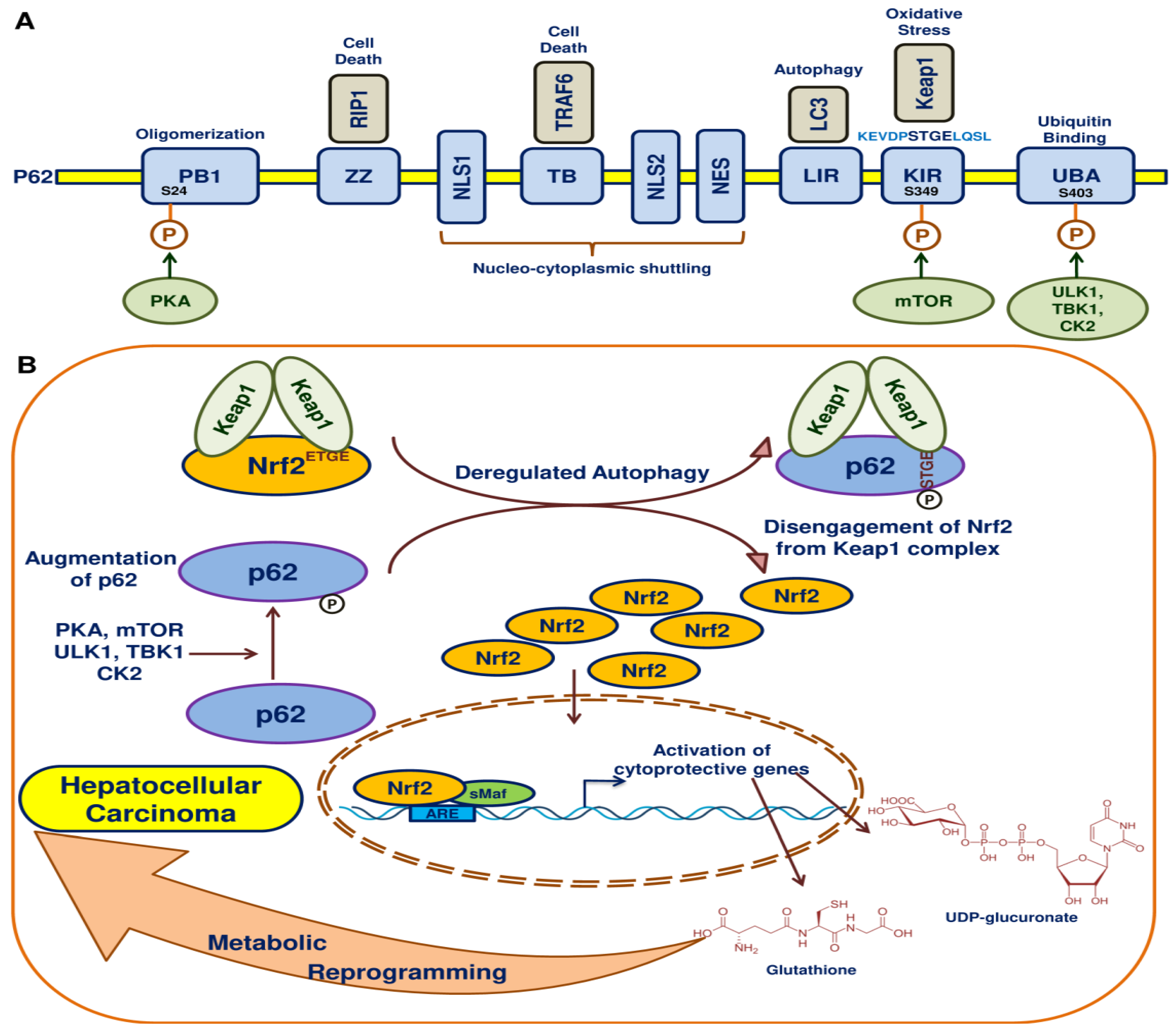
10. Autophagy, p62 and Nrf2 in HCC
11. Phytochemicals/Molecules Can Elicit Activation of Nrf2 in HCC
12. Phytochemicals/Molecules Sensitizing Resistant HCC through Nrf2 Signalling
13. Endoplasmic Reticulum Stress and Nrf2 Signalling in HCC
14. Metal Complexes and Nrf2 in HCC
15. miRs Regulation/Dysregulation of Nrf2 in HCC
16. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abebcassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Takaki, A.; Yamamoto, K. Control of oxidative stress in hepatocellular carcinoma: Helpful or harmful? World J. Hepatol. 2015, 7, 968–979. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Motohashi, H.; Yamamoto, M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells 2011, 16, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Manda, G.; Hassan, A.; Alcaraz, M.J.; Barbas, C.; Diaber, A.; Ghezzi, P.; Leon, R.; Lopez, M.G.; Oliva, B.; et al. Transcription factor NRF2 as a therapeutic target for chronic diseases: A systems medicine approach. Pharmacol. Rev. 2018, 70, 348–383. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Bosetti, C.; Turati, F.; La Vecchia, C. Hepatocellular carcinoma epidemiology. Best Pract. Res. Clin. Gastroenterol. 2014, 28, 753–770. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Cancer Facts and Figures; American Cancer Society Inc.: Atlanta, GA, USA, 2018. [Google Scholar]
- Ozakyol, A. Global epidemiology of hepatocellular carcinoma (HCC epidemiology). J. Gastrointest. Cancer 2017, 48, 238–240. [Google Scholar] [CrossRef]
- Tang, A.; Hallouch, O.; Chernyak, V.; Kamaya, A.; Sirlin, C.B. Epidemiology of hepatocellular carcinoma: Target population for surveillance and diagnosis. Abdom. Radiol. 2018, 43, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Zucman-Rossi, J.; Villanueva, A.; Nault, J.C.; Nault, J.C.; Llovet, J.M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015, 149, 1226–1239. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, K.A.; Petrick, J.L.; London, W.T. Global epidemiology of hepatocellular carcinoma: An emphasis on demographic and regional variability. Clin. Liver Dis. 2015, 19, 223–238. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Hepatocellular carcinoma. N. Engl. J. Med. 2011, 365, 1118–1127. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; El-Serag, H.B. Epidemiology of hepatocellular carcinoma: Consider the population. J. Clin. Gastroenterol. 2013, 47, S2–S6. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; El-Serag, H.B.; Sada, Y.H.; Kanwal, F.; Duan, Z.; Temple, S.; May, S.B.; Kramer, J.R.; Richardson, P.A.; Davila, J.A. Hepatocellular carcinoma in the absence of cirrhosis in united states veterans is associated with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2016, 14, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Han, Y.; Xu, C.; Xiao, T.; Wang, B. Effect of type 2 diabetes mellitus on the risk for hepatocellular carcinoma in chronic liver diseases: A meta-analysis of cohort studies. Eur. J. Cancer Prev. 2015, 24, 89–99. [Google Scholar] [CrossRef]
- Lee, Y.C.; Cohet, C.; Yang, Y.C.; Stayner, L.; Hashibe, M.; Straif, K. Meta-analysis of epidemiologic studies on cigarette smoking and liver cancer. Int. J. Epidemiol. 2009, 38, 1497–1511. [Google Scholar] [CrossRef]
- Nishida, N.; Nishimura, T.; Nagasaka, T.; Ikai, I.; Goael, A.; Boland, C.R. Extensive methylation is asscociated with beta-catenin mutations in hepatocellular carcinoma: Evidence for two distinct pathways of human hepatocarcinogenesis. Cancer Res. 2007, 67, 4586–4594. [Google Scholar] [CrossRef]
- Whittaker, S.; Marais, R.; Zhu, A.X. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 2010, 29, 4989–5005. [Google Scholar] [CrossRef]
- Calado, R.T.; Brudno, J.; Mehta, P.; Kovacs, J.J.; Zago, M.A.; Chanock, S.J.; Boyer, T.D.; Young, N.S. Constitutional telomerase mutations are genetis risk factors for cirrhosis. Hepatology 2011, 53, 1600–1607. [Google Scholar] [CrossRef]
- Cleary, S.P.; Jeck, W.R.; Zhao, X.; Chen, K.; Selitsky, S.R.; Savich, G.L.; Tan, T.X.; Wu, M.C.; Getz, G.; Lawrence, M.S.; et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013, 58, 1693–1702. [Google Scholar] [CrossRef]
- Chen, Y.W.; Klimstra, D.S.; Mongeau, M.E.; Tatem, J.L.; Boyartchuk, V.; Lewis, B.C. Loss of p53 and Ink4a/Arf cooperate in a cell autonomous fashion to induce metastasis of hepatocellular carcinoma cells. Cancer Res. 2007, 67, 7589–7596. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.B.; Nagae, G.; Midorikawa, Y.; Yagi, K.; Tsutsumi, S.; Yamamoto, S.; Hasegawa, K.; Kokudo, N.; Aburatani, H.; Kaneda, A. Identification of genes preferentially methylated in hepatitis C virus-related hepatocellular carcinoma. Cancer Sci. 2010, 101, 1501–1510. [Google Scholar] [CrossRef]
- Tischoff, I.; Tannapfe, A. DNA methylation in hepatocellular carcinoma. World J. Gastroenterol. 2008, 14, 1741–1748. [Google Scholar] [CrossRef] [PubMed]
- EI-Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- Moon, R.T.; Bowerman, B.; Boutros, M.; Perrimon, N. The promise and perils of Wnt signaling through beta-catenin. Science 2002, 296, 1644–1646. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A.; Newell, P.; Chiang, D.Y.; Friedman, S.L.; Llovet, J.M. Genomics and signaling pathways in hepatocellular carcinoma. Semin. Liver Dis. 2007, 27, 55–76. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.B.; Wakefield, L.M. The two faces of transforming growth factor beta in carcinogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 8621–8623. [Google Scholar] [CrossRef] [PubMed]
- Calvisi, D.F.; Ladu, S.; Gorden, A.; Farina, M.; Conner, EA.; Lee, J.S.; Factor, V.M.; Thorgeirsson, S.S. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 2006, 130, 117–1128. [Google Scholar] [CrossRef] [PubMed]
- Burdette, D.; Olivarez, M.; Waris, G. Activation of transcription factor Nrf2 by hepatitis C virus induces the cell survivial pathway. J. Gen. Virol. 2010, 91, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Schaedler, S.; Krause, J.; Himmelsbach, K.; Carvajal-Yepes, M.; Lieder, F.; Klingel, K.; Nassal, M.; Weiss, T.S.; Werner, S.; Hildt, E. Hepatitis B virus induces expression of antioxidant response element regulated genes by activation of Nrf2. J. Biol. Chem. 2010, 285, 41074–41086. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Cederbaum, A.I. Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatology 2006, 43, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.T.; Arjumand, W.; Nafees, S.; Seth, A.; Ali, N.; Rashid, S.; Sultana, S. Hesperidin alleviates acetaminophen induced toxicity in wistar rats by abrogation of oxidative stress, apoptosis and inflammation. Toxicol. Lett. 2012, 208, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role of STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.K.; Itoh, K.; Yamamoto, M.; Sutter, T.R.; Kensler, T.W. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol. Med. 2001, 7, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, C.; Zhang, L.; Yang, Q.; Zhou, S.; Wen, Q.; Wang, J. Nrf2 is a potential prognostic marker and promotes proliferation and invasion in human hepatocellular carcinoma. BMC Cancer 2015, 15, 531. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Yepes, M.; Himmelsbach, K.; Schaedler, S.; Ploen, D.; Krause, J.; Ludwig, L.; Weiss, T.; Klingel, K.; Hildt, E. Hepatitis C virus impairs the induction of cytoprotective Nrf2 target genes by delocalization of small Maf proteins. J. Biol. Chem. 2011, 286, 8941–8951. [Google Scholar] [CrossRef] [PubMed]
- Higgs, M.R.; Chouteau, P.; Lerat, H. Liver let die: Oxidative DNA damage and hepatotropic viruses. J. Gen. Virol. 2014, 95, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Tsai, W.L.; Shao, R.X.; Wu, G.; Peng, L.F.; Barlow, L.L.; Chung, W.J.; Zhang, L.; Zhao, H.; Jang, J.Y.; et al. Hepatitis C virus regulates transforming growth factor beta 1 production through the generation of reactive oxygen species in a nuclear factor kappaB-dependent manner. Gasteroenterology 2010, 138, 2509–2518. [Google Scholar] [CrossRef]
- Malik, A.N.; Czajka, A. Is mitochondria DNA content a potential biomarker of mitochondrial dysfunction. Mitochondrion 2013, 13, 481–492. [Google Scholar] [CrossRef]
- Lambert, M.P.; Paliwal, A.; Vaissiere, T.; Chemin, I.; Zoulum, F.; Tommasino, M.; Hainaut, P.; Sylla, B.; Scoazec, J.Y.; Tost, J.; et al. Aberrant DNA methylation distinguishes hepatocellular carcinoma associated with HBV and HCV infection and alcohol intake. J. Hepatol. 2011, 54, 705–715. [Google Scholar] [CrossRef]
- Hosel, M.; Quasdorff, M.; Wiegmann, K.; Webb, D.; Broxtermann, M.; Tedjokusumo, R.; Esser, K.; Arzberger, S.; Kirschning, C.J.; Langenkamp, A.; et al. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology 2009, 50, 1773–1782. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Harry, B.L.; Zhou, Q.; Skeen-Gaar, R.R.; Ge, X.; Lee, E.S.; Mitani, S.; Xue, D. Hepatitis B virus X protein targets the Bcl-2 protein CED-9 to induce intracellular Ca2+ increase and cell death in caenorhabditid elegans. Proc. Natl. Acad. Sci. USA 2012, 109, 18465–18470. [Google Scholar] [CrossRef] [PubMed]
- Clippinger, A.J.; Bouchard, M.J. Hepatitis B virus HBx protein localizes to mitochondria in primary rat hepatocytes and modulates mitochondrial membrane potential. J. Virol. 2008, 82, 6798–6811. [Google Scholar] [CrossRef] [PubMed]
- Tordjmann, T.; Soulie, A.; Guettier, C.; Schmidt, M.; Berthou, C.; Beaugrand, M.; Sasportes, M. Perforin and granzyme B lytic protein expression during chronic viral and autoimmune hepatitis. Liver 1998, 18, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Arzumanyan, A.; Reis, H.M.; Feitelson, M.A. Pathogenic mechanisms in HBV- and HCV associated hepatocellular carcinoma. Nat. Rev. Cancer 2013, 13, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Schwarzenbach, H.; Pantel, K. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 2012, 63, 199–215. [Google Scholar] [CrossRef]
- Ng, C.K.Y.; Di Costanzo, G.G.; Terracciano, L.M.; Piscuoglio, S. Circulating cell-free DNA in hepatocellular carcinoma: Current insights and outlook. Front. Med. 2018, 26, 78. [Google Scholar] [CrossRef]
- Gailhouste, L.; Gomez-Santos, L.; Ochiya, T. Potential applications of miRNAs as diagnostic and prognostic markers in liver cancer. Front. Biosci. 2013, 18, 199–223. [Google Scholar]
- Cardin, R.; Picocchi, M.; Sinigaglia, A.; Lavezzo, E.; Bortolami, M.; Kotsafti, A.; Cillo, U.; Zanus, G.; Mescoli, C.; Rugge, M.; et al. Oxidative DNA damage correlates with cell immortalization and mir-92 expression in hepatocellular carcinoma. BMC Cancer 2012, 12, 177. [Google Scholar] [CrossRef]
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Igarashi, K.; Hayashi, N.; Nishizawa, M.; Yamamoto, M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell Biol. 1995, 15, 4184–4193. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, H.; Katsuoka, F.; Engel, J.D.; Yamamoto, M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc. Natl. Acad. Sci. USA 2004, 27, 6379–6384. [Google Scholar] [CrossRef] [PubMed]
- Plafker, K.S.; Nguyen, L.; Barneche, M.; Mirza, S.; Crawford, D.; Plafker, S.M. The ubiquitin-conjugating enzyme UbcM2 can regulate the stability and activity of the antioxidant transcription factor Nrf2. J. Biol. Chem. 2010, 285, 23064–23074. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Katoh, Y.; Kusunoki, H.; Itoh, K.; Tanaka, T.; Yamamoto, M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: Characterization of the two-site molecular recognition model. Mol. Cell Biol. 2006, 26, 2887–2900. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Itoh, K.; Yoshida, E.; Miyagishi, M.; Fukamizu, A.; Yamamoto, M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 2001, 6, 857–868. [Google Scholar] [CrossRef]
- Nioi, P.; Nguyen, T.; Sherratt, P.J.; Pickett, C.B. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol. Cell Biol. 2005, 25, 10895–10906. [Google Scholar] [CrossRef]
- Kim, J.H.; Yu, S.; Chen, J.D.; Kong, A.N. The nuclear cofactor RAC3/AIB1/SRC-3 enhances Nrf2 signaling by interacting with transactivation domains. Oncogene 2013, 32, 514–527. [Google Scholar] [CrossRef]
- Chowdhry, S.; Zhang, Y.; McMahon, M.; Sutherland, C.; Cuadrado, A.; Hayes, J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013, 32, 3765–3781. [Google Scholar] [CrossRef]
- Rada, P.; Rojo, A.I.; Chowdhry, S.; McMahon, M.; Hayes, J.D.; Cuadrado, A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell Biol. 2011, 31, 1121–1133. [Google Scholar] [CrossRef]
- Rada, P.; Rojo, A.I.; Evrard-Todeschi, N.; Innamorato, N.G.; Cotte, A.; Jaworski, T.; Tobon-Velasco, J.C.; Devijver, H.; Garcia-Mayoral, M.F.; van Leuven, F.; et al. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/β-TrCP axis. Mol. Cell Biol. 2012, 32, 3486–3499. [Google Scholar] [CrossRef]
- Cuadrado, A. Structural and functional characterization of Nrf2 degradation by glycogen ynthase kinase 3/β-TrCP. Free Radic. Biol. Med. 2015, 88, 147–157. [Google Scholar] [CrossRef]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004, 279, 31556–31567. [Google Scholar] [CrossRef]
- Wang, H.; Liu, K.; Geng, M.; Gao, P.; Wu, X.; Hai, Y.; Li, Y.; Li, Y.; Luo, L.; Hayes, J.D.; et al. RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013, 73, 3097–3108. [Google Scholar] [CrossRef] [PubMed]
- Cullinan, S.B.; Gordan, J.D.; Jin, J.; Harper, J.W.; Diehl, J.A. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell Biol. 2004, 24, 8477–8486. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Kostov, R.V.; Canning, P. Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants. Arch. Biochem. Biophys. 2017, 617, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Bardwell, V.J.; Treisman, R. The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 1994, 8, 1664–1677. [Google Scholar] [CrossRef] [PubMed]
- Zipper, L.M.; Mulcahy, R.T. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 2002, 277, 36544–36552. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohstuji, J.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef] [PubMed]
- Canning, P.; Cooper, C.D.; Kroler, T.; Murray, J.W.; Pike, A.C.; Chaikuad, A.; Keates, T.; Thangaratnarajah, C.; Hoizan, V.; Ayinampudi, V.; et al. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J. Biol. Chem. 2013, 288, 7803–7814. [Google Scholar] [CrossRef]
- Li, X.; Zhang, D.; Hannink, M.; Beamer, L.J. Crystal structure of the Kelch domain of human Keap1. J. Biol. Chem. 2004, 279, 54750–54758. [Google Scholar] [CrossRef]
- Padmanabhan, B.; Tong, K.I.; Ohta, T.; Nakamura, Y.; Scharlock, M.; Ohstuji, M.; Kang, M.I.; Kobayashi, A.; Yokoyama, S.; Yamamoto, M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006, 21, 689–700. [Google Scholar] [CrossRef]
- Adams, J.; Kelso, R.; Cooley, L. The kelch repeat superfamily of proteins: Propellers of cell function. Trends Cell Biol. 2000, 10, 17–24. [Google Scholar] [CrossRef]
- Tong, K.I.; Kobayahi, A.; Katsuoka, F.; Yamamoto, M. Two-site substrate recognition model for the Keap1-Nrf2 system: A hinge and latch mechanism. Biol. Chem. 2006, 387, 1311–1320. [Google Scholar] [CrossRef]
- Lo, S.C.; Li, X.; Henzi, M.T.; Beamer, L.J.; Hannink, M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006, 25, 3605–3617. [Google Scholar] [CrossRef]
- Fukutomi, T.; Takagi, K.; Mizushima, T.; Ohuchi, N.; Yamamoto, M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell Biol. 2014, 34, 832–846. [Google Scholar] [CrossRef]
- Hayes, J.D.; McMahon, M. NRF2 and KEAP1 mutations: Permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 2009, 34, 176–188. [Google Scholar] [CrossRef]
- Hast, B.E.; Cloer, E.W.; Goldfarb, D.; Li, H.; Siesser, P.F.; Yan, F.; Walter, V.; Zheng, N.; Hayes, D.N.; Major, M.B. Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination. Cancer Res. 2014, 74, 808–817. [Google Scholar] [CrossRef]
- Eades, G.; Yang, M.; Yao, Y.; Zhang, Y.; Zhou, Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J. Biol. Chem. 2011, 286, 40725–40733. [Google Scholar] [CrossRef]
- Van Jaarsveld, M.T.; Helleman, J.; Boersma, A.W.; van Kujik, P.F.; van Ijcken, W.F.; Despierre, E.; Vergote, I.; Mathijssen, R.H.; Berns, E.M.; Verweij, J.; et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene 2013, 32, 4284–4293. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Wu, B.; Yan, J.; Li, X.; Sun, H.; Zhou, D. A possible gene silencing mechanism: Hypermethylation of the Keap1 promoter abrogates binding of the transcription factor Sp1 in lung cancer cells. Biochem. Biophys. Res. Commun. 2012, 428, 80–85. [Google Scholar] [CrossRef]
- Jeong, Y.; Hoang, N.T.; Lovejoy, A.; Stehr, H.; Newman, A.M.; Gentles, A.J.; Kong, W.; Truong, D.; Martin, S.; Chaudhuri, A.; et al. Role of KEAP1/NRF2 and TP53 mutations in lung squamus cell carcinoma development and radiation resistance. Cancer Discov. 2017, 7, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, H.; Yamamoto, M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 2004, 10, 549–557. [Google Scholar] [CrossRef]
- Zhang, D.D. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. 2006, 38, 769–789. [Google Scholar] [CrossRef]
- Itoch, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Iqarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifiying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef]
- Raghunath, A.; Sundarraj, K.; Nagarajan, R.; Arfuso, F.; Bian, J.; Kumar, A.P.; Sethi, G.; Perumal, E. Antioxidant response elements: Discovery, classes, regulation and potential applications. Redox Biol. 2018, 17, 297–314. [Google Scholar] [CrossRef]
- Menegon, S.; Columbano, A.; Giordano, S. The dual roles of NRF2 in cancer. Trends Mol. Med. 2016, 22, 578–593. [Google Scholar] [CrossRef]
- Hayes, J.D.; McMahon, M.; Chowdhry, S.; Dinkova-Kostova, A.T. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 2010, 13, 1713–1748. [Google Scholar] [CrossRef]
- Wang, X.J.; Sun, Z.; Villeneuve, N.F.; Zhang, S.; Zhao, F.; Li, Y.; Chen, W.; Yi, X.; Zheng, W.; Wondrak, G.T.; et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 2008, 29, 1235–1243. [Google Scholar] [CrossRef]
- Totoki, Y.; Tatsuno, K.; Covington, K.R.; Ueda, H.; Creighton, C.J.; Kato, M.; Tsuji, S.; Donehower, L.A.; Slagle, B.L.; Nakamura, H.; et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014, 46, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaki, H.; Kawakami, Y.; Ueno, M.; Gotoh, K.; et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Schulze, K.; Imbeaud, S.; Letouze, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Y.; Ji, T.; Ma, R.; Chen, M.; Li, G.; Li, F.; Ding, Q.; Kang, Q.; Huang, D.; et al. Clinical implication of Keap1 and phosphorylated Nrf2 expression in hepatocellular carcinoma. Cancer Med. 2016, 5, 2678–2687. [Google Scholar] [CrossRef] [PubMed]
- Zavattari, P.; Perra, A.; Menegon, S.; Kowalik, M.A.; Petrelli, A.; Angioni, M.M.; Follenzi, A.; Quagliata, L.; Ledda-Columbano, G.M.; Terracciano, L.; et al. Nrf2, but not β-catenin, mutation represents an early event in rat hepatocarcinogenesis. Hepatology 2015, 62, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Yamamoto, M. The KEAP1-NRF2 system in cancer. Front. Oncol. 2017, 7, 85. [Google Scholar] [CrossRef] [PubMed]
- Guichard, C.; Amaddeo, G.; Imbeaud, S.; Ladeiro, Y.; Pelletier, L.; Maad, I.B.; Calderaro, J.; Bioulac-Sage, P.; Letexier, M.; Degos, F.; et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012, 44, 694–698. [Google Scholar] [CrossRef] [PubMed]
- Nault, J.C.; Zucman-Rossi, J. Genetics of hepatocellular carcinoma: The next generation. J. Hepatol. 2014, 60, 224–226. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.K.C.; Kim, D.H.; Cha, Y.N.; Na, H.K.; Surh, Y.J. Nrf2 mutagenic activation drives hepatocarcinogenesis. Cancer Res. 2017, 77, 4797–4808. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Dhar, D. Liver carcinogenesis: From naughty chemicals to soothing fat and the surprising role of NRF2. Carcinogenesis 2016, 37, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Niture, S.K.; Jaiswal, A.K. Nrf2-induced antiapoptotic Bcl-xL protein enhances cell survival and drug resistance. Free Radic. Biol. Med. 2013, 57, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Ma-On, C.; Sanpavat, A.; Whongsiri, P.; Suwannasin, S.; Hirankarn, N.; Tangkijvanich, P.; Boonla, C. Oxidative stress indicated by elevated expression of Nrf2 and 8-OHdG promotes hepatocellular carcinoma progression. Med. Oncol. 2017, 34, 57. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, M.; Kuroiwa, Y.; Inoue, T.; Hibi, D.; Matsushita, K.; Kijima, A.; Maruyama, S.; Nishikawa, A.; Umemura, T. Lack of nrf2 results in progression of proliferative lesions to neoplasms induced by long-term exposure to non-genotoxic hepatocarcinogens involving oxidative stress. Exp. Toxicol. Pathol. 2014, 66, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Nishi, S.; Sakai, M. Transcription factor Nrf2/MafK regulates rat placental glutathione S-transferase gene during hepatocarcinogenesis. Biochem. J. 2004, 380, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Inoue, K.; Hachiva, H.; Shibuya, N.; Aoki, T.; Kubota, K. Accumulation of phosphorylated p62 is associated with NF-E2-related factor 2 activation in hepatocellular carcinoma. J. Hepatobiliary Pancreat Sci. 2016, 23, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ou, Z.; Chen, R.; Niu, X.; Chen, D.; Kang, R.; Tang, D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016, 63, 173–184. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantely, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Inami, Y.; Waguri, S.; Sakamoto, A.; Kouno, T.; Nakada, K.; Hino, O.; Watanabe, S.; Ando, J.; Iwadate, M.; Yamamoto, M.; et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 2011, 193, 275–284. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Umemura, A.; He, F.; Taniquchi, K.; Nakagawa, H.; Yamachika, S.; Font-Burgada, J.; Zhong, Z.; Subramaniam, S.; Raghunandan, S.; Duran, A.; et al. p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell 2016, 29, 935–948. [Google Scholar] [CrossRef]
- Rojo de la Vega, M.; Dodson, M.; Chapman, E.; Zhang, D.D. NRF2-targeted therapeutics: New targets and modes of NRF2 regulation. Curr. Opin. Toxicol. 2016, 1, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Bartolini, D.; Dallaglio, K.; Torguato, P.; Piroddi, M.; Galli, F. Nrf2-p62 autophagy pathway and its response to oxidative stress in hepatocellular carcinoma. Transl. Res. 2018, 193, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Ichimura, Y.; Taguchi, K.; Suzuki, T.; Mizushima, T.; Takagi, H.; Hirose, Y.; Nagahashi, M.; Iso, T.; Fukutomi, T.; et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat. Commun. 2016, 7, 12030. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Lu, Z.N.; Guo, X.L. Regulation and role of nuclear factor-E2-related factor 2 (Nrf2) in multidrug resistance of hepatocellular carcinoma. Chem. Biol. Interact. 2018, 280, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Kato, S.; Saeki, M.; DeMartino, G.N.; Slaughter, C.A.; Takeuchi, J.; Toh-e, A.; Tanaka, K. cDNA cloning and functional analysis of p28 (Nas6p) and p40.5 (Nas7p), two novel regulatory subunits of the 26S proteasome. Gene 1998, 216, 113–122. [Google Scholar] [CrossRef]
- Higasthitsuji, H.; Iton, K.; Nagao, T.; Dawson, S.; Nonoguchi, K.; Kido, T.; Maver, R.J.; Arii, S.; Fujita, J. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat. Med. 2000, 6, 96–99. [Google Scholar] [CrossRef]
- Fu, X.Y.; Wang, H.Y.; Tan, L.; Liu, S.Q.; Cao, H.F.; Wu, M.C. Overexpression of p28/gankyrin in human hepatocellular carcinoma and its clinical significance. World J. Gastroenterol. 2002, 8, 638–643. [Google Scholar] [CrossRef]
- Yang, C.; Tan, Y.X.; Yang, G.Z.; Zhang, J.; Pan, Y.F.; Liu, C.; Fu, J.; Chen, Y.; Ding, Z.W.; Dong, L.W.; et al. Gankyrin has an antioxidative role through the feedback regulation of Nrf2 in hepatocellular carcinoma. J. Exp. Med. 2016, 213, 859–875. [Google Scholar] [CrossRef]
- Corton, J.M.; Gillespie, J.G.; Hawley, S.A.; Hardie, D.G. 5-aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem. 1995, 229, 558–565. [Google Scholar] [CrossRef]
- Sid, B.; Glorieux, C.; Valenzuela, M.; Rommelaere, G.; Najimi, M.; Dejeans, N.; Renard, P.; Verrax, J.; Calderon, P.B. AICAR induces Nrf2 activation by an AMPK-independent mechanism in hepatocarcinoma cells. Biochem. Pharmacol. 2014, 91, 168–180. [Google Scholar] [CrossRef]
- Yokomizo, T.; Izumi, T.; Takahashi, T.; Kasama, T.; Kobayashi, Y.; Sato, F.; Taketni, Y.; Shimizu, T. Enzymatic inactivation of leukotriene B4 by a novel enzyme found in the porcine kidney. Purification and properties of leukotriene B4 12-hydroxydehydrogenase. J. Biol. Chem. 1993, 268, 18128–18135. [Google Scholar] [PubMed]
- Sanchez-Rodriguez, R.; Torres-Mena, J.E.; De-la-Luz-Crus, M.; Bernal-Ramos, G.A.; Villa-Trevino, S.; Chagoya-Hazas, L.; Landero-Lopez, L.; Garcia-Roman, R.; Rouimi, P.; Perez-Carreon, J.I.; et al. Increased expression of prostaglandin reductase 1 in hepatocellular carcinomas from clinical cases and experimental tumors in rats. Int. J. Biochem. Cell Biol. 2014, 53, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Rodriguez, R.; Torres-Mena, J.E.; Quintanar-Jurado, V.; Chagoya-Hazas, V.; Rojas Del Castillo, E.; Del Pozo Yauner, L.; Villa-Trevino, S.; Perez-Carreon, J.I. Ptgr1 expression is regulated by NRF2 in rat hepatocarcinogenesis and promotes cell proliferation and resistance to oxidative stress. Free Radic. Biol. Med. 2017, 102, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Surh, Y.J. More than spice: Capsaicin in hot chili peppers makes tumor cells commit suicide. J. Natl. Cancer Inst. 2002, 94, 1263–1265. [Google Scholar] [CrossRef]
- Joung, E.J.; Li, M.H.; Lee, H.G.; Somparn, N.; Jung, Y.S.; Na, H.K.; Kim, S.H.; Cha, Y.N.; Surh, Y.J. Capsaicin induces heme oxygenase-1 expression in HepG2 cells via activation of PI3K-Nrf2 signaling: NAD(P)H: Quinone oxidoreductase as a potential target. Antioxid. Redox Signal. 2007, 9, 2087–2098. [Google Scholar] [CrossRef] [PubMed]
- Feron, V.J.; Til, H.P.; de Vrijer, F.; Woutersen, R.A.; Cassee, F.R.; van Bladeren, P.J. Aldehydes: Occurrence, carcinogenic potential, mechanism of action and risk assessment. Mutat. Res. 1991, 259, 363–385. [Google Scholar] [CrossRef]
- Lee, S.E.; Yang, H.; Jeong, S.I.; Jin, Y.H.; Park, C.S.; Park, Y.S. Induction of heme oxygenase-1 inhibits cell death in crotonaldehyde-stimulated HepG2 cells via the PKC-δ-p38-Nrf2 pathway. PLoS ONE. 2012, 7, e41676. [Google Scholar] [CrossRef]
- Lee, S.B.; Kim, C.Y.; Lee, H.J.; Yun, J.H.; Nho, C.W. Induction of the phase II detoxification enzyme NQO1 in hepatocarcinoma cells by lignans from the fruit of Schisandra chinensis through nuclear accumulation of Nrf2. Planta Med. 2009, 75, 1314–1318. [Google Scholar] [CrossRef]
- Puri, M.; Sharma, D.; Tiwari, A.K. Downstream processing of stevioside and its potential applications. Biotechnol. Adv. 2011, 29, 781–791. [Google Scholar] [CrossRef]
- Ceunen, S.; Geuns, J.M. Steviol glycosides: Chemical diversity, metabolism, and function. J. Nat. Prod. 2013, 76, 1201–1228. [Google Scholar] [CrossRef] [PubMed]
- Momtazi-Borojeni, A.A.; Esmaeili, S.A.; Abdollah, E.; Sahebkar, A. A review on the pharmacology and toxicology of steviol glycosides extracted from Stevia rebaudiana. Curr. Pharm. Des. 2017, 23, 1616–1622. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, L.; Wang, Y.; Zhu, X.; Jiang, M.; Song, E.; Song, Y. New application of the commercial sweetener rebaudioside a as a hepatoprotective candidate: Induction of the Nrf2 signaling pathway. Eur. J. Pharmacol. 2018, 822, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Blanc, P.J.; Laussac, J.P.; Le Bars, J.; Le Bars, P.; Loret, M.O.; Pareilleux, A.; Prome, D.; Prome, J.C.; Santerre, A.L.; Goma, G. Characterization of monascidin A from Monascus as citrinin. Int. J. Food Microbiol. 1995, 27, 201–213. [Google Scholar] [CrossRef]
- Sharath Babu, G.R.; Anand, T.; Ilaiyaraja, N.; Khanum, F.; Gopalan, N. Peargonidin modulates Keap1/Nrf2 parhway gene expression and ameliorates citrinin induced oxidative stress in HepG2 cells. Front. Pharmacol. 2017, 8, 868. [Google Scholar] [CrossRef] [PubMed]
- Giusti, M.M.; Wrolstad, R.E. Characterization of red radish anthocyanins. J. Food Sci. 1996, 61, 322–326. [Google Scholar] [CrossRef]
- Gil, M.I.; Tomas-Barberan, F.A.; Hess-Pierce, B.; Holcroft, D.M.; Kader, A.A. Antioxidant activity of pomegranate juice and its relationship with phenolic composition and processing. J. Agric. Food. Chem. 2000, 48, 4581–4589. [Google Scholar] [PubMed]
- Faria, A.; Monteiro, R.; Mateus, N.; Azevedo, I.; Calhau, C. Effect of pomegranate (Punica granatum) juice intake on hepatic oxidative stress. Eur. J. Nutr. 2007, 46, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Bishayee, A.; Bhatia, D.; Thoppil, R.T.; Darvesh, A.S.; Nevo, E.; Lansky, E.P. Pomegranate-mediated chemoprevention of experimental hepatocarcinogenesis involves Nrf2 regulated antioxidant mechanisms. Carcinogenesis 2011, 32, 888–896. [Google Scholar] [PubMed]
- Krajka-Kuzniak, V.; Paluszczak, J.; Szaefer, H.; Baer-Dubowska, W. The activation of the Nrf2/ARE pathway in HepG2 hepatoma cells by phytochemicals and subsequent modulation of phase II and antioxidant enzyme expression. J. Physiol. Biochem. 2015, 71, 227–238. [Google Scholar] [CrossRef]
- Wall, M.E.; Wani, M.C. Camptothecin and taxol: Discovery to clinic—Thirteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 1995, 55, 753–760. [Google Scholar] [PubMed]
- Chen, F.; Wang, H.; Zhu, J.; Zhao, R.; Xue, P.; Zhang, Q.; Bud Nelson, M.; Qu, W.; Feng, B.; Pi, J. Camptothecin suppresses NRF2-ARE activity and sensitises hepatocellular carcinoma cells to anticancer drugs. Br. J. Cancer 2017, 117, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Yin, S.; Huo, Y.; Liang, M.; Fan, L.; Ye, M.; Hu, H. Glycycoumarin ameliorates alcohol-induced hepatotoxicity via activation of Nrf2 and autophagy. Free Radic. Biol. Med. 2015, 89, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Shen, T.; Yang, H.; Gu, W. Ruthenium complexes induce HepG2 human hepatocellular carcinoma cell apoptosis and inhibit cell migration and invasion through regulation of the Nrf2 pathway. Int. J. Mol. Sci. 2016, 17, 775. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.M.; Ke, Z.P.; Wang, J.N.; Yang, J.Y.; Chen, S.Y.; Chen, H. Apigenin sensitizes doxorubicin-resistant hepatocellular carcinoma BEL-7402/ADM cells to doxorubicin via inhibiting PI3K/Akt/Nrf2 pathway. Carcinogenesis 2013, 34, 1806–1814. [Google Scholar] [CrossRef]
- Gao, A.M.; Ke, Z.P.; Shi, F.; Sun, G.C.; Chen, H. Chrysin enhances sensitivity of BEL-7402/ADM cells to doxorubicin by suppressing PI3K/Akt/Nrf2 and ERK/Nrf2 pathway. Chem. Biol. Interact. 2013, 206, 100–108. [Google Scholar] [CrossRef]
- Ye, C.L.; Lu, Y.H.; Wei, D.Z. Flavonoids from Cleistocalyx operculatus. Phytochemistry 2004, 65, 445–447. [Google Scholar] [CrossRef]
- Yu, W.G.; Qian, J.; Lu, Y.H. Hepatoprotective effects of 2′,4′-dihydroxy-6′-methoxy-3′,5′-dimethylchalcone on CCl4-induced acute liver injury in mice. J. Agric. Food Chem. 2011, 59, 12821–12829. [Google Scholar] [CrossRef]
- Ye, C.L.; Liu, J.W.; Wei, D.Z.; Lu, Y.H.; Qian, F. In vivo antitumor activity by 2′,4′-dihydroxy-6′-methoxy-3′,5′-dimethylchalcone in a solid human carcinoma xenograft model. Cancer Chemother. Pharmacol. 2005, 56, 70–74. [Google Scholar] [CrossRef]
- Wei, X.; Mo, X.; An, F.; Ji, X.; Lu, Y. 2′,4′-Dihydroxy-6′-methoxy-3′,5′-dimethylchalcone, a potent Nrf2/ARE pathway inhibitor, reverses drug resistance by decreasing glutathione synthesis and drug efflux in BEL-7402/5-FU cells. Food Chem. Toxicol. 2018, 119, 252–259. [Google Scholar] [CrossRef]
- Keating, G.M.; Santoro, A. Sorafenib: A review of its use in advanced hepatocellular carcinoma. Drugs 2009, 69, 223–240. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Ye, W.; Duan, X.; Zhang, M.; Wang, J. The noncytotoxic dose of sorafenib sensitizes Bel-7402/5-FU cells to 5-FU by down regulating 5-FU induced Nrf2 expression. Dig. Dis. Sci. 2013, 58, 1615–1626. [Google Scholar] [CrossRef] [PubMed]
- Jager, S.; Trojan, H.; Kopp, T.; Laszcyzck, M.N.; Scheffler, A. Pentacyclic triterpene distribution in various plants-rich sources for a new group of multi-potent plant extracts. Molecules 2009, 14, 2016–2031. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, T.; Du, J. Urosolic acid sensitizes cisplatin-resistant HepG2/DDP cells to cisplatin via inhibiting Nrf2/ARE pathway. Drug Des. Dev. Ther. 2016, 10, 3471–3481. [Google Scholar] [CrossRef]
- Terbach, N.; Williams, R.S. Structure-function studies for the panacea, valproic acid. Biochem. Soc. Trans. 2009, 37, 1126–1132. [Google Scholar] [PubMed]
- Tan, J.; Cang, S.; Ma, Y.; Petrillo, R.L.; Liu, D. Novel histone deacetylase inhibitors in clinical trials as anti-cancer agents. J. Hematol. Oncol. 2010, 3, 5. [Google Scholar] [CrossRef]
- Yu, J.I.; Choi, C.; Shin, S.W.; Son, A.; Lee, G.H.; Kim, S.Y.; Park, H.C. Valproic Acid Sensitizes Hepatocellular Carcinoma Cells to Proton Therapy by Suppressing NRF2 Activation. Sci. Rep. 2018, 8, 7597. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Blower, M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell Mol. Life Sci. 2016, 73, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Lyssiotis, C.A.; Kimmelman, A.C. Metabolic interactions in the tumor microenvironment. Trends Cell Biol. 2017, 27, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Moenner, M.; Pluguet, O.; Bouchecareilh, M.; Chevet, E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007, 67, 10631–10634. [Google Scholar] [CrossRef]
- Martinon, F. Targeting endoplasmic reticulum signaling pathways in cancer. Acta Oncol. 2012, 51, 822–830. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Zhao, H.; Gao, L.; Zhang, W.; Shuli, A.Y.; Shay, C. FGF19 protect hepatocellular carcinoma cells against endoplasmic reticulum stress via activation of FGFR4-GSK3β-Nrf2 Signaling. Cancer Res. 2017, 77, 6215–6225. [Google Scholar] [CrossRef] [PubMed]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Hobert, O. Gene regulation by transcription factors and microRNAs. Science 2008, 319, 1785–1786. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.M.; Zhang, X.Y.; Ke, Z.P. Apigenin sensitizes BEL-7402/ADM cells to doxorubicin through inhibiting miR-101/Nrf2 pathway. Oncotarget 2017, 8, 82085–82091. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Ye, W.; Zhang, Y.; Yu, D.; Shao, Q.; Liang, J.; Zhang, M. miR-144 reverses chemoresistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Am. J. Transl. Res. 2016, 8, 2992–3002. [Google Scholar]
- Shi, L.; Chen, Z.G.; Wu, L.L.; Zheng, J.J.; Yang, J.R.; Chen, X.F.; Chen, X.F.; Chen, Z.Q.; Liu, C.L.; Chi, S.Y.; et al. miR-340 reverses cisplatin resistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Asian Pac. J. Cancer Prev. 2014, 15, 10439–10444. [Google Scholar] [CrossRef]
- Petrelli, A.; Perra, A.; Cora, D.; Sulas, P.; Menegon, C.; Manca, C.; Migliore, C.; Kowalik, M.A.; Ledda-Columbano, G.M.; Giordano, S.; et al. MicroRNA/gene profiling unveils early molecular changes and nuclear factor erythroid related factor 2 (NRF2) activation in a rat model recapitulating human hepatocellular carcinoma (HCC). Hepatology 2014, 59, 228–241. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Shi, L.; Wu, L.; Chen, Z.; Yang, J.; Yu, F.; Zheng, F.; Lin, X. MiR-141 activates Nrf2-dependent antioxidant pathway via down-regulating the expression of Keap1 conferring the resistance of hepatocellular carcinoma cells to 5-fluorouracil. Cell Physiol. Biochem. 2015, 35, 2333–2348. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Chen, J.; Liu, X.M.; Zhao, R.; Zhe, H. Nrf2-mediated metabolic reprogramming in cancer. Oxid. Med. Cell Longev. 2018, 2018, 9304091. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Happel, C.; Manna, S.K.; Acquaah-Mensah, G.; Carrerero, J.; Kumar, S.; Nasipuri, P.; Krausz, K.W.; Wakabayashi, N.; Dewi, R. Transcription factor NRF2 regulates miR-1 and miR-201 to drive tumorigenesis. J. Clin. Investig. 2013, 123, 2921–2934. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yang, P.; Li, H.; Cheng, P.; Zhang, L.; Wei, D.; Su, X.; Peng, J.; Gao, H.; Tan, Y.; et al. MicroRNA-1 inhibits proliferation of hepatocarcinoma cells by targeting endothelin-1. Life Sci. 2012, 91, 440–447. [Google Scholar] [CrossRef] [PubMed]
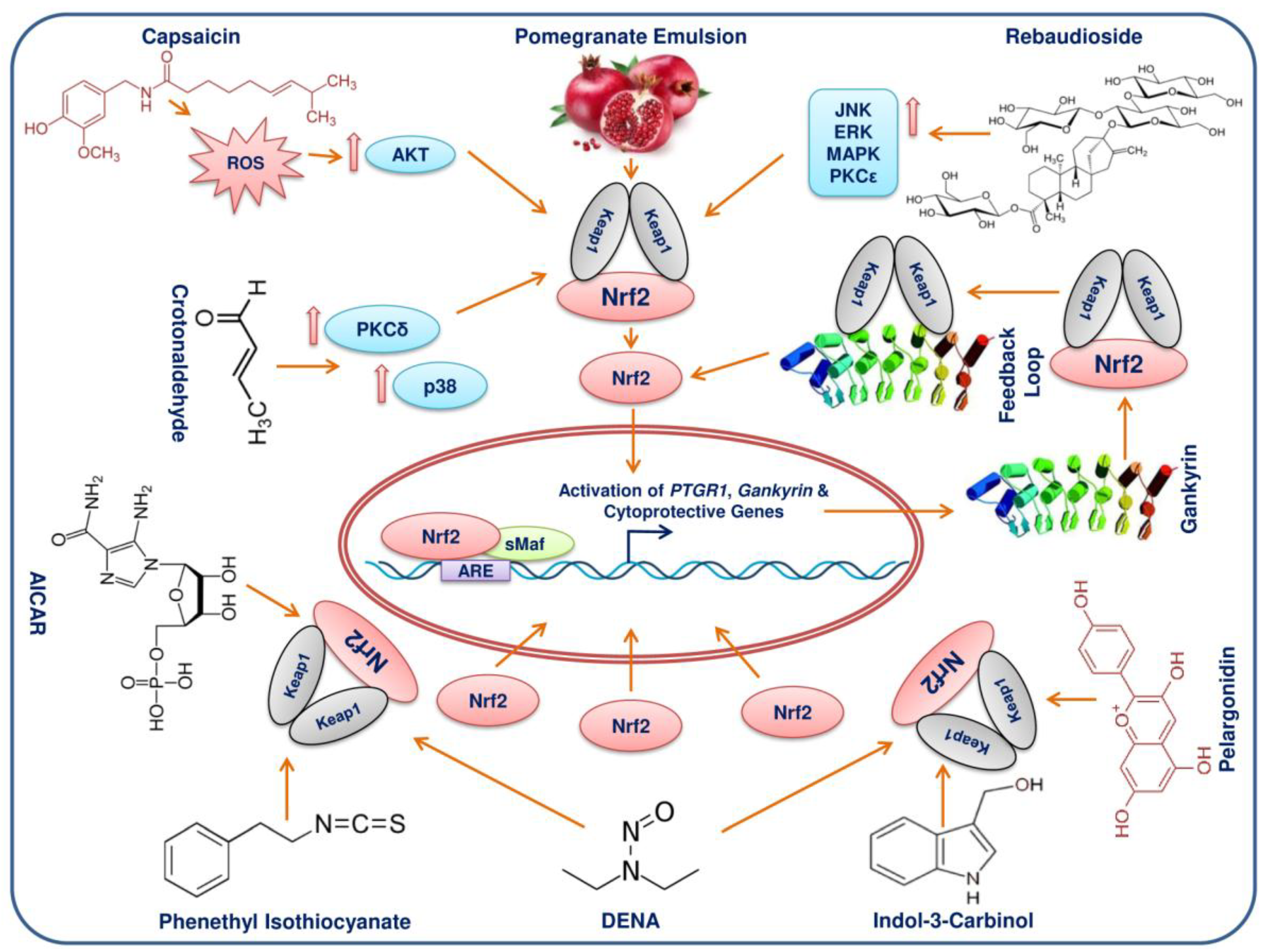
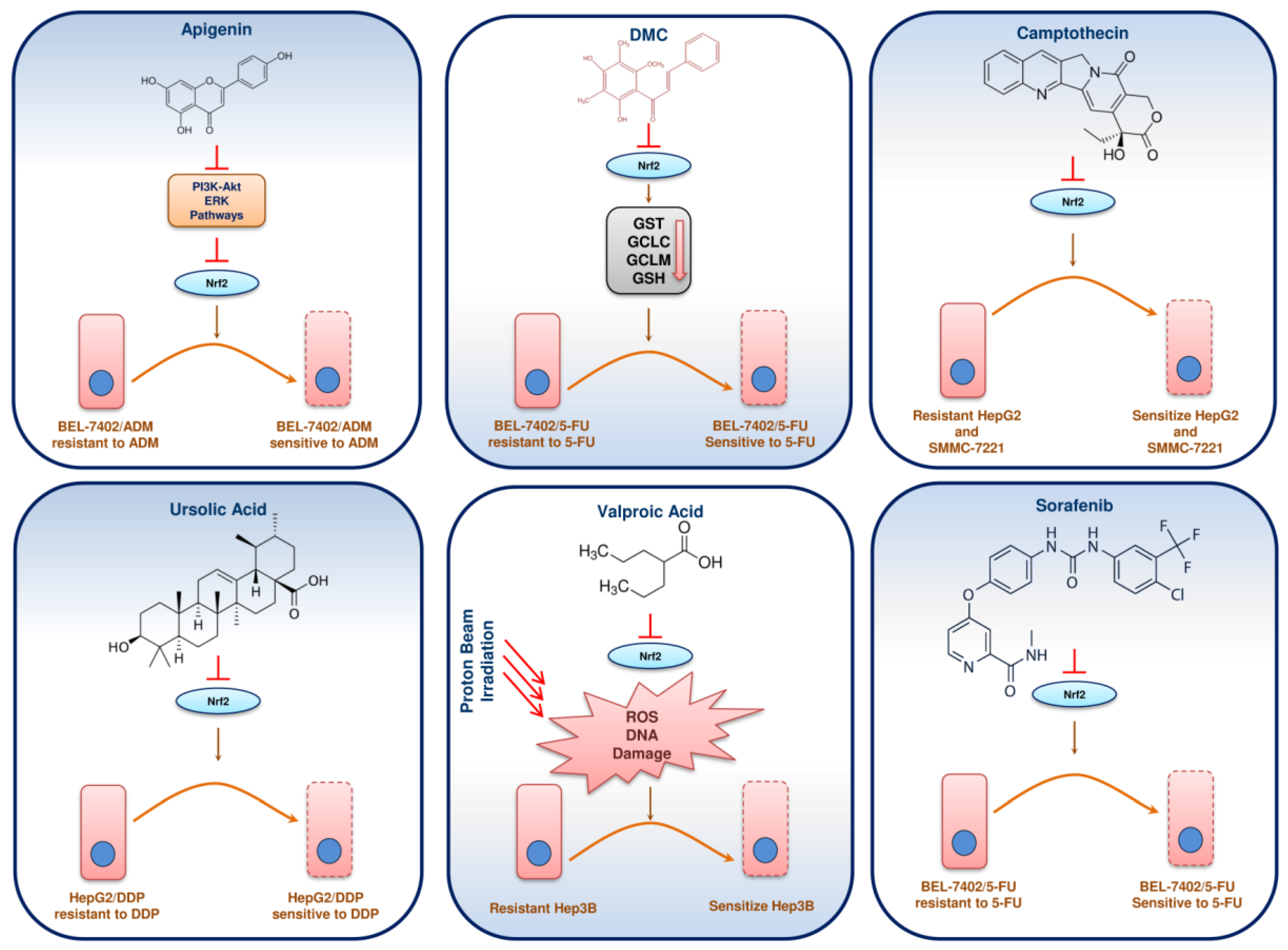
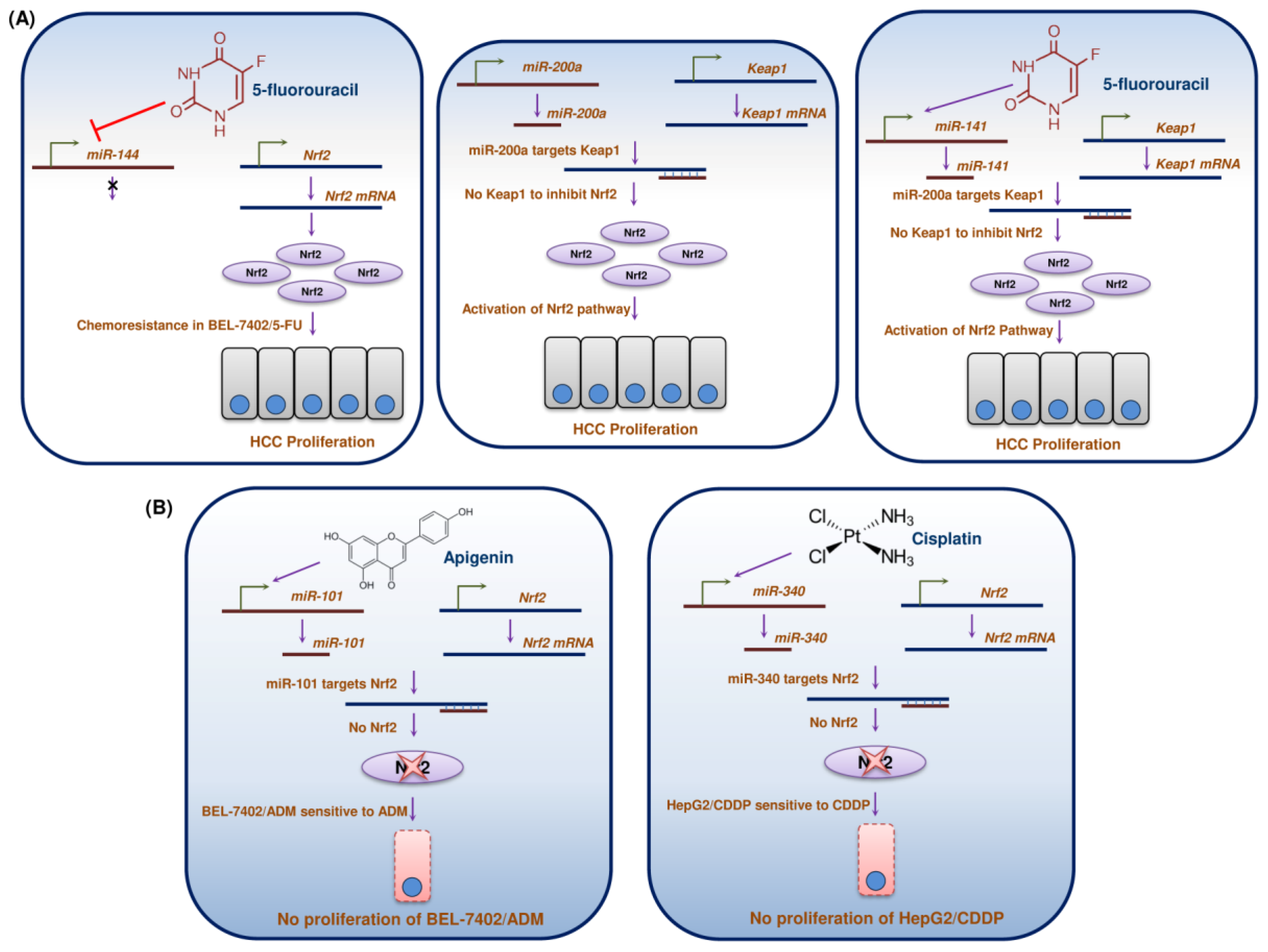
| Compounds | Dose and Duration | Cell Lines/Animal Model | Molecular Targets | Molecular Mechanism | Reference |
|---|---|---|---|---|---|
| Camptothecin | 0.1 and 0.5 µM for 24 h | HepG2 | ↓GCLC, ↓GCLM, ↓NQO1, ↓HMOX-1, ↓AKR1C1, ↓AKR1C2, ↓AKR1C3 | Down regulation of NRF2 suppression of ARE- dependent genes | [143] |
| Capsaicin | 200 µM for 24 h | HepG2 | ↑HO-1, ↓NQO1, ↑p-AKT, ↑p-ERK, ↑NRF2 | Down regulation of NQO1 triggers the production of ROS, leading to phosphorylation of AKT, ERK and ARE binding of NRF2 | [127] |
| Glycycoumarin | 10 mg and 20 mg/kg for once a day for 3 weeks | C57BL/6 mice | ↑NRF2, ↑HO-1, ↑GCLC | Glycycoumarin activates NRF2 and induces autophagy via up regulation of p62 and p38 | [144] |
| Glycycoumarin | 50 µM for 24 h | HepG2 | ↑Nrf2, ↑HO-1, ↑GCLC, ↑p38, ↑p-ERK1/2, ↑p62, ↓KEAP1, ↑LC3-II | Glycycoumarin activates NRF2 | [144] |
| Crotonaldehyde | 50 µM for 24 h | HepG2 | ↑HO-1, ↑p38, ↑p-PKC-δ, ↑NRF2 | Anti-apoptotic effect of crotonaldehyde induced by HO-1 through the PKC-δ-p38 MAPK-NRF2 signalling pathway | [129] |
| Pelargonidin Chloride | 50 and 100 µM for 2 h | HepG2 | ↑NRF2, ↑HO-1, ↑GST, ↑NQO1, ↑CAT, ↑SOD1, ↑GPX1 | Up regulation of detoxification enzymes genes through the KEAP1/NRF2 signalling pathway | [136] |
| Pomegranate emulsion | 1 g and 10 g/kg for three times a week | Male Sprague-Dawley rats | ↑GSTA2, ↑GSTA5, ↑GSTM1, ↑GSTM7, ↑GSTT1, ↑NQO1, ↑UGT1A1, ↑UGT2B17, ↑NRF2 | Induction of antioxidant and phase 2 xenobiotic enzymes leading to up regulation of NRF2 | [140] |
| Ruthenium complex | 2 and 4 µM for 24 h | HepG2 | ↓NRF2, ↓NQO1, ↓HO-1 | Suppression of NQO1 and HO-1 expression through down regulation of the Nrf2 signalling pathway | [145] |
| Compounds | Drug Sensitized | Dose and Duration | Cell Lines | Mode of Nrf2 Inhibition | Molecular Targets | Reference |
|---|---|---|---|---|---|---|
| Apigenin | Doxorubicin | Apigenin-20 μM for 24 h Doxorubicin-2 μM for 24 h | BEL-7402/ADM | NRF2 expression was inhibited by down regulation of the PI3K/AKT pathway | ↓NRF2, ↓HO-1, ↓AKR1B10, ↓MRP5 | [147] |
| Chrysin | Doxorubicin | Chrysin-20 μM for 24 h | BEL-7402/ADM | Chrysin suppressed the activation of NRF2 and its downstream genes through inhibition of the PI3K/AKT and ERK signalling pathway | ↓NRF2, ↓HO-1, ↓AKR1B10, ↓MRP5, ↓p-Akt, ↓p-ERK1/2 | [148] |
| DMC | 5-FU | DMD-5,10 and 20 μM for 24 h | BEL-7402/5-FU | NRF2 suppression, prevented NRF2 translocation and inhibited the ARE binding | ↓NRF2, ↓GCLC, ↓GCLM, ↓GST, ↓GSH | [151] |
| Sorafenib | 5-FU | Sorafenib-2 µM for 24 h 5-flurouracil-1000 μg/mL for 24 h | Bel-7402/5-FU | Sorafenib inhibited the expression of NRF2 induced by 5-flurouracil | ↓NRF2, ↓MRP1, ↓MRP2, ↓MRP3 | [153] |
| Ursolic acid | Cisplatin | Ursolic acid-2.25 μg/mL for 48 h | HepG2/DDP | Ursolic acid highly induced ROS and reduced mitochondrial membrane potential, leading to suppression of NRF2 expression and its downstream genes | ↓NRF2, ↓HO-1, ↓NQO1, ↓GST | [155] |
| Valproic acid | Proton therapy | Valproic acid-1 mM for 2 h and 24 h | Hep3B | NRF2 expression was suppressed by NADPH oxidase activation through increased intracellular ROS level | ↑PARP cleavage, ↑caspase-3 cleavage, ↓NRF2, ↓HO-1 | [158] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raghunath, A.; Sundarraj, K.; Arfuso, F.; Sethi, G.; Perumal, E. Dysregulation of Nrf2 in Hepatocellular Carcinoma: Role in Cancer Progression and Chemoresistance. Cancers 2018, 10, 481. https://doi.org/10.3390/cancers10120481
Raghunath A, Sundarraj K, Arfuso F, Sethi G, Perumal E. Dysregulation of Nrf2 in Hepatocellular Carcinoma: Role in Cancer Progression and Chemoresistance. Cancers. 2018; 10(12):481. https://doi.org/10.3390/cancers10120481
Chicago/Turabian StyleRaghunath, Azhwar, Kiruthika Sundarraj, Frank Arfuso, Gautam Sethi, and Ekambaram Perumal. 2018. "Dysregulation of Nrf2 in Hepatocellular Carcinoma: Role in Cancer Progression and Chemoresistance" Cancers 10, no. 12: 481. https://doi.org/10.3390/cancers10120481
APA StyleRaghunath, A., Sundarraj, K., Arfuso, F., Sethi, G., & Perumal, E. (2018). Dysregulation of Nrf2 in Hepatocellular Carcinoma: Role in Cancer Progression and Chemoresistance. Cancers, 10(12), 481. https://doi.org/10.3390/cancers10120481






