Analyzing Developing Brain-On-Chip Cultures with the CALIMA Calcium Imaging Tool
Abstract
1. Introduction
2. Materials and Methods
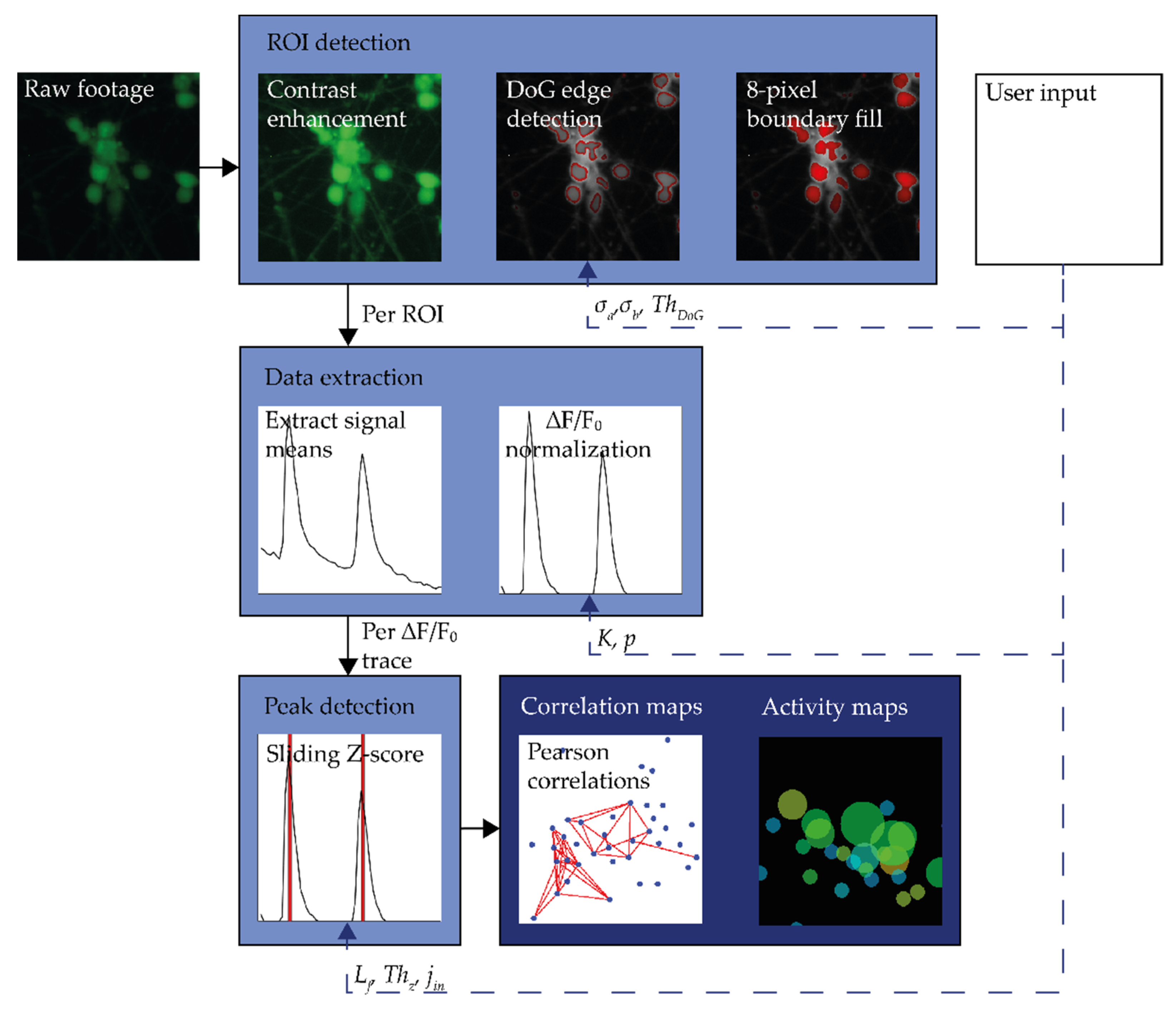
2.1. Processing Workflow
2.2. Processing Changes Compared to CALIMA 1.0
2.2.1. ROI Detection
2.2.2. Signal Extraction
2.2.3. Peak Detection
2.3. Algorithms and Parameter Space
2.3.1. ROI Detection
2.3.2. Signal Extraction
2.3.3. Peak Detection
2.3.4. Parameter Space Summary
2.4. Data Acquisition and Management
3. Results
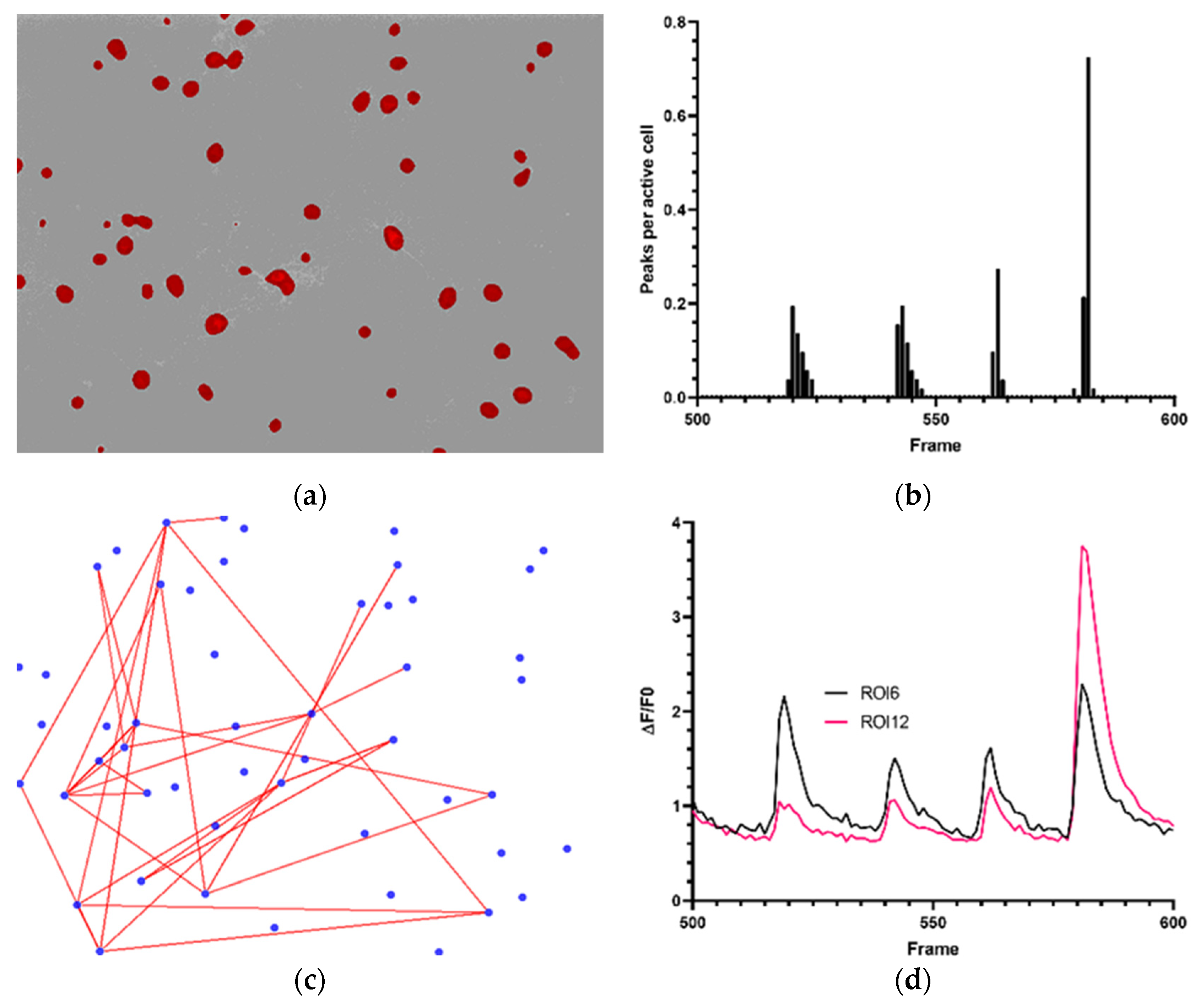
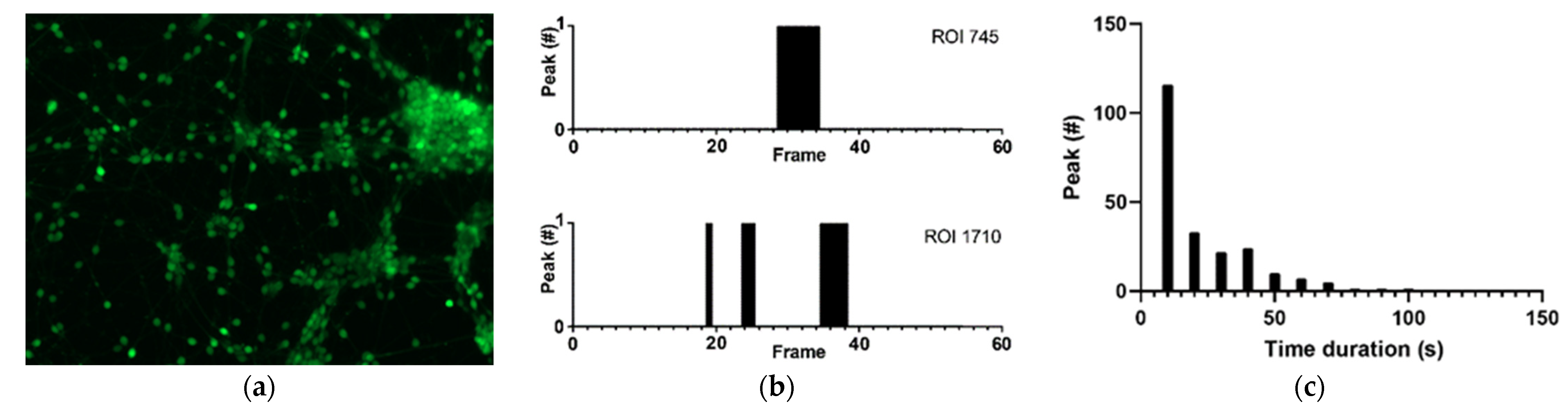
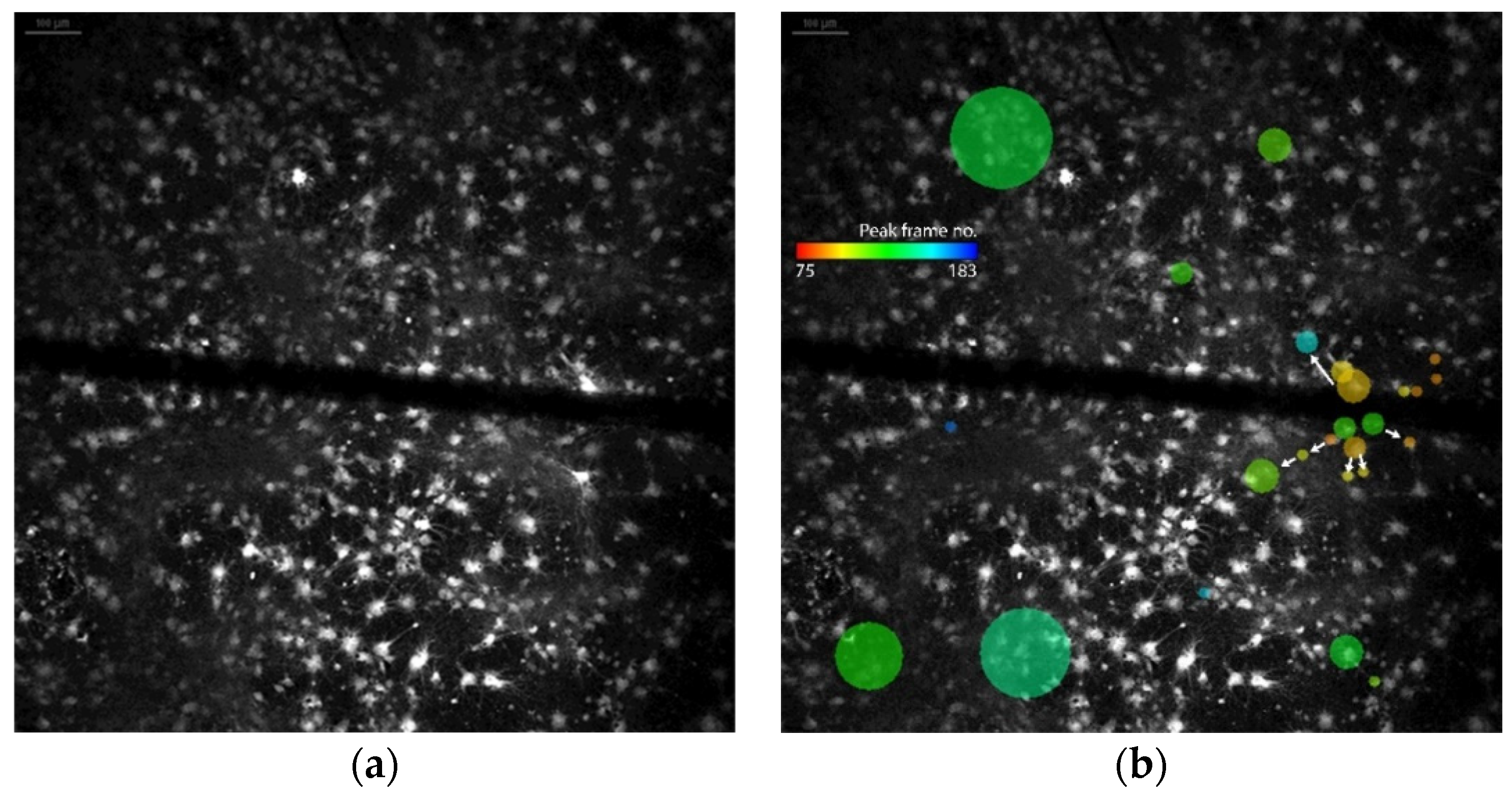
3.1. Physiological Behavior
3.2. Processing Speed
3.3. Processing Speed Validation
4. Discussion
4.1. CALIMA 2.0 as a Valuable Tool for Calcium Spike Detection in BoC-Devices
4.2. CALIMA 2.0 Operation Speed
4.3. Outlook
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bang, S.; Jeong, S.; Choi, N.; Kim, H.N. Brain-on-a-Chip: A History of Development and Future Perspective. Biomicrofluidics 2019, 13, 051301. [Google Scholar] [CrossRef]
- Frimat, J.-P.; Luttge, R. The Need for Physiological Micro-Nanofluidic Systems of the Brain. Front. Bioeng. Biotechnol. 2019, 7, 100. [Google Scholar] [CrossRef]
- Van Der Velden, L.; Vinck, M.A.; Werkman, T.R.; Wadman, W.J. Modulation of Functional Connectivity Between Dopamine Neurons of the Rat Ventral Tegmental Area in Vitro. Front. Integr. Neurosci. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Bastiaens, A.; Frimat, J.-P.; Van Nunen, T.; Luttge, R. Exploiting Nanogroove-Induced Cell Culture Anisotropy to Advance in Vitro Brain Models. J. Vac. Sci. Technol. B 2019, 37, 61802. [Google Scholar] [CrossRef]
- Xie, S.; E Gardeniers, J.G.; Luttge, R.; E Gardeniers, H.J.G. Nanoscale Membrane Actuator for in Vitro Mechano-Stimuli Responsive Studies of Neuronal Cell Networks on Chip. J. Micromech. Microeng. 2018, 28, 85011. [Google Scholar] [CrossRef]
- Mitani, A.; Komiyama, T. Real-Time Processing of Two-Photon Calcium Imaging Data Including Lateral Motion Artifact Correction. Front. Neuroinform. 2018, 12, 98. [Google Scholar] [CrossRef] [PubMed]
- Giovannucci, A.; Friedrich, J.; Kaufman, M.; Churchland, A.; Chklovskii, D.; Paninski, L.; Pnevmatikakis, E.A. OnACID: Online Analysis of Calcium Imaging Data in Real Time. BiorXiv 2017, 193383. [Google Scholar] [CrossRef]
- Knot, H.J.; Laher, I.; Sobie, E.A.; Guatimosim, S.; Gomez-Viquez, L.; Hartmann, H.; Song, L.-S.; Lederer, W.; Graier, W.F.; Malli, R.; et al. Twenty Years of Calcium Imaging: Cell Physiology to Dye For. Mol. Interv. 2005, 5, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Picht, E.; Zima, A.V.; Blatter, L.A.; Bers, D.M. SparkMaster: Automated Calcium Spark Analysis With ImageJ. Am. J. Physiol. Physiol. 2007, 293, C1073–C1081. [Google Scholar] [CrossRef]
- Kaifosh, P.; Zaremba, J.D.; Danielson, N.B.; Elosonczy, A. SIMA: Python Software for Analysis of Dynamic Fluorescence Imaging Data. Front. Neuroinform. 2014, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Cantu, D.A.; Wang, B.; Gongwer, M.W.; He, C.X.; Goel, A.; Suresh, A.; Kourdougli, N.; Arroyo, E.D.; Zeiger, W.; Portera-Cailliau, C. EZcalcium: Open-Source Toolbox for Analysis of Calcium Imaging Data. Front. Neural Circuits 2020, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.P.; Man, K.; Firestein, B.L.; Meaney, D.F. Automated Quantification of Neuronal Networks and Single-Cell Calcium Dynamics Using Calcium Imaging. J. Neurosci. Methods 2015, 243, 26–38. [Google Scholar] [CrossRef] [PubMed]
- del Rio, J.A.; Ferrer, I. Potential of Microfluidics and Lab-on-Chip Platforms to Improve Understanding of ‘prion-like’ Protein Assembly and Behavior. Front. Bioeng. Biotechnol. 2020, 8, 1057. [Google Scholar] [CrossRef] [PubMed]
- Musafargani, S. Blood brain barrier: A tissue engineered microfluidic chip. J. Neurosci. Methods 2020, 331, 108525. [Google Scholar] [CrossRef] [PubMed]
- Pamies, D.; Hartung, T.; Hogberg, H.T. Biological and medical applications of a brain-on-a-chip. Exp. Biol. Med. 2014, 239, 1096–1107. [Google Scholar] [CrossRef] [PubMed]
- Radstake, F.; Raaijmakers, E.; Luttge, R.; Zinger, S.; Frimat, J. CALIMA: The Semi-Automated Open-Source Calcium Imaging Analyzer. Comput. Methods Programs Biomed. 2019, 179, 104991. [Google Scholar] [CrossRef]
- Moonen, E.; Luttge, R.; Frimat, J.-P. Single Cell Trapping by Capillary Pumping Using NOA81 Replica Moulded Stencils. Microelectron. Eng. 2018, 197, 1–7. [Google Scholar] [CrossRef]
- Raaijmakers, E.A.L.; Wanders, N.; Mestrom, R.M.C.; Luttge, R. “CALIMA 2.0,” 2021. Available online: https://github.com/EALRaaijmakers/CALIMA-2.0 (accessed on 30 March 2021).
- Winnemöller, H.; Kyprianidis, J.E.; Olsen, S.C. XDoG: An EXtended Difference-of-Gaussians Compendium Including Advanced Image Stylization. Comput. Graph. 2012, 36, 740–753. [Google Scholar] [CrossRef]
- Bruton, J.; Cheng, A.J.; Westerblad, H. Methods to Detect Ca2+ in Living Cells in Calcium Signaling, 2nd ed.; Springer: Cham, Switzerland, 2012. [Google Scholar]
- Monteiro Rocha Lima, B.; Sampaio, L.; De Oliveira, T.E.; Fonseca, V.; Petriu, E. Heart Rate Detection Using a Multimodal Tactile Sensor and a Z-Score Based Peak Detection Algorithm. In Proceedings of the 42th Conference of The Canadian Medical and Biological Engineering Society, Ottawa, ON, Canada, 21–24 May 2019. [Google Scholar]
- Patel, T.P. Fluorescence Single Neuron and Network Analysis Package. University of Pennsylvania, 2013. Available online: https://www.seas.upenn.edu/~molneuro/software.Html (accessed on 2 March 2018).
- Marr, D.; Hildreth, E. Theory of Edge Detection. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1980, 207, 187–217. [Google Scholar] [CrossRef]
- Lock, J.T.; Parker, I.; Smith, I.F. A Comparison of Fluorescent Ca2+ Indicators for Imaging Local Ca2+ Signals in Cultured Cells. Cell Calcium 2015, 58, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Olson, E.C.; Spitzer, N.C. Spontaneous Neuronal Calcium Spikes and Waves During Early Differentiation. J. Neurosci. 1994, 14, 6325–6335. [Google Scholar] [CrossRef]
- Bastiaens, A.; Akcay, G.; Fransen, M.; Xie, S.; Luttge, R. Mems Actuation Promotes in Vitro Brain-on-Chip maturation. In Proceedings of the MicroTAS 2020—24th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Online Conference. 4–9 October 2020; pp. 957–958. [Google Scholar]
- Tada, M.; Takeuchi, A.; Hashizume, M.; Kitamura, K.; Kano, M. A Highly Sensitive Fluorescent Indicator Dye for Calcium Imaging of Neural Activity in Vitro and in Vivo. Eur. J. Neurosci. 2014, 39, 1720–1728. [Google Scholar] [CrossRef]
- Sprinthall, R.C. Basic Statistical Analysis, 9th ed.; Pearson Education: London, UK, 2011. [Google Scholar]
- Life Technologies. Fluo-4 Calcium Imaging Kit; Catalog No. F10489; Life Technologies: Carlsbad, CA, USA, 2013. [Google Scholar]
- Sironi, A.; Tekin, B.; Rigamonti, R.; Lepetit, V.; Fua, P. Learning Separable Filters. IEEE Trans. Pattern Anal. Mach. Intell. 2014, 37, 94–106. [Google Scholar] [CrossRef]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium Signalling: Dynamics, Homeostasis and Remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef]
- Grienberger, C.; Konnerth, A. Imaging Calcium in Neurons. Neuron 2012, 73, 862–885. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Kwan, A.C. Interpreting in Vivo Calcium Signals from Neuronal Cell Bodies, Axons, and Dendrites: A Review. Neurophotonics 2019, 7, 011402. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.S.; Nicholas, C.S. Calcium signaling in neuronal development. Cold Spring Harbor Perspect. Biol. 2011, 3, a004259. [Google Scholar] [CrossRef] [PubMed]
- Sood, D.; Cairns, D.M.; Dabbi, J.M.; Ramakrishnan, C.; Deisseroth, K.; Black, L.D.; Santaniello, S.; Kaplan, D.L. Functional Maturation of Human Neural Stem Cells in a 3D Bioengineered Brain Model Enriched With Fetal Brain-Derived Matrix. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Baylor, S.M.; Hollingworth, S. Calcium Indicators and Calcium Signalling in Skeletal Muscle Fibres During excitation–contraction Coupling. Prog. Biophys. Mol. Biol. 2011, 105, 162–179. [Google Scholar] [CrossRef]
- Vogelstein, J.T.; Packer, A.M.; Machado, T.A.; Sippy, T.; Babadi, B.; Yuste, R.; Paninski, L. Fast Nonnegative Deconvolution for Spike Train Inference From Population Calcium Imaging. J. Neurophysiol. 2010, 104, 3691–3704. [Google Scholar] [CrossRef]
- Lamprecht, M.R.; Sabatini, D.M.; Carpenter, A.E. CellProfiler™: Free, Versatile Software for Automated Biological Image Analysis. Biotechniques 2007, 42, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Akram, F.; Garcia, M.A.; Puig, D. Active Contours Driven by Difference of Gaussians. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
| Operation | ROI Detection | Signal Extraction | Peak Detection | |||||
|---|---|---|---|---|---|---|---|---|
| Parameter | ||||||||
| Unit | Pixels | Pixels | - | Frames | % | Frames | - | - |
| Value | 3.0 to 10.0 | >calcium spike duration | 10–20 | >calcium spike duration | ≥3.0 | 0.1–0.2 | ||
| Operation | ROI Detection | Signal Extraction | Peak Detection | |||||
|---|---|---|---|---|---|---|---|---|
| Parameter | ||||||||
| Unit | Pixels | Pixels | - | Frames | % | Frames | - | - |
| Value | 6.6 | 10.6 | 0.0030 | 25 | 10 | 10 | 5.0 | 0.2 |
| Operation | ROI Detection | Signal Extraction | Peak Detection | |||||
|---|---|---|---|---|---|---|---|---|
| Parameter | ||||||||
| Unit | Pixels | Pixels | - | Frames | % | Frames | - | - |
| Value | 1.0 | 2.5 | 0.0040 | 10 | 30 | 8 | 5.0 | 0.2 |
| Operation | Signal Extraction | Peak Detection | |||
|---|---|---|---|---|---|
| Parameter | |||||
| Unit | Frames | % | Frames | - | - |
| Value | 10 | 30 | 20 | 5.0 | 0.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raaijmakers, E.A.L.; Wanders, N.; Mestrom, R.M.C.; Luttge, R. Analyzing Developing Brain-On-Chip Cultures with the CALIMA Calcium Imaging Tool. Micromachines 2021, 12, 412. https://doi.org/10.3390/mi12040412
Raaijmakers EAL, Wanders N, Mestrom RMC, Luttge R. Analyzing Developing Brain-On-Chip Cultures with the CALIMA Calcium Imaging Tool. Micromachines. 2021; 12(4):412. https://doi.org/10.3390/mi12040412
Chicago/Turabian StyleRaaijmakers, Elles A. L., Nikki Wanders, Rob M. C. Mestrom, and Regina Luttge. 2021. "Analyzing Developing Brain-On-Chip Cultures with the CALIMA Calcium Imaging Tool" Micromachines 12, no. 4: 412. https://doi.org/10.3390/mi12040412
APA StyleRaaijmakers, E. A. L., Wanders, N., Mestrom, R. M. C., & Luttge, R. (2021). Analyzing Developing Brain-On-Chip Cultures with the CALIMA Calcium Imaging Tool. Micromachines, 12(4), 412. https://doi.org/10.3390/mi12040412






