A Parallelized Nanofluidic Device for High-Throughput Optical DNA Mapping of Bacterial Plasmids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Nanofluidics and System Automation
2.2. Sample Preparation
2.2.1. λ-DNA at Different Ionic Strengths
2.2.2. Plasmid Samples for ODM
2.2.3. Clinical Fecal Samples
2.3. Data Analysis
3. Results and Discussion
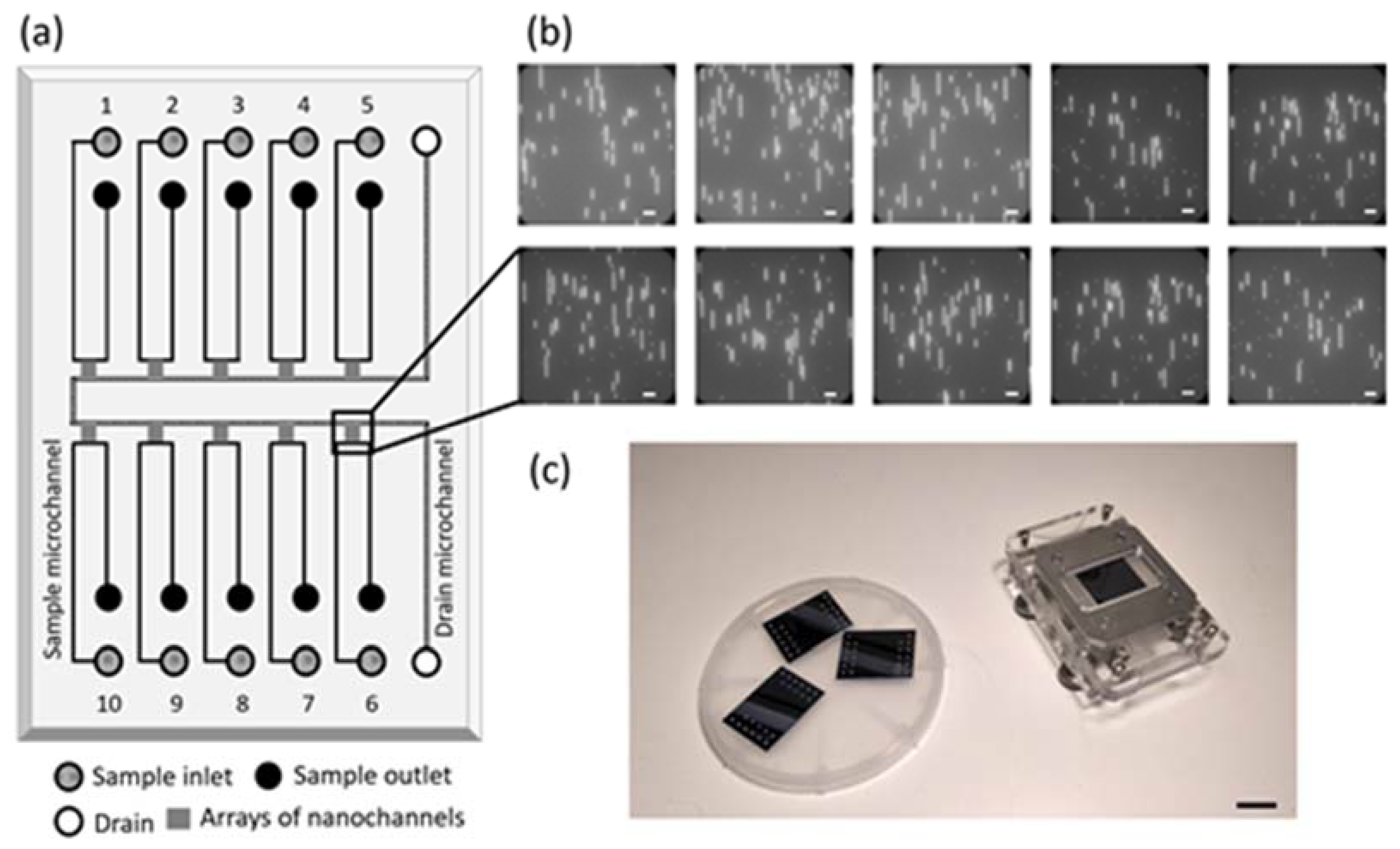
3.1. Nanofluidic Device
3.2. Automation of Pressure-Driven Flow and Imaging
3.3. DNA Stretching at Varying Ionic Strengths
3.4. Proof-of-Concept ODM Experiment
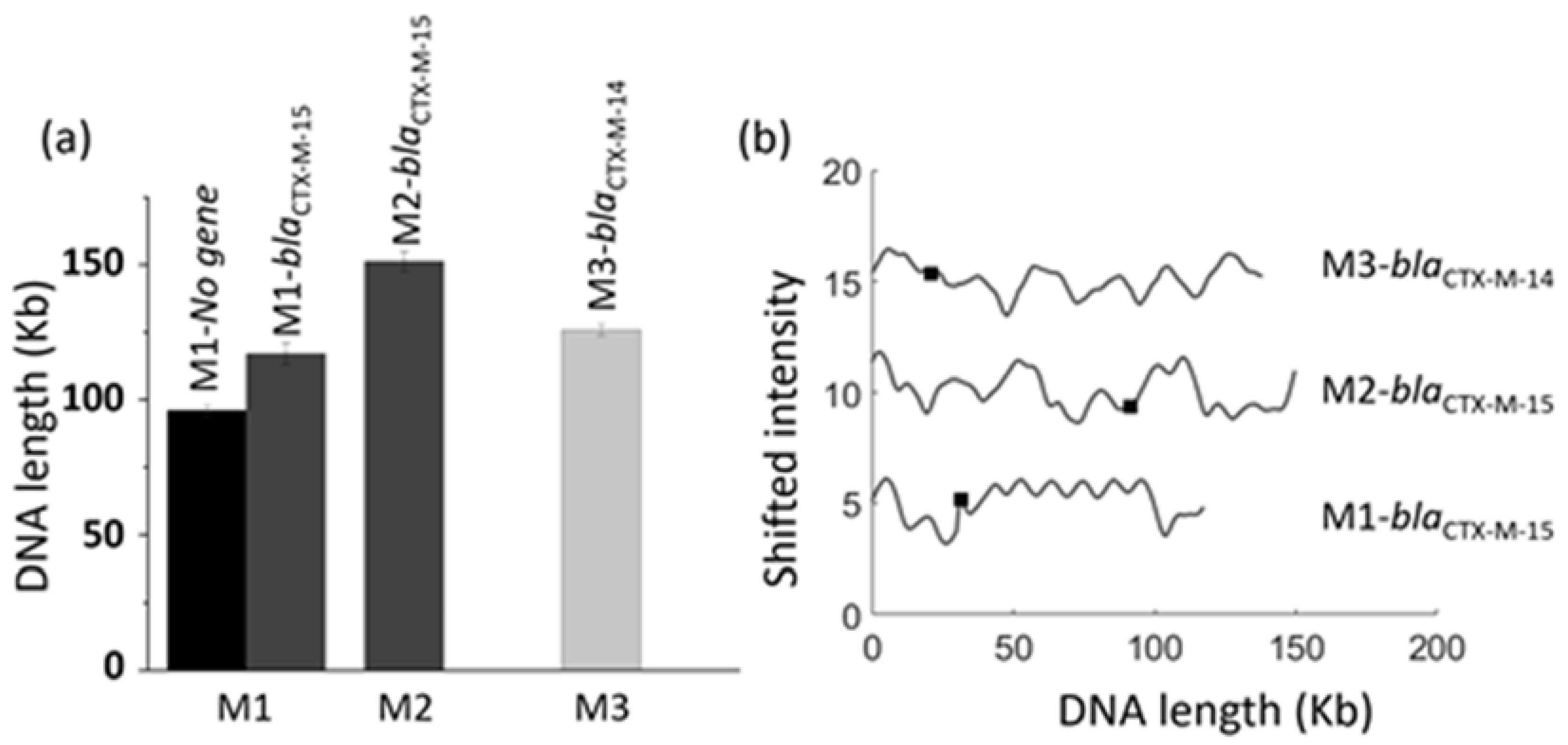
3.5. Identification of Multiple Resistance Genes on Plasmids
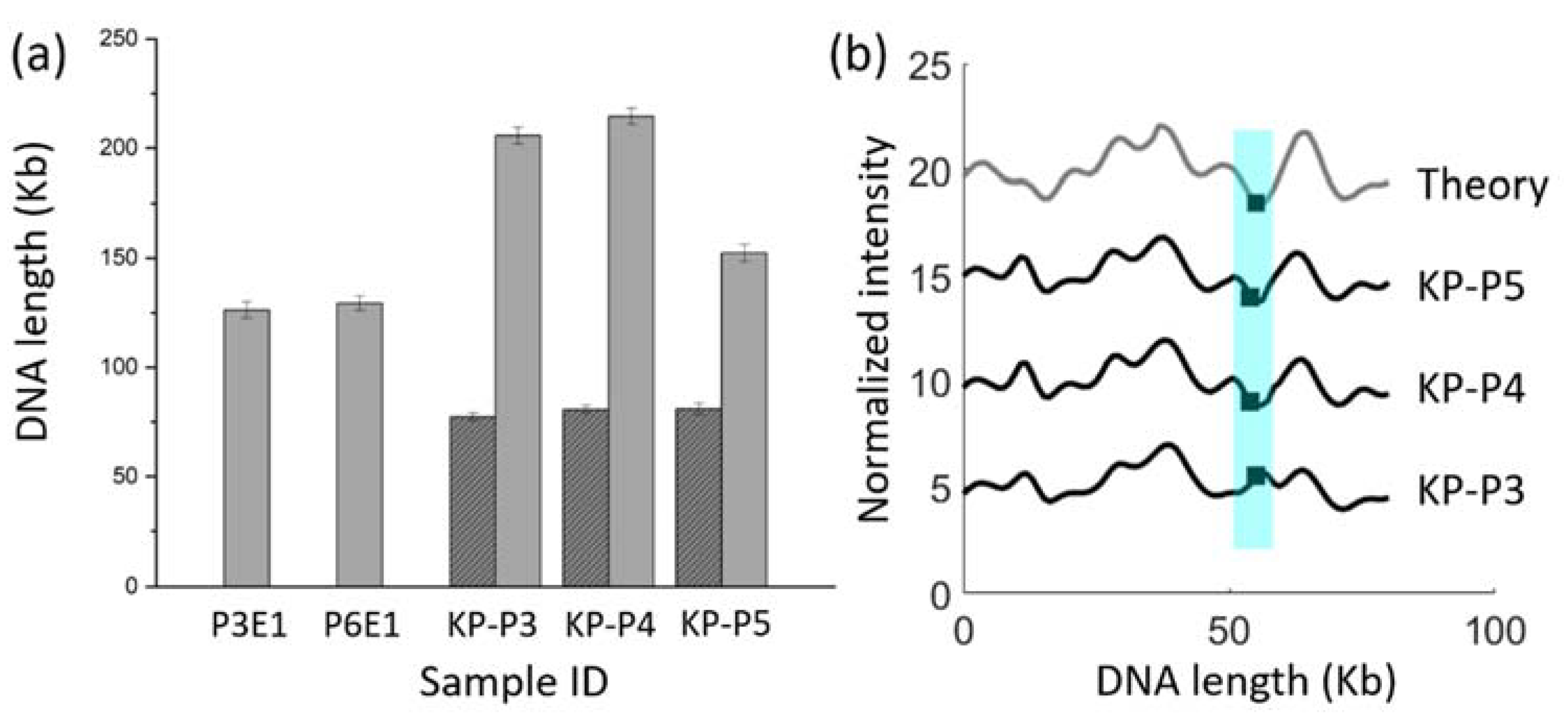
3.6. Antibiotic Resistance Gene Identification in Clinical Fecal Isolates
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meng, X.; Benson, K.; Chada, K.; Huff, E.J.; Schwartz, D.C. Optical mapping of lambda bacteriophage clones using restriction endonucleases. Nat. Genet. 1995, 9, 432–438. [Google Scholar] [CrossRef]
- Samad, A.H.; Cai, W.W.; Hu, X.; Irvin, B.; Jing, J.; Reed, J.; Meng, X.; Huang, J.; Huff, E.; Porter, B.; et al. Mapping the genome one molecule at a time—Optical mapping. Nat. Cell Biol. 1995, 378, 516–517. [Google Scholar] [CrossRef]
- Ebenstein, Y.; Gassman, N.; Kim, S.; Antelman, J.; Kim, Y.; Ho, S.; Samuel, R.; Michalet, X.; Weiss, S. Lighting up individual DNA binding proteins with quantum dots. Nano Lett. 2009, 9, 1598–1603. [Google Scholar] [CrossRef] [Green Version]
- Jing, J.; Reed, J.; Huang, J.; Hu, X.; Clarke, V.; Edington, J.; Housman, D.; Anantharaman, T.S.; Huff, E.J.; Mishra, B.; et al. Automated high resolution optical mapping using arrayed, fluid-fixed DNA molecules. Proc. Natl. Acad. Sci. USA 1998, 95, 8046–8051. [Google Scholar] [CrossRef] [Green Version]
- Labit, H.; Goldar, A.; Guilbaud, G.; Douarche, C.; Hyrien, O.; Marheineke, K. A simple and optimized method of producing silanized surfaces for FISH and replication mapping on combed DNA fibers. BioTechniques 2008, 45, 649–658. [Google Scholar] [CrossRef]
- Müller, V.; Westerlund, F. Optical DNA mapping in nanofluidic devices: Principles and applications. Lab Chip 2017, 17, 579–590. [Google Scholar] [CrossRef]
- Sriram, K.K.; Yeh, J.-W.; Lin, Y.-L.; Chang, Y.-R.; Chou, C.-F. Direct optical mapping of transcription factor binding sites on field-stretched λ-DNA in nanofluidic devices. Nucleic Acids Res. 2014, 42, e85. [Google Scholar] [CrossRef] [PubMed]
- Tegenfeldt, J.; Prinz, C.; Cao, H.; Chou, S.; Reisner, W.W.; Riehn, R.; Wang, Y.M.; Cox, E.C.; Sturm, J.C.; Silberzan, P.; et al. The dynamics of genomic-length DNA molecules in 100-nm channels. Proc. Natl. Acad. Sci. USA 2004, 101, 10979–10983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Zhang, F.; A van Kan, J.; van der Maarel, J.R. Effects of electrostatic screening on the conformation of single DNA molecules confined in a nanochannel. J. Chem. Phys. 2008, 128, 225109. [Google Scholar] [CrossRef]
- Persson, F.; Tegenfeldt, J.O. DNA in nanochannels—Directly visualizing genomic information. Chem. Soc. Rev. 2010, 39, 985. [Google Scholar] [CrossRef] [PubMed]
- Reisner, W.; Pedersen, J.N.; Austin, R.H. DNA confinement in nanochannels: Physics and biological applications. Rep. Prog. Phys. 2012, 75, 106601. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Tegenfeldt, J.O.; Reisner, W.; Riehn, R.; Guan, X.-J.; Guo, L.; Golding, I.; Cox, E.C.; Sturm, J.; Austin, R.H. Single-molecule studies of repressor-DNA interactions show long-range interactions. Proc. Natl. Acad. Sci. USA 2005, 102, 9796–9801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyberg, L.; Persson, F.; Åkerman, B.; Westerlund, F. Heterogeneous staining: A tool for studies of how fluorescent dyes affect the physical properties of DNA. Nucleic Acids Res. 2013, 41, e184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyberg, L.K.; Persson, F.; Berg, J.; Bergström, J.; Fransson, E.; Olsson, L.; Persson, M.; Stålnacke, A.; Wigenius, J.; Tegenfeldt, J.O.; et al. A single-step competitive binding assay for mapping of single DNA molecules. Biochem. Biophys. Res. Commun. 2012, 417, 404–408. [Google Scholar] [CrossRef]
- Gabrieli, T.; Sharim, H.; Nifker, G.; Jeffet, J.; Shahal, T.; Arielly, R.; Levi-Sakin, M.; Hoch, L.; Arbib, N.; Michaeli, Y.; et al. Epigenetic optical mapping of 5-hydroxymethylcytosine in nanochannel arrays. ACS Nano 2018, 12, 7148–7158. [Google Scholar] [CrossRef]
- Das, S.K.; Austin, M.D.; Akana, M.C.; Deshpande, P.; Cao, H.; Xiao, M. Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes. Nucleic Acids Res. 2010, 38, e177. [Google Scholar] [CrossRef]
- Grunwald, A.; Dahan, M.; Giesbertz, A.; Nilsson, A.; Nyberg, L.K.; Weinhold, E.; Ambjörnsson, T.; Westerlund, F.; Ebenstein, Y. Bacteriophage strain typing by rapid single molecule analysis. Nucleic Acids Res. 2015, 43, e117. [Google Scholar] [CrossRef]
- Nilsson, A.N.; Emilsson, G.; Nyberg, L.K.; Noble, C.; Stadler, L.S.; Fritzsche, J.; Moore, E.R.B.; Tegenfeldt, J.O.; Ambjörnsson, T.; Westerlund, F. Competitive binding-based optical DNA mapping for fast identification of bacteria—Multi-ligand transfer matrix theory and experimental applications on Escherichia coli. Nucleic Acids Res. 2014, 42, e118. [Google Scholar] [CrossRef] [Green Version]
- Bikkarolla, S.K.; Nordberg, V.; Rajer, F.; Müller, V.; Kabir, M.H.; Kk, S.; Dvirnas, A.; Ambjörnsson, T.; Giske, C.G.; Navér, L.; et al. Optical DNA mapping combined with Cas9-targeted resistance gene identification for rapid tracking of resistance plasmids in a neonatal intensive care unit outbreak. mBio 2019, 10, e00347-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, V.; Rajer, F.; Frykholm, K.; Nyberg, L.K.; Quaderi, S.; Fritzsche, J.; Kristiansson, E.; Ambjörnsson, T.; Sandegren, L.; Westerlund, F. Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Sci. Rep. 2016, 6, 37938. [Google Scholar] [CrossRef] [PubMed]
- Müller, V.; Nyblom, M.; Johnning, A.; Wrande, M.; Dvirnas, A.; Kk, S.; Giske, C.G.; Ambjornsson, T.; Sandegren, L.; Kristiansson, E.; et al. Cultivation-free typing of bacteria using optical DNA mapping. ACS Infect. Dis. 2020, 6, 1076–1084. [Google Scholar] [CrossRef] [PubMed]
- Müller, V.; Dvirnas, A.; Andersson, J.; Singh, V.; Kk, S.; Johansson, P.; Ebenstein, Y.; Ambjörnsson, T.; Westerlund, F. Enzyme-free optical DNA mapping of the human genome using competitive binding. Nucleic Acids Res. 2019, 47, e89. [Google Scholar] [CrossRef]
- World Health Organization. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Karami, N.; Kk, S.; Yazdanshenas, S.; Lin, Y.-L.; Jaén-Luchoro, D.; Ekedahl, E.; Parameshwaran, S.; Lindblom, A.; Åhrén, C.; Westerlund, F. Identity of blaCTX-M carrying plasmids in sequential ESBL-E. coli isolates from patients with recurrent urinary tract infections. Microorganisms 2021, 9, 1138. [Google Scholar] [CrossRef] [PubMed]
- Kk, S.; Sewunet, T.; Wangchinda, W.; Tangkoskul, T.; Thamlikitkul, V.; Giske, C.G.; Westerlund, F. Optical DNA mapping of plasmids reveals clonal spread of carbapenem-resistant Klebsiella pneumoniae in a Large Thai hospital. Antibiottics 2021, 10, 1029. [Google Scholar] [CrossRef]
- Lindblom, A.; Kk, S.; Müller, V.; Öz, R.; Sandström, H.; Åhrén, C.; Westerlund, F.; Karami, N. Interspecies Plasmid Transfer Appears Rare in Sequential Infections with Extended-Spectrum Β-Lactamase (Esbl)-Producing Enterobacteriaceae; Elsevier: Amsterdam, The Netherlands, 2019; Volume 93, p. 6. [Google Scholar]
- Lin, Y.-L.; Sewunet, T.; Kk, S.; Giske, C.G.; Westerlund, F. Optical maps of plasmids as a proxy for clonal spread of MDR bacteria: A case study of an outbreak in a rural Ethiopian hospital. J. Antimicrob. Chemother. 2020, 75, 2804–2811. [Google Scholar] [CrossRef]
- Öz, R.; Kk, S.; Westerlund, F. A nanofluidic device for real-time visualization of DNA—Protein interactions on the single DNA molecule level. Nanoscale 2019, 11, 2071–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dvirnas, A.; Pichler, C.; Stewart, C.L.; Quaderi, S.; Nyberg, L.K.; Müller, V.; Bikkarolla, S.K.; Kristiansson, E.; Sandegren, L.; Westerlund, F.; et al. Facilitated sequence assembly using densely labeled optical DNA barcodes: A combinatorial auction approach. PLoS ONE 2018, 13, e0193900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alizadehheidari, M.; Werner, E.; Noble, C.; Reiter-Schad, M.; Nyberg, L.K.; Fritzsche, J.; Mehlig, B.; Tegenfeldt, J.O.; Ambjörnsson, T.; Persson, F.; et al. Nanoconfined circular and linear DNA: Equilibrium conformations and unfolding kinetics. Macromolecules 2015, 48, 871–878. [Google Scholar] [CrossRef] [Green Version]
- Iarko, V.; Werner, E.; Nyberg, L.K.; Müller, V.; Fritzsche, J.; Ambjörnsson, T.; Beech, J.; Tegenfeldt, J.; Mehlig, K.; Westerlund, F.; et al. Extension of nanoconfined DNA: Quantitative comparison between experiment and theory. Phys. Rev. E 2015, 92, 062701. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Lee, Y.; Kim, Y.; Wang, C.; Park, J.; Jung, G.Y.; Chen, Y.; Chang, R.; Ikeda, S.; Sugiyama, H.; et al. Nanochannel-confined TAMRA-polypyrrole stained DNA stretching by varying the ionic strength from micromolar to millimolar concentrations. Polymers 2018, 11, 15. [Google Scholar] [CrossRef] [Green Version]
- Reisner, W.; Morton, K.J.; Riehn, R.; Wang, Y.M.; Yu, Z.; Rosen, M.; Sturm, J.C.; Chou, S.Y.; Frey, E.; Austin, R.H. Statics and dynamics of single DNA molecules confined in nanochannels. Phys. Rev. Lett. 2005, 94, 196101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brolund, A.; Rajer, F.; Giske, C.G.; Melefors, Ö.; Titelman, E.; Sandegren, L. Dynamics of resistance plasmids in extended-spectrum-β-lactamase-producing enterobacteriaceae during postinfection colonization. Antimicrob. Agents Chemother. 2019, 63, e02201-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
KK, S.; Lin, Y.-L.; Sewunet, T.; Wrande, M.; Sandegren, L.; Giske, C.G.; Westerlund, F. A Parallelized Nanofluidic Device for High-Throughput Optical DNA Mapping of Bacterial Plasmids. Micromachines 2021, 12, 1234. https://doi.org/10.3390/mi12101234
KK S, Lin Y-L, Sewunet T, Wrande M, Sandegren L, Giske CG, Westerlund F. A Parallelized Nanofluidic Device for High-Throughput Optical DNA Mapping of Bacterial Plasmids. Micromachines. 2021; 12(10):1234. https://doi.org/10.3390/mi12101234
Chicago/Turabian StyleKK, Sriram, Yii-Lih Lin, Tsegaye Sewunet, Marie Wrande, Linus Sandegren, Christian G. Giske, and Fredrik Westerlund. 2021. "A Parallelized Nanofluidic Device for High-Throughput Optical DNA Mapping of Bacterial Plasmids" Micromachines 12, no. 10: 1234. https://doi.org/10.3390/mi12101234
APA StyleKK, S., Lin, Y.-L., Sewunet, T., Wrande, M., Sandegren, L., Giske, C. G., & Westerlund, F. (2021). A Parallelized Nanofluidic Device for High-Throughput Optical DNA Mapping of Bacterial Plasmids. Micromachines, 12(10), 1234. https://doi.org/10.3390/mi12101234





