Analysis of Single Nucleotide-Mutated Single-Cancer Cells Using the Combined Technologies of Single-Cell Microarray Chips and Peptide Nucleic Acid-DNA Probes
Abstract
1. Introduction
2. Materials and Method
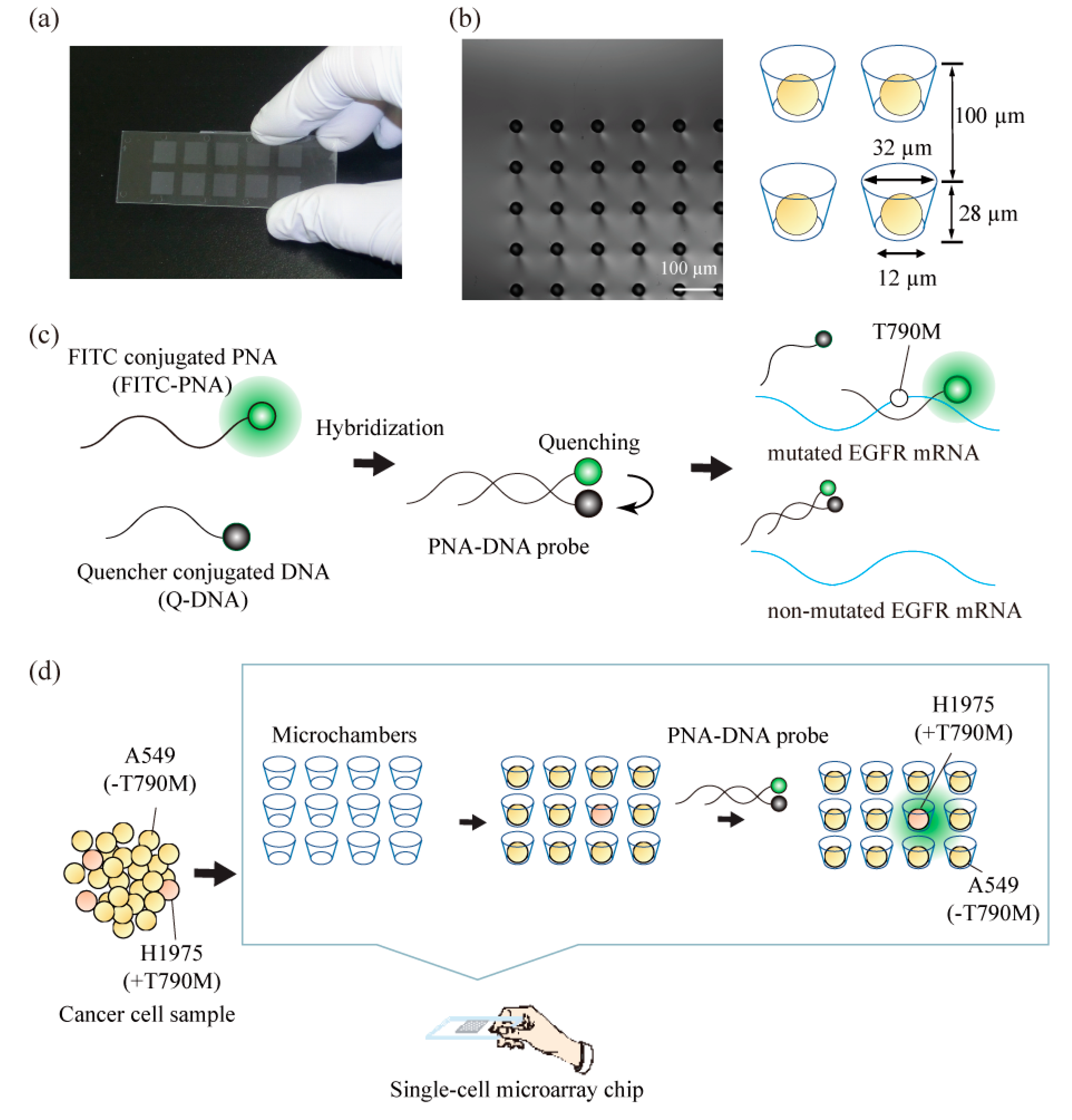
2.1. Construction of a Single-Cell Microarray Chip
2.2. Chip Surface Treatment
2.3. Construction of PNA-DNA Probes
2.4. Cell Culture and Preparation
2.5. Single-Cell Separation, Staining, and Analysis
3. Results and Discussion
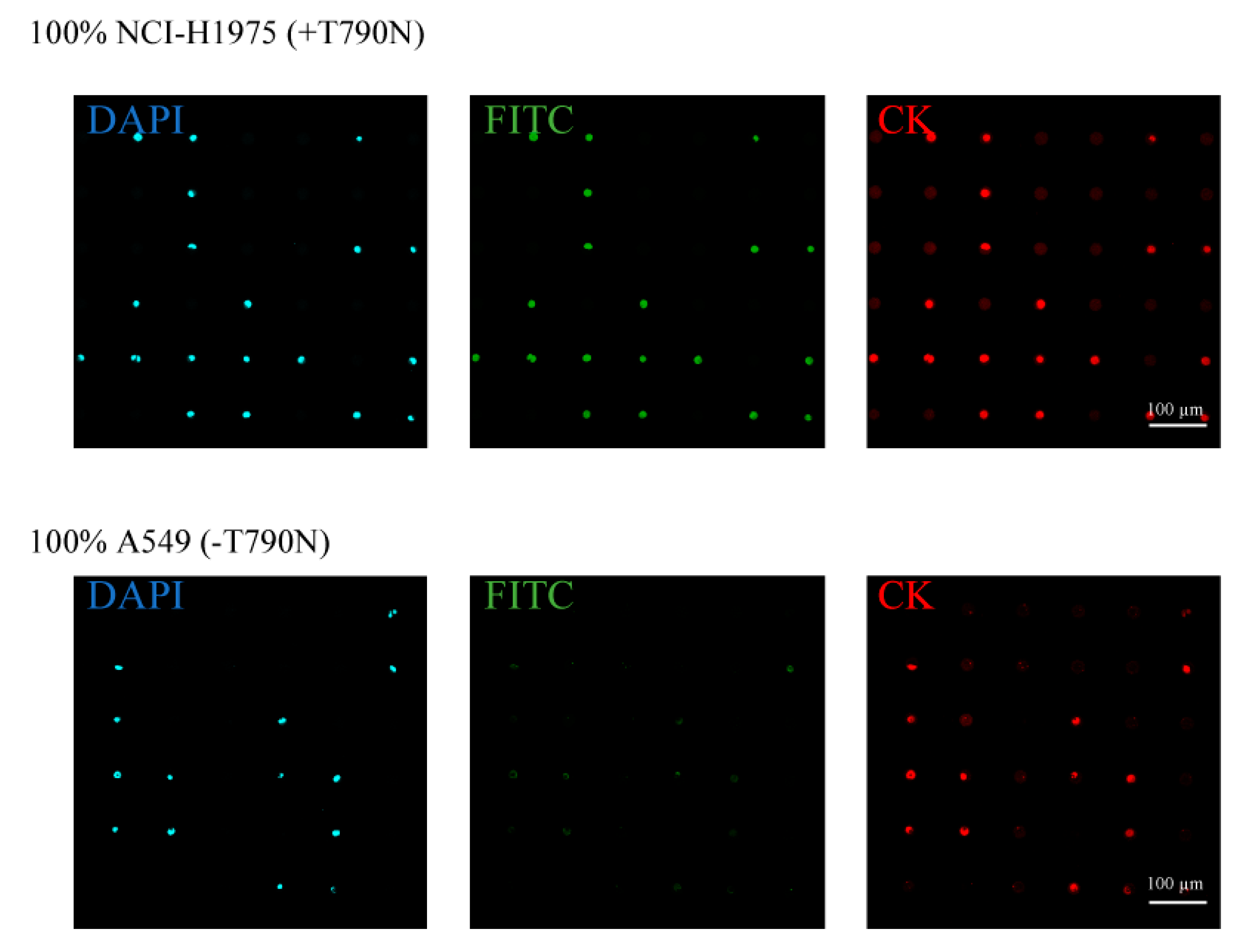
3.1. Separation and Analysis of the Different Lung Cancer Cell Lines on the Single-Cell Microarray Chip
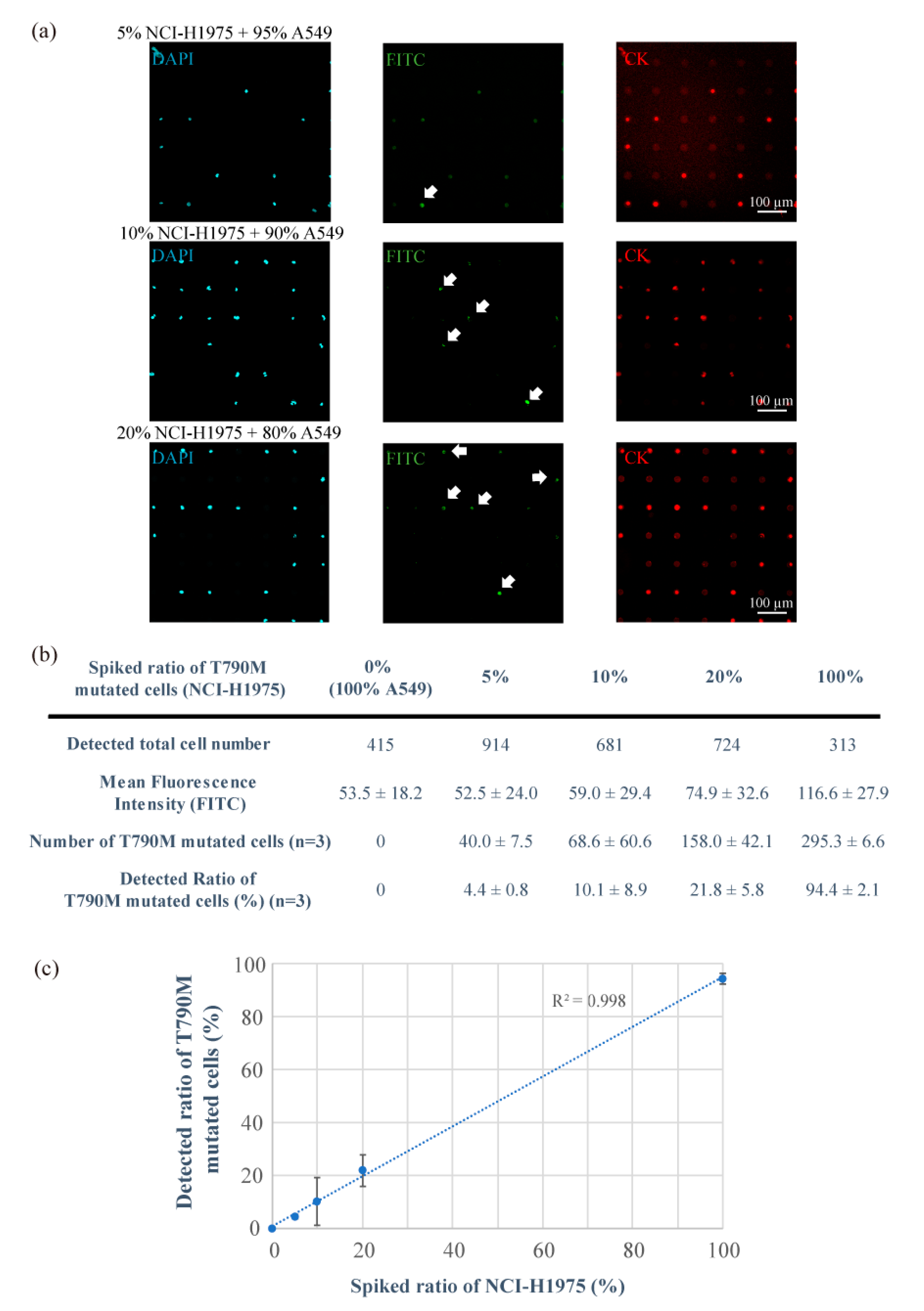
3.2. Detection of T790M-Mutated Cancer Cells Using the Combined Technologies of the Single-Cell Microarray Chip and the PNA-DNA Probe
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Di Carlo, D.; Lee, L.P. Dynamic single-cell analysis for quantitative biology. Anal. Chem. 2006, 78, 7918–7925. [Google Scholar] [CrossRef] [PubMed]
- Shigeto, H.; Yamamura, S. Bioluminescence resonance energy transfer (BRET)-based biosensing probes using novel luminescent and fluorescent protein pairs. Sens. Mater. 2019, 31, 71–78. [Google Scholar] [CrossRef]
- Shigeto, H.; Ono, T.; Ikeda, T.; Hirota, R.; Ishida, T.; Kuroda, A.; Funabashi, H. Insulin sensor cells for the analysis of insulin secretion responses in single living pancreatic β cells. Analyst 2019, 144, 3765–3772. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, S.; Kishi, H.; Tokimitsu, Y.; Kondo, S.; Honda, R.; Ramachandra Rao, S.; Omori, M.; Tamiya, E.; Muraguchi, A. Single-cell microarray for analyzing cellular response. Anal. Chem. 2005, 77, 8050–8056. [Google Scholar] [CrossRef]
- Yatsushiro, S.; Yamamura, S.; Yamaguchi, Y.; Shinohara, Y.; Tamiya, E.; Horii, T.; Baba, Y.; Kataoka, M. Rapid and highly sensitive detection of malaria-infected erythrocytes using a cell microarray chip. PLoS ONE 2010, 5, e13179. [Google Scholar] [CrossRef]
- Yamamura, S.; Yatsushiro, S.; Yamaguchi, Y.; Abe, K.; Shinohara, Y.; Tamiya, E.; Baba, Y.; Kataoka, M. Accurate detection of carcinoma cells by use of a cell microarray chip. PLoS ONE 2012, 7, e32370. [Google Scholar] [CrossRef]
- Yamamura, S.; Yamada, E.; Kimura, F.; Miyajima, K.; Shigeto, H. Separation and analysis of adherent and non-adherent cancer cells using a single-cell microarray chip. Sensors 2017, 17, 2410. [Google Scholar] [CrossRef]
- Sawada, T.; Araki, J.; Yamashita, T.; Masubuchi, M.; Chiyoda, T.; Yunokawa, M.; Hoshi, K.; Tao, S.; Yamamura, S.; Yatsushiro, S.; et al. Prognostic Impact of Circulating Tumor Cell Detected Using a Novel Fluidic Cell Microarray Chip System in Patients with Breast Cancer. EBioMedicine 2016, 11, 173–182. [Google Scholar] [CrossRef]
- Yatsushiro, S.; Yamamoto, T.; Yamamura, S.; Abe, K.; Obana, E.; Nogami, T.; Hayashi, T.; Sesei, T.; Oka, H.; Okello-Onen, J.; et al. Application of a cell microarray chip system for accurate, highly sensitive, and rapid diagnosis for malaria in Uganda. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Thorsen, T.; Maerkl, S.J.; Quake, S.R. Microfluidic Large-Scale Integration. Science 2002, 298, 580–585. [Google Scholar] [CrossRef]
- Rao, S.R.; Yamamura, S.; Takamura, Y.; Tamiya, E. Multiplexed microfluidic devices for single-cell manipulation and analysis. In Proceedings of the Micro Total Analysis Systems 2004, Malmö, Sweden, 26–30 September 2004; pp. 61–63. [Google Scholar]
- Kung, Y.C.; Huang, K.W.; Fan, Y.J.; Chiou, P.Y. Fabrication of 3D high aspect ratio PDMS microfluidic networks with a hybrid stamp. Lab. Chip 2015, 15, 1861–1868. [Google Scholar] [CrossRef] [PubMed]
- Kung, Y.C.; Huang, K.W.; Chong, W.; Chiou, P.Y. Tunnel Dielectrophoresis for Tunable, Single-Stream Cell Focusing in Physiological Buffers in High-Speed Microfluidic Flows. Small 2016, 12, 4343–4348. [Google Scholar] [CrossRef] [PubMed]
- Brevet, M.; Arcila, M.; Ladanyi, M. Assessment of EGFR mutation status in lung adenocarcinoma by immunohistochemistry using antibodies specific to the two major forms of mutant EGFR. J. Mol. Diagn. 2010, 12, 169–176. [Google Scholar] [CrossRef]
- Mellors, R.C. The Application of Labeled Antibody Technics in Studying Cell Antigens. Cancer Res. 1968, 28, 1372–1381. [Google Scholar] [PubMed]
- Hicks, J.M. Fluorescence immunoassay. Hum. Pathol. 1984, 15, 112–116. [Google Scholar] [CrossRef]
- Tawa, K.; Yamamura, S.; Sasakawa, C.; Shibata, I.; Kataoka, M. Sensitive Detection of Cell Surface Membrane Proteins in Living Breast Cancer Cells Using Multicolor Fluorescence Microscopy with a Plasmonic Chip. ACS Appl. Mater. Interfaces 2016, 8, 29893–29898. [Google Scholar] [CrossRef]
- Izumi, S.; Yamamura, S.; Hayashi, N.; Toma, M.; Tawa, K. Dual-color fluorescence imaging of EpCAM and EGFR in breast cancer cells with a bull’s eye-type plasmonic chip. Sensors 2017, 17, 2942. [Google Scholar] [CrossRef]
- Tyagi, S.; Kramer, F.R. Molecular Beacons: Probes that Fluoresce upon Hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef]
- Tan, W.; Wang, K.; Drake, T.J. Molecular beacons. Curr. Opin. Chem. Biol. 2004, 8, 547–553. [Google Scholar] [CrossRef]
- Santangelo, P.; Nitin, N.; Bao, G. Nanostructured probes for RNA detection in living cells. Ann. Biomed. Eng. 2006, 34, 39–50. [Google Scholar] [CrossRef]
- Funabashi, H.; Shigeto, H.; Nakatsuka, K.; Kuroda, A. A FRET-based DNA nano-tweezer technique for the imaging analysis of specific mRNA. Analyst 2015, 140, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Shigeto, H.; Nakatsuka, K.; Ikeda, T.; Hirota, R.; Kuroda, A.; Funabashi, H. Continuous monitoring of specific mRNA expression responses with a fluorescence resonance energy transfer-based DNA nano-tweezer technique that does not require gene recombination. Anal. Chem. 2016, 88, 7894–7898. [Google Scholar] [CrossRef] [PubMed]
- Tay, C.Y.; Yuan, L.; Leong, D.T. Nature-inspired DNA nanosensor for real-time in situ detection of mRNA in living cells. ACS Nano 2015, 9, 5609–5617. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Hou, X.; Wiraja, C.; Sun, L.; Xu, Z.J.; Xu, C. Smart Magnetic Nanosensors Synthesized through Layer-by-Layer Deposition of Molecular Beacons for Noninvasive and Longitudinal Monitoring of Cellular mRNA. ACS Appl. Mater. Interfaces 2016, 8, 5877–5886. [Google Scholar] [CrossRef]
- Nakatsuka, K.; Shigeto, H.; Kuroda, A.; Funabashi, H. A split G-quadruplex-based DNA nano-tweezers structure as a signal-transducing molecule for the homogeneous detection of specific nucleic acids. Biosens. Bioelectron. 2015, 74, 222–226. [Google Scholar] [CrossRef]
- Sander, C.; Wallenborn, M.; Brandt, V.P.; Ahnert, P.; Reuschel, V.; Eisenlöffel, C.; Krupp, W.; Meixensberger, J.; Holland, H. Central neurocytoma: SNP array analyses, subtel FISH, and review of the literature. Pathol. Res. Pract. 2019, 215, 152397. [Google Scholar] [CrossRef]
- Heinrichs, S.; Look, A.T. Identification of structural aberrations in cancer by SNP array analysis. Genome Biol. 2007, 8. [Google Scholar] [CrossRef]
- Wang, Y.; Armstrong, S.A. Genome-Wide SNP Analysis in Cancer: Leukemia Shows the Way. Cancer Cell 2007, 11, 308–309. [Google Scholar] [CrossRef][Green Version]
- Cella, D.; Herbst, R.S.; Lynch, T.J.; Prager, D.; Belani, C.P.; Schiller, J.H.; Heyes, A.; Ochs, J.S.; Wolf, M.K.; Kay, A.C.; et al. Clinically meaningful improvement in symptoms and quality of life for patients with non-small-cell lung cancer receiving gefitinib in a randomized controlled trial. J. Clin. Oncol. 2005, 23, 2946–2954. [Google Scholar] [CrossRef]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non–Small-Cell Lung Cancer to Gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef]
- Yun, C.-H.; Mengwasser, K.E.; Toms, A.V.; Woo, M.S.; Greulich, H.; Wong, K.-K.; Meyerson, M.; Eck, M.J. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. USA 2008, 105, 2070–2075. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Mitsudomi, T. Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualized treatment strategy. Cancer Sci. 2016, 107, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung, cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, S.; Kukimoto-Niino, M.; Parker, L.; Handa, N.; Terada, T.; Fujimoto, T.; Terazawa, Y.; Wakiyama, M.; Sato, M.; Sano, S.; et al. Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor. Oncogene 2013, 32, 27–38. [Google Scholar] [CrossRef]
- Stewart, E.L.; Tan, S.Z.; Liu, G.; Tsao, M.-S. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations—A review. Transl. Lung Cancer Res. 2015, 4, 67–81. [Google Scholar] [CrossRef]
- Denis, M.G.; Vallée, A.; Théoleyre, S. EGFR T790M resistance mutation in non small-cell lung carcinoma. Clin. Chim. Acta 2015, 444, 81–85. [Google Scholar] [CrossRef]
- Pao, W.; Miller, V.A.; Politi, K.A.; Riely, G.J.; Somwar, R.; Zakowski, M.F.; Kris, M.G.; Varmus, H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005, 2, 0225–0235. [Google Scholar] [CrossRef]
- Sordella, R.; Vanni, I.; Rossella, V.; Truini, A.; Lazarevic, D.; Bello, M.G.D.; Alama, A.; Mora, M.; Rijavec, E.; Genova, C.; et al. Gefitinib-Sensitizing EGFR Mutations in Lung Cancer Activate Anti-Apoptotic Pathways. Science 2004, 305, 1163–1167. [Google Scholar] [CrossRef]
- Kobayashi, S.; Boggon, T.J.; Dayaram, T.; Jänne, P.A.; Kocher, O.; Meyerson, M.; Johnson, B.E.; Eck, M.J.; Tenen, D.G.; Halmos, B. EGFR Mutation and Resistance of Non–Small-Cell Lung Cancer to Gefitinib. N. Engl. J. Med. 2005, 352, 786–792. [Google Scholar] [CrossRef]
- De Biase, D.; Visani, M.; Malapelle, U.; Simonato, F.; Cesari, V.; Bellevicine, C.; Pession, A.; Troncone, G.; Fassina, A.; Tallini, G. Next-generation sequencing of lung cancer EGFR exons 18-21 allows effective molecular diagnosis of small routine samples (cytology and biopsy). PLoS ONE 2013, 8, e83607. [Google Scholar] [CrossRef]
- Sholl, L. Molecular diagnostics of lung cancer in the clinic. Transl. Lung Cancer Res. 2017, 6, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Bahassi, E.M.; Stambrook, P.J. Next-generation sequencing technologies: Breaking the sound barrier of human genetics. Mutagenesis 2014, 29, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.J.; Howard, M.; Andrews, D.F.; Agura, E.; Radich, J. Rare occurrence of N-ras point mutations in Philadelphia chromosome positive chronic myeloid leukemia. Blood 1989, 73, 1028–1032. [Google Scholar] [CrossRef]
- Bar-Eli, M.; Ahuja, H.; Gonzalez-Cadavid, N.; Foti, A.; Cline, M.J. Analysis of N-RAS exon-1 mutations in myelodysplastic syndromes by polymerase chain reaction and direct sequencing. Blood 1989, 73, 281–283. [Google Scholar] [CrossRef] [PubMed]
- Jarry, A.; Masson, D.; Cassagnau, E.; Parois, S.; Laboisse, C.; Denis, M.G. Real-time allele-specific amplification for sensitive detection of the BRAF mutation V600E. Mol. Cell. Probes 2004, 18, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Balic, M.; Stadelmeyer, E.; Ausch, C.; Wild, M.; Guelly, C.; Bauernhofer, T.; Samonigg, H.; Hoefler, G.; Dandachi, N. Evaluation of high-resolution melting analysis as a diagnostic tool to detect the BRAF V600E mutation in colorectal tumors. J. Mol. Diagn. 2009, 11, 140–147. [Google Scholar] [CrossRef]
- Shigeto, H.; Ohtsuki, T.; Iizuka, A.; Akiyama, Y.; Yamamura, S. Imaging analysis of EGFR mutated cancer cells using peptide nucleic acid (PNA)-DNA probes. Analyst 2019, 144, 4613–4621. [Google Scholar] [CrossRef]
- Nielsen, P.E. PNA technology. Appl. Biochem. Biotechnol. Part. B Mol. Biotechnol. 2004, 26, 233–248. [Google Scholar] [CrossRef]
- Pellestor, F.; Paulasova, P. The peptide nucleic acids (PNAs), powerful tools for molecular genetics and cytogenetics. Eur. J. Hum. Genet. 2004, 12, 694–700. [Google Scholar] [CrossRef]
- Wang, J. DNA biosensors based on peptide nucleic acid (PNA) recognition layers. A review. Biosens. Bioelectron. 1998, 13, 757–762. [Google Scholar] [CrossRef]
- Vilaivan, T. Fluorogenic PNA probes. Beilstein J. Org. Chem. 2018, 14, 253–281. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shigeto, H.; Yamada, E.; Kitamatsu, M.; Ohtsuki, T.; Iizuka, A.; Akiyama, Y.; Yamamura, S. Analysis of Single Nucleotide-Mutated Single-Cancer Cells Using the Combined Technologies of Single-Cell Microarray Chips and Peptide Nucleic Acid-DNA Probes. Micromachines 2020, 11, 628. https://doi.org/10.3390/mi11070628
Shigeto H, Yamada E, Kitamatsu M, Ohtsuki T, Iizuka A, Akiyama Y, Yamamura S. Analysis of Single Nucleotide-Mutated Single-Cancer Cells Using the Combined Technologies of Single-Cell Microarray Chips and Peptide Nucleic Acid-DNA Probes. Micromachines. 2020; 11(7):628. https://doi.org/10.3390/mi11070628
Chicago/Turabian StyleShigeto, Hajime, Eriko Yamada, Mizuki Kitamatsu, Takashi Ohtsuki, Akira Iizuka, Yasuto Akiyama, and Shohei Yamamura. 2020. "Analysis of Single Nucleotide-Mutated Single-Cancer Cells Using the Combined Technologies of Single-Cell Microarray Chips and Peptide Nucleic Acid-DNA Probes" Micromachines 11, no. 7: 628. https://doi.org/10.3390/mi11070628
APA StyleShigeto, H., Yamada, E., Kitamatsu, M., Ohtsuki, T., Iizuka, A., Akiyama, Y., & Yamamura, S. (2020). Analysis of Single Nucleotide-Mutated Single-Cancer Cells Using the Combined Technologies of Single-Cell Microarray Chips and Peptide Nucleic Acid-DNA Probes. Micromachines, 11(7), 628. https://doi.org/10.3390/mi11070628





