Methacrylate Coatings for Titanium Surfaces to Optimize Biocompatibility †
Abstract
1. Introduction
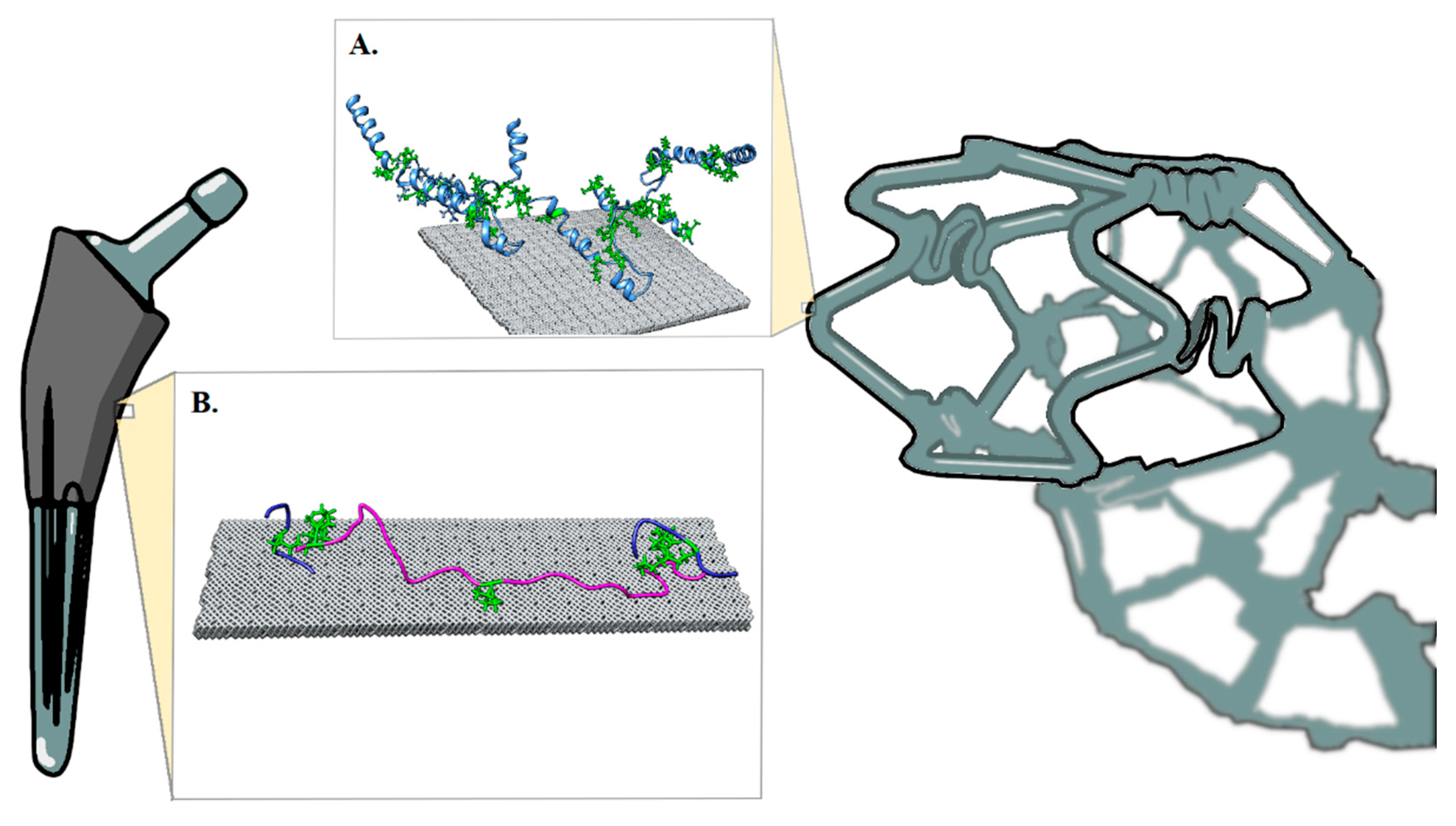
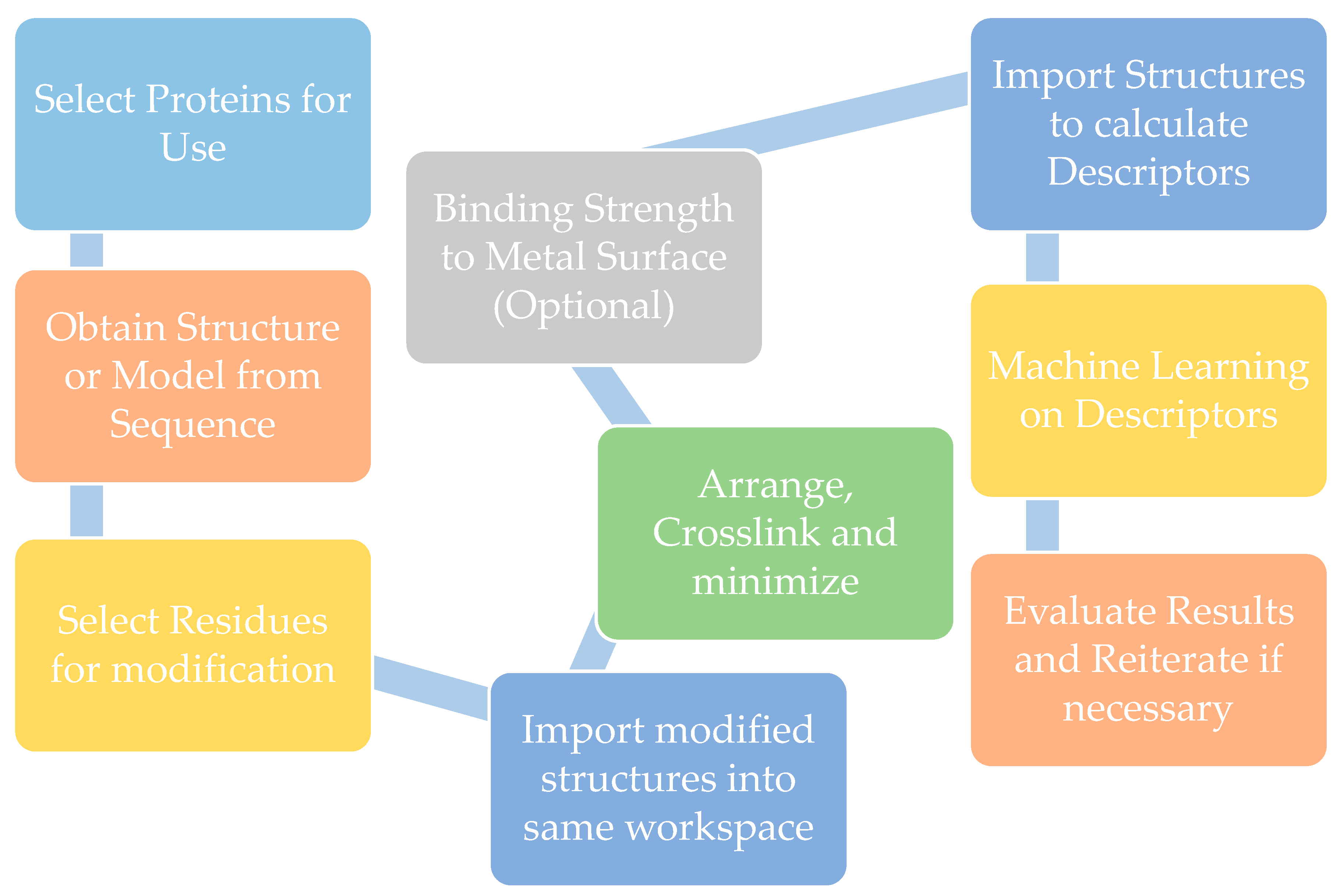
2. Biomaterial Modeling
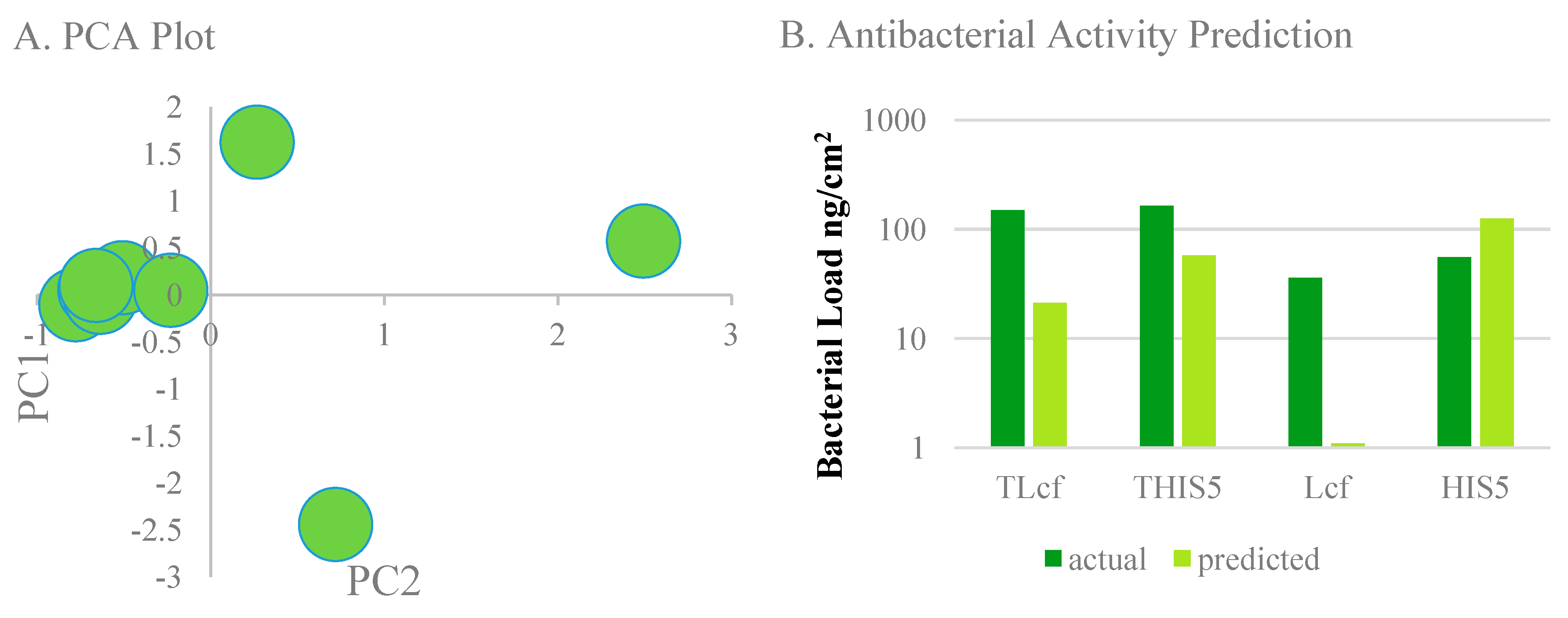
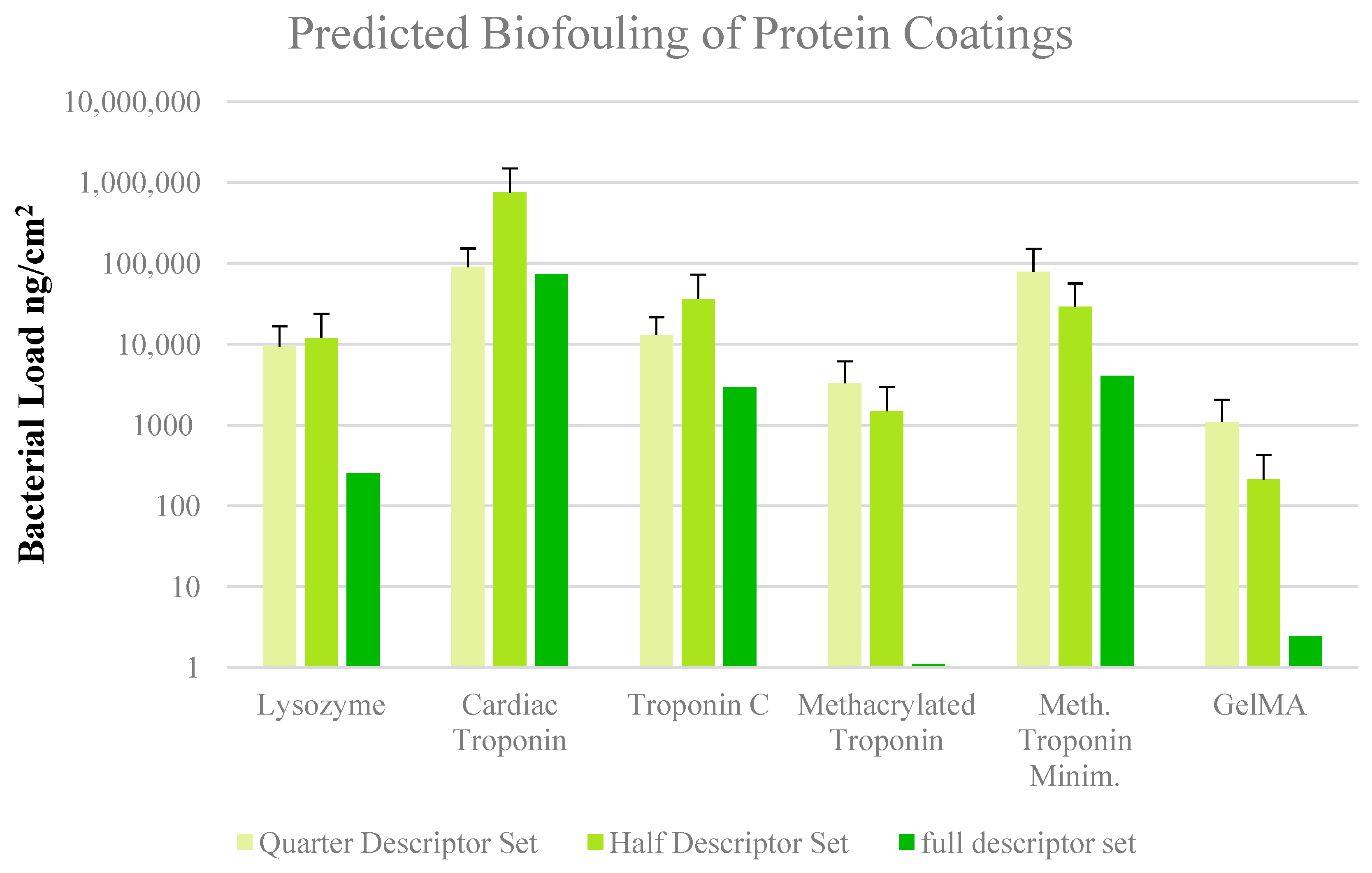
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fang, C.-H.; Tsai, P.-I.; Huang, S.-W.; Sun, J.-S.; Chang, J.Z.-C.; Shen, H.-H.; Chen, S.-Y.; Lin, F.H.; Hsu, L.-T.; Chen, Y.-C. Magnetic hyperthermia enhance the treatment efficacy of peri-implant osteomyelitis. BMC Infect. Dis. 2017, 17, 516. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Ong, K.; Lau, E.; Mowat, F.; Halpern, M. Projections of Primary and Revision Hip and Knee Arthroplasty in the United States from 2005 to 2030. J. Bone Jt. Surg. 2007, 89, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R. Advances in Bearing Materials for Total Artificial Hip Arthroplasty. In Orthopedic Biomaterials; Springer International Publishing: Cham, Switzerland, 2017; pp. 467–494. [Google Scholar]
- Davidson, D.J.; Spratt, D.; Liddle, A.D. Implant materials and prosthetic joint infection: The battle with the biofilm. EFORT Open Rev. 2019, 4, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Suhardi, V.J.; Bichara, D.A.; Kwok, S.J.J.; Freiberg, A.A.; Rubash, H.; Malchau, H.; Yun, S.H.; Muratoglu, O.K.; Oral, E. A fully functional drug-eluting joint implant. Nat. Biomed. Eng. 2017, 1, 0080. [Google Scholar] [CrossRef]
- Inzana, J.A.; Schwarz, E.M.; Kates, S.L.; Awad, H.A. Biomaterials approaches to treating implant-associated osteomyelitis. Biomaterials 2016, 81, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, C.; Liu, W.; He, X.; Zhou, N.; Zhang, D.; Gu, H.; Li, J.; Jiang, J.; Huang, W. Levofloxacin loaded mesoporous silica microspheres/nano-hydroxyapatite/polyurethane composite scaffold for the treatment of chronic osteomyelitis with bone defects. Sci. Rep. 2017, 7, 41808. [Google Scholar] [CrossRef]
- Learmonth, I.D. Biocompatibility: A biomechanical and biological concept in total hip replacement. Surgeon 2003, 1, 1–8. [Google Scholar] [CrossRef]
- Matsushita, T.; Fujibayashi, S.; Kokubo, T. Titanium foam for bone tissue engineering. In Metallic Foam Bone; Elsevier: Amsterdam, The Netherlands, 2017; pp. 111–130. [Google Scholar]
- Fujisawa, S.; Kadoma, Y. Relationships between base-catalyzed hydrolysis rates or glutathione reactivity for acrylates and methacrylates and their NMR spectra or heat of formation. Int. J. Mol. Sci. 2012, 13, 5789–5800. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, W.G.; Chung, B.G.; Park, J.H.; Khademhosseini, A. Rapid formation of acrylated microstructures by microwave-induced thermal crosslinking. Macromol. Rapid Commun. 2009, 30, 1382–1386. [Google Scholar] [CrossRef]
- Kang, S.M.; Kong, B.; Oh, E.; Choi, J.S.; Choi, I.S. Osteoconductive conjugation of bone morphogenetic protein-2 onto titanium/titanium oxide surfaces coated with non-biofouling poly(poly(ethylene glycol) methacrylate). Colloids Surf. B Biointerfaces 2010, 75, 385–389. [Google Scholar] [CrossRef]
- Bian, C.; Zhou, Y.N.; Guo, J.K.; Luo, Z.H. Aqueous Metal-Free Atom Transfer Radical Polymerization: Experiments and Model-Based Approach for Mechanistic Understanding. Macromolecules 2018, 51, 2367–2376. [Google Scholar] [CrossRef]
- Zhang, T.; Gieseler, D.; Jordan, R. Lights on! A significant photoenhancement effect on ATRP by ambient laboratory light. Polym. Chem. 2016, 7, 775–779. [Google Scholar] [CrossRef]
- Kutahya, C.; Aykac, F.S.; Yilmaz, G.; Yagci, Y. LED and visible light-induced metal free ATRP using reducible dyes in the presence of amines. Polym. Chem. 2016, 7, 6094–6098. [Google Scholar] [CrossRef]
- Pan, X.; Fantin, M.; Yuan, F.; Matyjaszewski, K. Externally controlled atom transfer radical polymerization. Chem. Soc. Rev. 2018, 47, 5457–5490. [Google Scholar] [CrossRef] [PubMed]
- Le, T.C.; Penna, M.; Winkler, D.A.; Yarovsky, I. Quantitative design rules for protein-resistant surface coatings using machine learning. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.; Guvendiren, M.; Freeman, O.; Mao, Y.; Kohn, J. Optimization of Polymer-ECM Composite Scaffolds for Tissue Engineering: Effect of Cells and Culture Conditions on Polymeric Nanofiber Mats. J. Funct. Biomater. 2017, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Yoshinari, M.; Kato, T.; Matsuzaka, K.; Hayakawa, T.; Shiba, K. Prevention of biofilm formation on titanium surfaces modified with conjugated molecules comprised of antimicrobial and titanium-binding peptides. Biofouling 2010, 26, 103–110. [Google Scholar] [CrossRef]
- Dodo, C.G.; Senna, P.M.; Custodio, W.; Paes Leme, A.F.; Del Bel Cury, A.A. Proteome analysis of the plasma protein layer adsorbed to a rough titanium surface. Biofouling 2013, 29, 549–557. [Google Scholar] [CrossRef]
- Kohavi, D.; Badihi, L.; Rosen, G.; Steinberg, D.; Sela, M.N. An in vivo method for measuring the adsorption of plasma proteins to titanium in humans. Biofouling 2013, 29, 1215–1224. [Google Scholar] [CrossRef]
- Huang, B.; Lou, Y.; Li, T.; Lin, Z.; Sun, S.; Yuan, Y.; Liu, C.; Gu, Y. Molecular dynamics simulations of adsorption and desorption of bone morphogenetic protein-2 on textured hydroxyapatite surfaces. Acta Biomater. 2018, 80, 121–130. [Google Scholar] [CrossRef]
- Yeo, J.; Jung, G.S.; Tarakanova, A.; Martín-Martínez, F.J.; Qin, Z.; Cheng, Y.; Zhang, Y.W.; Buehler, M.J. Multiscale modeling of keratin, collagen, elastin and related human diseases: Perspectives from atomistic to coarse-grained molecular dynamics simulations. Extrem. Mech. Lett. 2018, 20, 112–124. [Google Scholar] [CrossRef]
- Gautieri, A.; Russo, A.; Vesentini, S.; Redaelli, A.; Buehler, M.J. Coarse-grained model of collagen molecules using an extended MARTINI force field. J. Chem. Theory Comput. 2010, 6, 1210–1218. [Google Scholar] [CrossRef]
- Ma, R.; Huang, D.; Zhang, T.; Luo, T. Determining influential descriptors for polymer chain conformation based on empirical force-fields and molecular dynamics simulations. Chem. Phys. Lett. 2018, 704, 49–54. [Google Scholar] [CrossRef]
- Mauri, A.; Consonni, V.; Todeschini, R. Recent Advances in QSAR Studies; Springer Science & Business Media: Berlin, Germany, 2010. [Google Scholar]
- Schneider, P.; Müller, A.T.; Gabernet, G.; Button, A.L.; Posselt, G.; Wessler, S.; Hiss, J.A.; Schneider, G. Hybrid Network Model for “Deep Learning” of Chemical Data: Application to Antimicrobial Peptides. Mol. Inform. 2017, 36, 1–7. [Google Scholar] [CrossRef]
- Wallach, I.; Dzamba, M.; Heifets, A. AtomNet: A Deep Convolutional Neural Network for Bioactivity Prediction in Structure-based Drug Discovery. arXiv 2015, arXiv:1510.02855. [Google Scholar]
- Wenzel, J.; Matter, H.; Schmidt, F. Predictive Multitask Deep Neural Network Models for ADME-Tox Properties: Learning from Large Data Sets. J. Chem. Inf. Model. 2019, 59, 1253–1268. [Google Scholar] [CrossRef]
- Hu, Q.; Feng, M.; Lai, L.; Pei, J. Prediction of Drug-Likeness Using Deep Autoencoder Neural Networks. Front. Genet. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Kong, D.; Yan, X. Industrial process deep feature representation by regularization strategy autoencoders for process monitoring. Meas. Sci. Technol. 2019, 31, 025104. [Google Scholar] [CrossRef]
- Bjerrum, E.J. SMILES Enumeration as Data Augmentation for Neural Network Modeling of Molecules. arXiv 2017, arXiv:1703.07076. [Google Scholar]
- Li, X.; Fourches, D. Inductive Transfer Learning for Molecular Activity Prediction: Next-Gen QSAR Models with MolPMoFiT. ChemRxiv 2019. [Google Scholar] [CrossRef]

| Molecule | Mass (kDa) | pI seq | pI 3D | r g | Hydrodynamic Radius |
|---|---|---|---|---|---|
| Lysozyme (253L) | 18.57 | 10.18 | 10.17 | 16.64 | 20.84 |
| Fibrinogen (3GHG) | 225.36 | 6.24 | 6.74 | 153.87 | 50.970001 |
| Troponin-C (1NCX) | 18.44 | 3.65 | 3.47 | 22.55 | 21.35 |
| Collagen (1BKV) | 7.96 | 12.6 | 10.29 | 24.8 | 13.92 |
| Troponin-T (4Y99) | 9.13 | 10.01 | 9.95 | 20.73 | 20.309999 |
| Methacrylated-Troponin monomer | 9.67 | 10.01 | 5.26 | 20.62 | 20.83 |
| Methacrylated Troponin trimer | 28.95 | 10.11 | 6.89 | 40.53 | 41.099998 |
| GelMA trimer 210aa fragment | 59.53 | 9.96 | 7.17 | 251 | 200.78999 |
| GelMA trimer 30aa fragment | 4.45 | 10.46 | 8.34 | 23.89 | 23.889999 |
| Molecule | Mobility | Net Charge | Dipole Moment | Zeta Potential |
|---|---|---|---|---|
| Lysozyme (253L) | 17 | 10.58 | 310.07001 | 30.82 |
| Fibrinogen (3GHG) | −50 | −10.87 | 1442.53 | −82.110001 |
| Troponin-C (1NCX) | −66 | −33.23 | 267.32999 | −119.47 |
| Collagen (1BKV) | 15 | 3.9300001 | 1209.37 | 28.559999 |
| Troponin-T (4Y99) | 6.5 | 5.1100001 | 707.23999 | 11.75 |
| Methacrylated Troponin monomer | −1.5 | −0.07 | 631.57001 | −2.73 |
| Methacrylated Troponin trimer | −3.7 | −5.2399998 | 722.01001 | −6.3400002 |
| GelMA trimer 210aa fragment | −0.12 | −1.3200001 | 14932.37 | −0.17 |
| GelMA trimer 30aa fragment | 5.5 | 3.01 | 597.71002 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, A.; Ashammakhi, N.; Dokmeci, M.R. Methacrylate Coatings for Titanium Surfaces to Optimize Biocompatibility. Micromachines 2020, 11, 87. https://doi.org/10.3390/mi11010087
Sun A, Ashammakhi N, Dokmeci MR. Methacrylate Coatings for Titanium Surfaces to Optimize Biocompatibility. Micromachines. 2020; 11(1):87. https://doi.org/10.3390/mi11010087
Chicago/Turabian StyleSun, Argus, Nureddin Ashammakhi, and Mehmet R. Dokmeci. 2020. "Methacrylate Coatings for Titanium Surfaces to Optimize Biocompatibility" Micromachines 11, no. 1: 87. https://doi.org/10.3390/mi11010087
APA StyleSun, A., Ashammakhi, N., & Dokmeci, M. R. (2020). Methacrylate Coatings for Titanium Surfaces to Optimize Biocompatibility. Micromachines, 11(1), 87. https://doi.org/10.3390/mi11010087







