A Cancer Spheroid Array Chip for Selecting Effective Drug
Abstract
1. Introduction
2. Materials and Methods
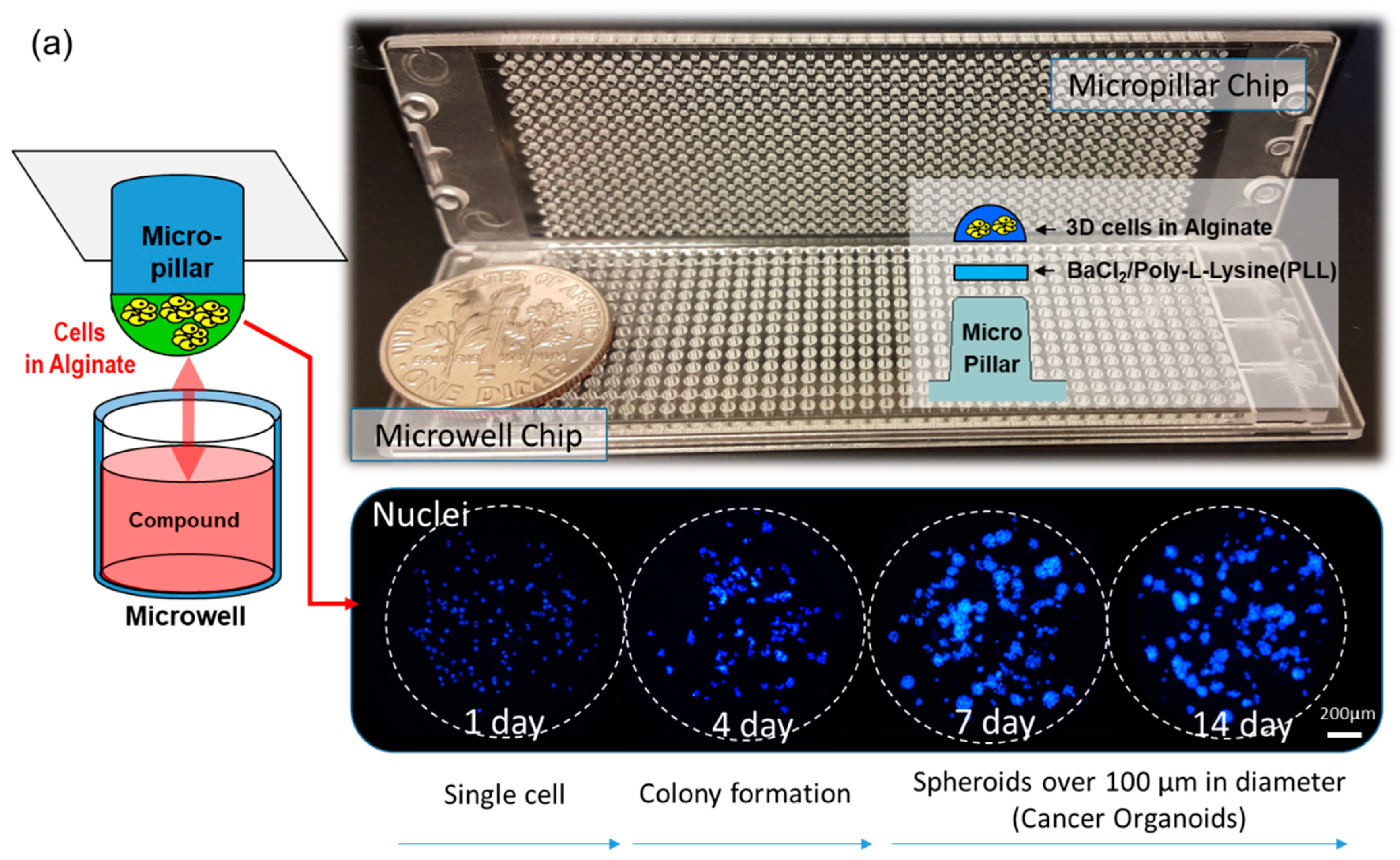
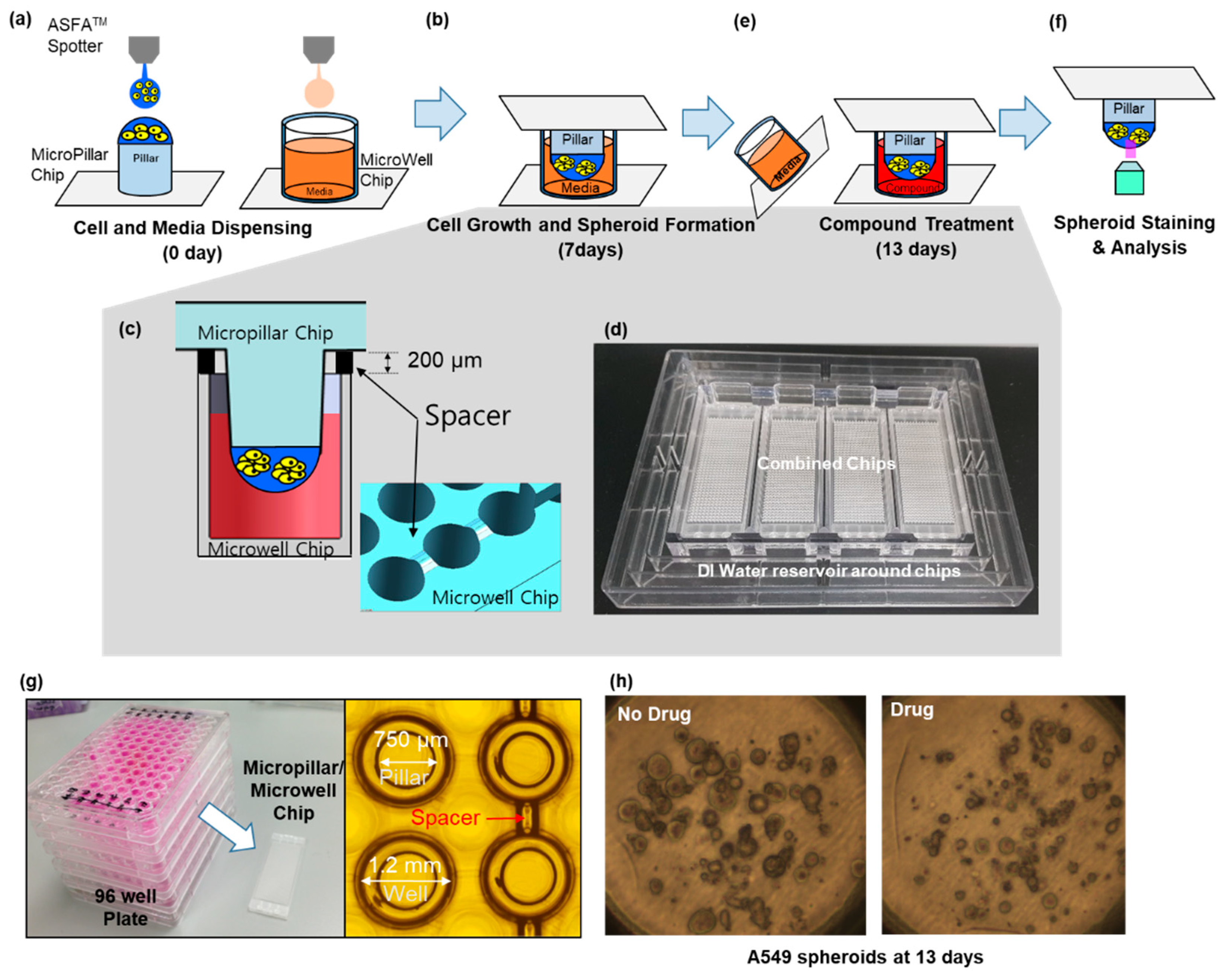
2.1. Fabrication of Micropillar/Microwell Chips and Incubation Chamber for the Spheroid Array
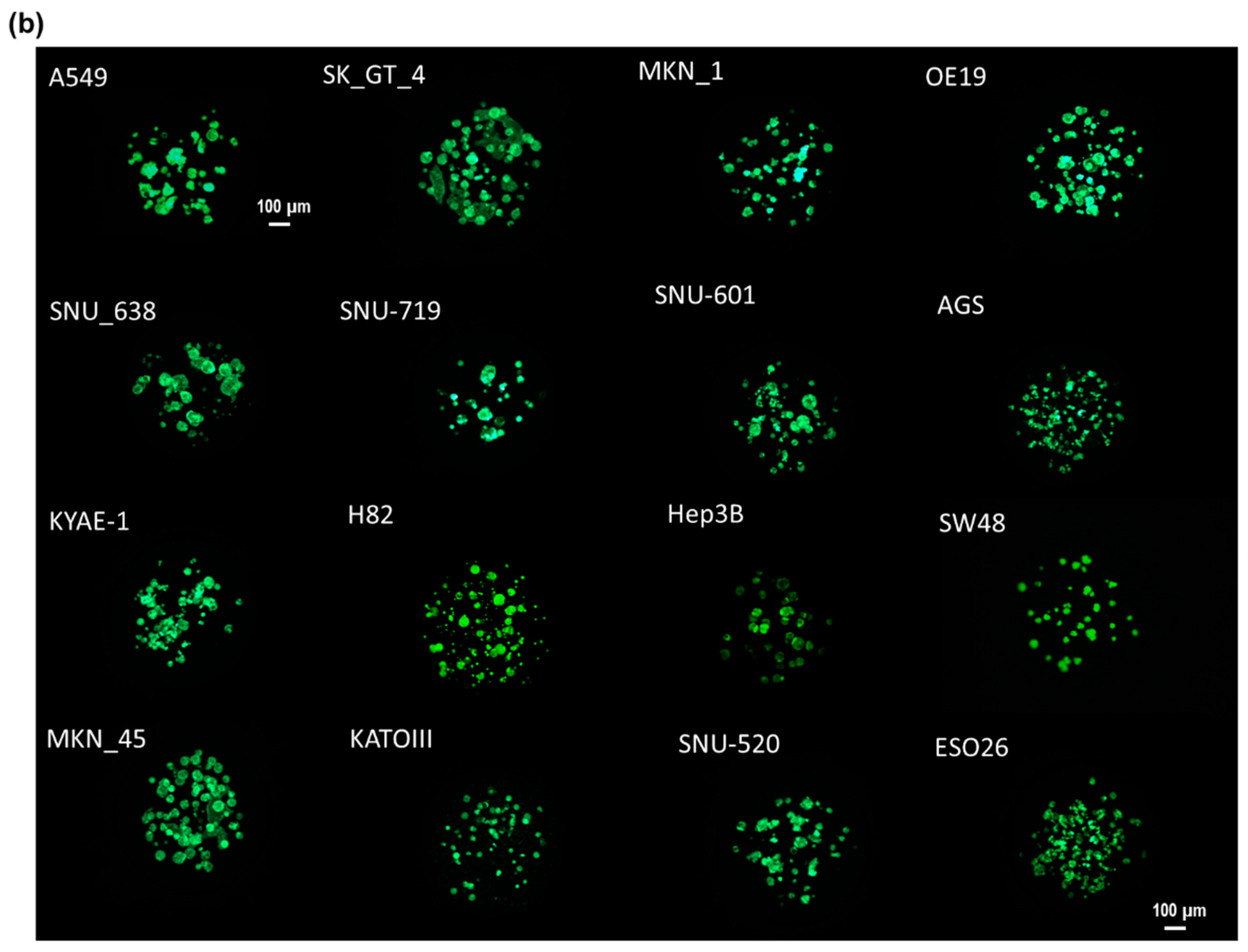
2.2. Cell Line Culture
2.3. Experimental Procedure
2.4. Comparison of Drug Response between the Single-Cell and Spheroid Models
2.5. p-EGFR Measurement
2.6. Viability Measurement
2.7. Western Blot Assay
3. Results and Discussion
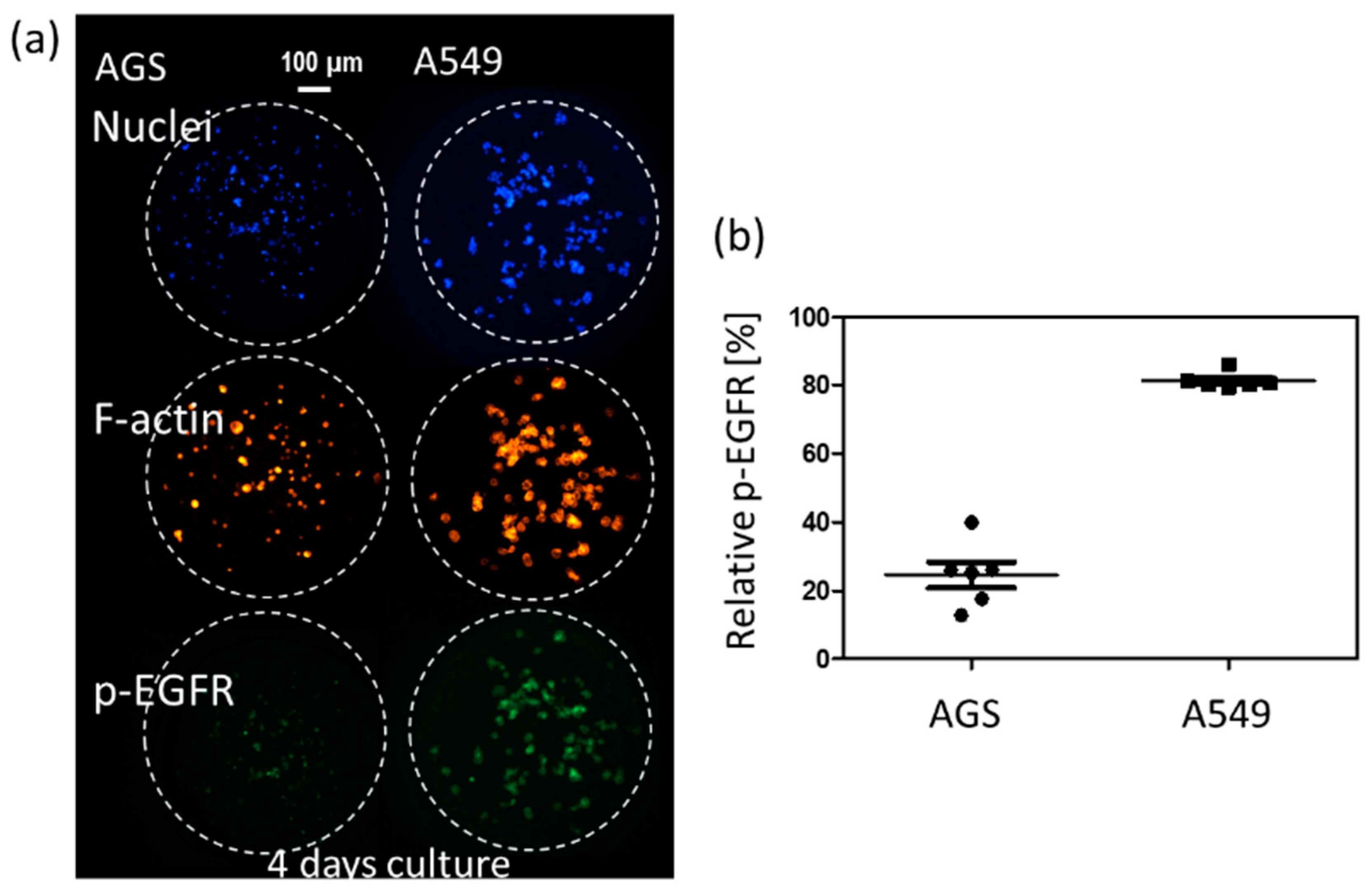
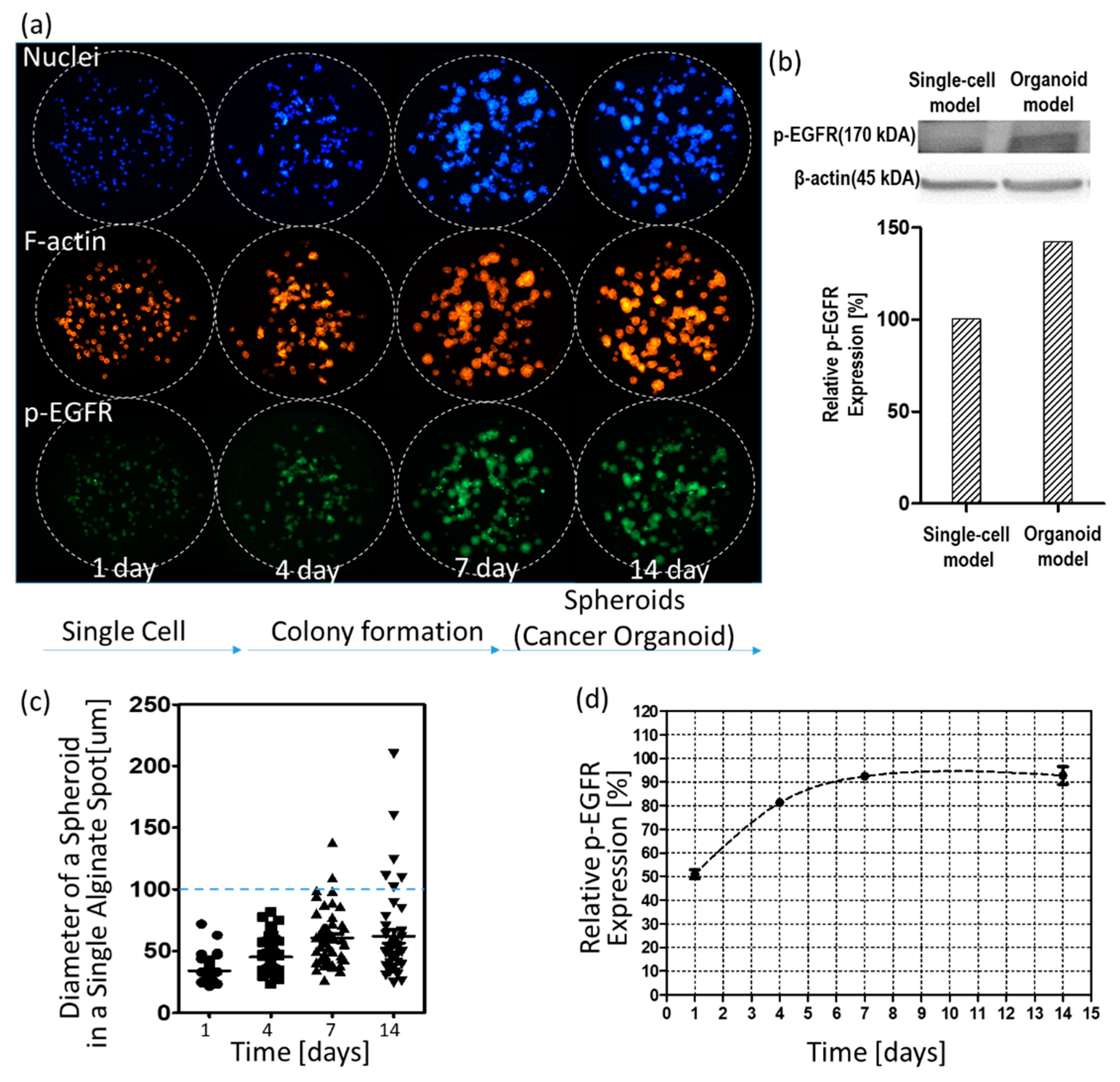
3.1. p-EGFR Expression in Spheroids
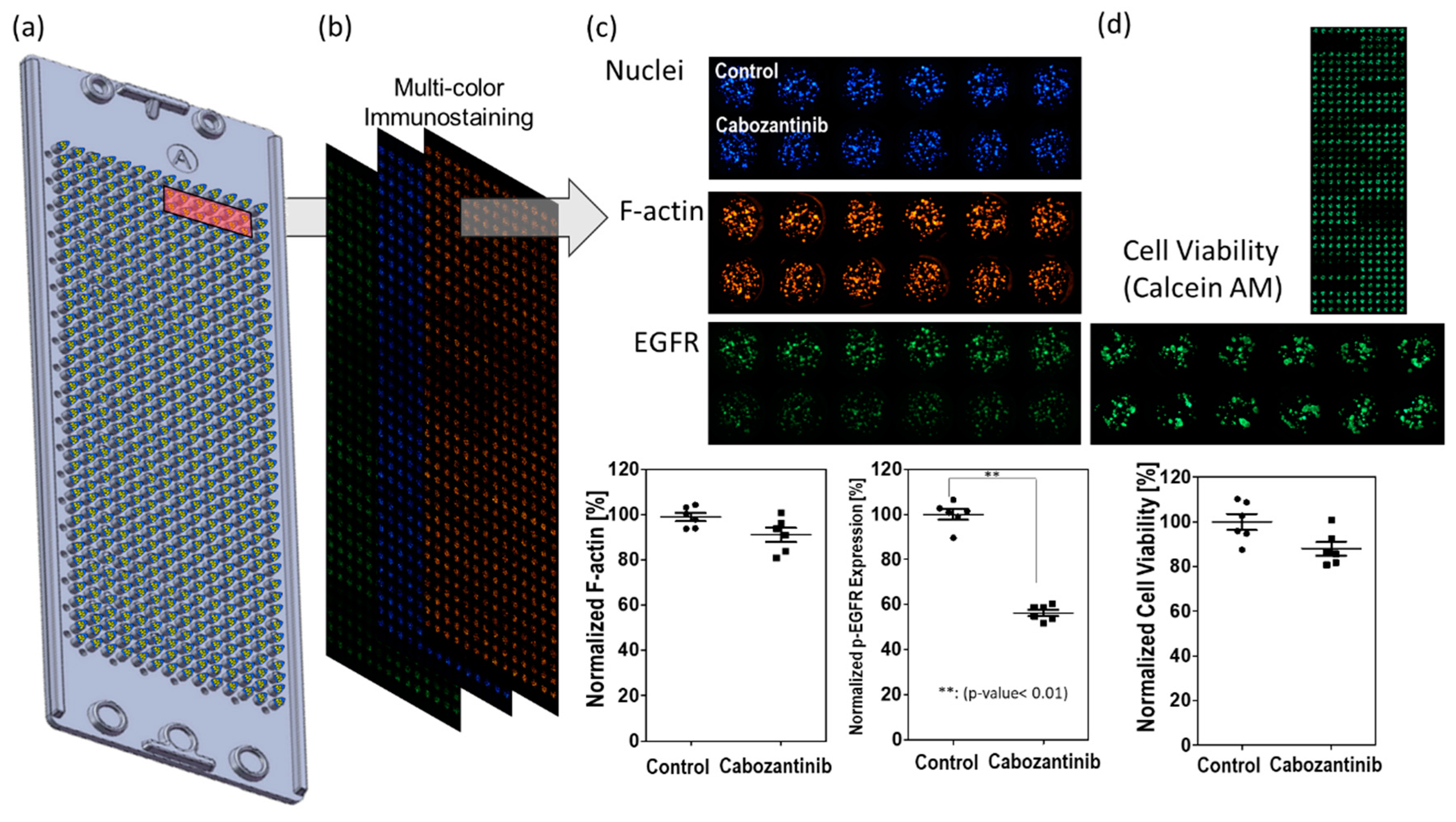
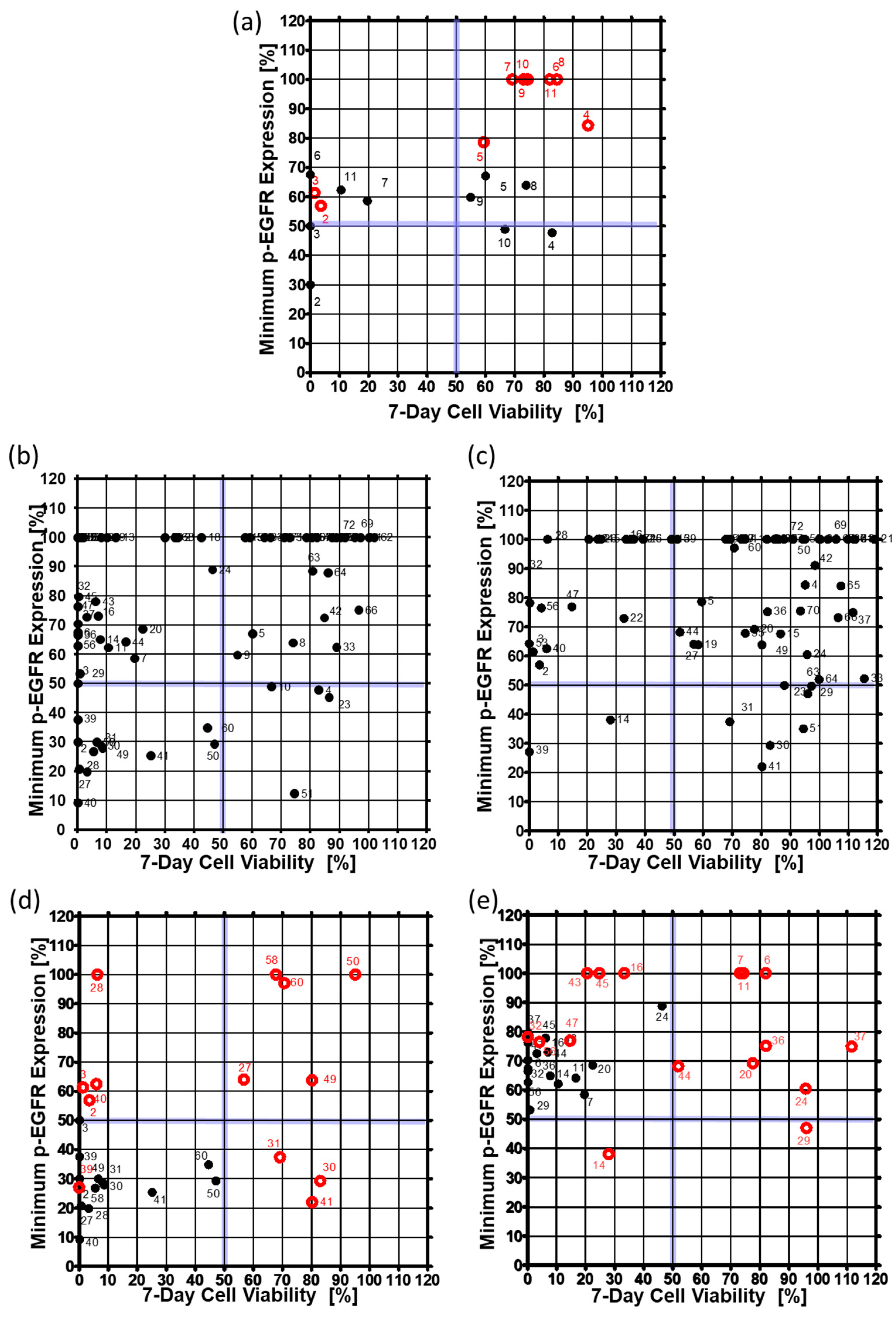
3.2. Drug Selection of Targeting p-EGFR Based on the Spheroid Model
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Li, G.N.; Livi, L.L.; Gourd, C.M.; Deweerd, E.S.; Hoffman-Kim, D. Genomic and Morphological Changes of Neuroblastoma Cells in Response to Three-Dimensional Matrices. Tissue Eng. 2007, 13, 1035–1047. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three‑Dimensional Cell Culture, A Powerful Tool in Tumor Research and Drug Discovery. Oncol. Lett. 2017, 14, 6999–7010. [Google Scholar] [CrossRef] [PubMed]
- Bokhari, M.; Carnachan, R.J.; Cameron, N.R.; Przyborski, S.A. Culture of HepG2 Liver Cells on Three Dimensional Polystyrene Scaffolds Enhances Cell Structure and Function during Toxicological Challenge. J. Anat. 2007, 211, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Elkayam, T.; Amitay-Shaprut, S.; Dvir-Ginzberg, M.; Harel, T.; Cohen, S. Enhancing the Drug Metabolism Activities of C3a—A Human Hepatocyte Cell Line—By Tissue Engineering within Alginate Scaffolds. Tissue Eng. 2006, 12, 1357–1368. [Google Scholar] [CrossRef] [PubMed]
- Gurski, L.A.; Jha, A.K.; Zhang, C.; Jia, X.; Farach-Carson, M.C. Hyaluronic Acid-Based Hydrogels as 3D Matrices for in Vitro Evaluation of Chemotherapeutic Drugs Using Poorly Adherent Prostate Cancer Cells. Biomaterials 2009, 30, 6076–6085. [Google Scholar] [CrossRef] [PubMed]
- Schyschka, L.; Sánchez, J.J.M.; Wang, Z.; Burkhardt, B.; Müller-Vieira, U.; Zeilinger, K.; Bachmann, A.; Nadalin, S.; Damm, G.; Nussler, A.K. Hepatic 3D Cultures but Not 2D Cultures Preserve Specific Transporter Activity for Acetaminophen-Induced Hepatotoxicity. Arch. Toxicol. 2013, 87, 1581–1593. [Google Scholar] [CrossRef] [PubMed]
- Es, E.; Aboulkheyr, H.; Montazeri, L.; Aref, A.R.; Vosough, M.; Baharvand, H. Personalized Cancer Medicine, an Organoid Approach. Trends Biotechnol. 2018, 36, 358–371. [Google Scholar]
- Jabs, J.; Zickgraf, F.M.; Park, J.; Wagner, S.; Jiang, X.; Jechow, K.; Kleinheinz, K.; Toprak, U.H.; Schneider, M.A.; Meister, M. Screening Drug Effects in Patient-Derived Cancer Cells Links Organoid Responses to Genome Alterations. Mol. Syst. Biol. 2017, 13, 955. [Google Scholar] [CrossRef]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L. Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Berkers, G.; Kruisselbrink, E.; Vonk, A.; de Jonge, H.R.; Janssens, H.M.; Bronsveld, I.; van de Graaf, E.A.; Nieuwenhuis, E.E.S.; Houwen, R.H.J. Characterizing Responses to CFTR-Modulating Drugs Using Rectal Organoids Derived from Subjects with Cystic Fibrosis. Sci. Transl. Med. 2016, 8, 344ra84. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Wiegerinck, C.L.; de Jonge, H.R.; Bronsveld, I.; Janssens, H.M.; de Groot, K.M.; Brandsma, A.M.; de Jong, N.W.M.; Bijvelds, M.J.C.; Scholte, B.J. A Functional Cftr Assay Using Primary Cystic Fibrosis Intestinal Organoids. Nat. Med. 2013, 19, 939. [Google Scholar] [CrossRef] [PubMed]
- Freedman, B.S.; Brooks, C.R.; Lam, A.Q.; Fu, H.; Morizane, R.; Agrawal, V.; Saad, A.F.; Li, M.K.; Hughes, M.R.; Werff, R.V. Modelling Kidney Disease with Crispr-Mutant Kidney Organoids Derived from Human Pluripotent Epiblast Spheroids. Nat. Commun. 2015, 6, 8715. [Google Scholar] [CrossRef] [PubMed]
- Xinaris, C.; Brizi, V.; Remuzzi, G. Organoid Models and Applications in Biomedical Research. Nephron 2015, 130, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Boj, S.F.; Hwang, C.I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Clevers, H. Organoids in Cancer Research. Nat. Rev. Cancer 2018, 18, 407. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.A.; Tiriac, H.; Clevers, H.; Tuveson, D.A. Modeling Pancreatic Cancer with Organoids. Trends Cancer 2016, 2, 176–190. [Google Scholar] [CrossRef] [PubMed]
- King, S.M.; Quartuccio, S.; Hilliard, T.S.; Inoue, K.; Burdette, J.E. Alginate Hydrogels for Three-Dimensional Organ Culture of Ovaries and Oviducts. J. Vis. Exp. 2011, 52, e2804. [Google Scholar] [CrossRef] [PubMed]
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor Organoids as a Pre-Clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 2017, 24, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Astashkina, A.; Grainger, D.W. Critical Analysis of 3-D Organoid in Vitro Cell Culture Models for High-Throughput Drug Candidate Toxicity Assessments. Adv. Drug Deliv. Rev. 2014, 69, 1–18. [Google Scholar] [CrossRef]
- Verissimo, C.S.; Overmeer, R.M.; Ponsioen, B.; Drost, J.; Mertens, S.; Verlaan-Klink, I.; van Gerwen, B.; van der Ven, M.; van de Wetering, M.; Egan, D.A. Targeting Mutant Ras in Patient-Derived Colorectal Cancer Organoids by Combinatorial Drug Screening. Elife 2016, 5, e18489. [Google Scholar] [CrossRef]
- Du, G.; Fang, Q.; den Toonder, J.M.J. Microfluidics for Cell-Based High Throughput Screening Platforms—A Review. Anal. Chim. Acta 2016, 903, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Lee, J.H.; Lee, C.S.; Nam, Y. Agarose Microwell Based Neuronal Micro-Circuit Arrays on Microelectrode Arrays for High Throughput Drug Testing. Lab Chip 2009, 9, 3236–3242. [Google Scholar] [CrossRef]
- Tung, Y.C.; Hsiao, A.Y.; Allen, S.G.; Torisawa, Y.; Ho, M.; Takayama, S. High-Throughput 3D Spheroid Culture and Drug Testing Using a 384 Hanging Drop Array. Analyst 2011, 136, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Lee, D.W.; Hwang, H.J.; Yeon, S.; Lee, M.; Kuh, H. Mini-Pillar Array for Hydrogel-Supported 3D Culture and High-Content Histologic Analysis of Human Tumor Spheroids. Lab Chip 2016, 16, 2265–2276. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.; Woo, D.; Choi, Y.; Seo, Y.J.; Lee, M.; Jeon, S.Y.; Ku, B.; Kim, S.; Yi, S.H.; Nam, D. High-Throughput Screening (HTS) of Anticancer Drug Efficacy on a Micropillar/Microwell Chip Platform. Anal. Chem. 2013, 86, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Doh, I.; Nam, D.; Lee, D.W. 3D Cell-Based High-Content Screening (HCS) Using a Micropillar and Microwell Chip Platform. Anal. Chem. 2018, 90, 8354–8361. [Google Scholar] [CrossRef] [PubMed]
- Doh, I.; Kwon, Y.; Ku, B.; Lee, D.W. Drug Efficacy Comparison of 3D Forming and Preforming Sphere Models with a Micropillar and Microwell Chip Platform. SLAS Discov. Adv. Life Sci. RD 2019, 24, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.; Woo, D.; Doh, I.L.; Nam, D. Unified 2D and 3D Cell-Based High-Throughput Screening Platform Using a Micropillar/Microwell Chip. Sens. Actuators B Chem. 2016, 228, 523–528. [Google Scholar] [CrossRef]
- Wang, D.; Wang, B.; Wang, R.; Zhang, Z.; Lin, Y.; Huang, G.; Lin, S.; Jiang, Y.; Wang, W.; Wang, L. High Expression of Egfr Predicts Poor Survival in Patients with Resected T3 Stage Gastric Adenocarcinoma and Promotes Cancer Cell Survival. Oncol. Lett. 2017, 13, 3003–3013. [Google Scholar] [CrossRef][Green Version]
- Hung, M.S.; Chen, I.; Lung, J.; Lin, P.; Li, Y.; Tsai, Y. Epidermal Growth Factor Receptor Mutation Enhances Expression of Cadherin-5 in Lung Cancer Cells. PLoS ONE 2016, 11, e0158395. [Google Scholar] [CrossRef]
| Drug | Target | Single-Cell Model | Cancer Spheroid Model | Drug | Target | Single-Cell Model | Cancer Spheroid Model | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-EGFR Expression [%] | Cell Viability [%] | p-EGFR Expression [%] | Cell Viability [%] | p-EGFR Expression [%] | Cell Viability [%] | p-EGFR Expression [%] | Cell Viability [%] | ||||||||||||
| Average | SD | Average | SD | Average | SD | Average | SD | Average | SD | Average | SD | Average | SD | Average | SD | ||||
| 1_DMSO | - | 100.0 | 0.0 | 100.2 | 6.5 | 100.0 | 0.0 | 100.0 | 8.8 | 37_AZD4547 | FGFR1/2/3 | 76.4 | 9.5 | 0.0 | 0.0 | 74.9 | 22.6 | 111.6 | 20.6 |
| 2_AEE788 | EGFR | 30.0 | 8.5 | 0.0 | 0.1 | 56.9 | 16.1 | 3.6 | 1.6 | 38_BGJ398 | FGFR1/2/3 | 100.0 | 0.0 | 66.2 | 12.7 | 100.0 | 0.0 | 109.8 | 11.7 |
| 3_Afatinib | EGFR | 50.0 | 8.4 | 0.0 | 0.0 | 61.3 | 20.9 | 1.4 | 2.3 | 39_Dovitinib | Flt3, c-Kit, FGFR1/3, VEGFR1/2/3, PDGFRα/β | 37.6 | 6.9 | 0.0 | 0.0 | 27.1 | 3.6 | 0.1 | 0.1 |
| 4_BMS-599626 | EGFR | 47.7 | 8.1 | 82.8 | 7.6 | 84.3 | 6.3 | 95.1 | 15.1 | 40_Bosutinib | dual Src/Abl | 9.2 | 3.2 | 0.0 | 0.0 | 62.5 | 8.9 | 6.0 | 3.1 |
| 5_Erlotinib HCl | HER1/EGFR | 67.1 | 12.7 | 60.0 | 10.1 | 78.6 | 3.9 | 59.4 | 7.1 | 41_Dasatinib | Bcr-Abl | 25.4 | 6.8 | 25.0 | 11.1 | 22.0 | 2.6 | 80.3 | 8.0 |
| 6_Dacomitinib | EGFR | 67.5 | 16.0 | 0.0 | 0.0 | 100.0 | 0.0 | 82.0 | 9.2 | 42_Nilotinib | Bcr-Abl | 72.5 | 13.8 | 84.8 | 9.5 | 91.1 | 5.8 | 98.5 | 10.6 |
| 7_Gefitinib | EGFR | 58.6 | 6.0 | 19.5 | 19.3 | 100.0 | 0.0 | 73.2 | 13.5 | 43_AZD6244 | MEK1 | 78.0 | 20.6 | 6.1 | 1.4 | 100.0 | 0.0 | 20.6 | 6.3 |
| 8_Lapatinib | EGFR | 63.9 | 12.3 | 73.9 | 5.8 | 100.0 | 0.0 | 84.5 | 8.5 | 44_Trametinib | MEK1/2 | 64.3 | 11.7 | 16.5 | 5.4 | 68.1 | 32.0 | 52.0 | 5.4 |
| 9_Neratinib | EGFR | 59.8 | 7.2 | 54.9 | 5.0 | 100.0 | 0.0 | 73.1 | 6.0 | 45_Bortezomib | Proteasome | 79.7 | 12.1 | 0.3 | 0.3 | 100.0 | 0.0 | 24.8 | 11.7 |
| 10_CI-1033 | EGFR, HER2 | 48.9 | 12.9 | 66.6 | 6.8 | 100.0 | 0.0 | 69.2 | 10.7 | 46_Carfilzomib | Proteasome | 100.0 | 0.0 | 0.2 | 0.2 | 100.0 | 0.0 | 39.2 | 9.5 |
| 11_CO-1686 | EGFR | 62.3 | 14.3 | 10.5 | 10.7 | 100.0 | 0.0 | 74.4 | 12.5 | 47_ABT-199 | Bcl-2 | 72.7 | 8.7 | 3.1 | 0.9 | 76.8 | 13.4 | 14.7 | 3.8 |
| 12_BKM120 | PI3K | 100.0 | 0.0 | 1.9 | 0.8 | 100.0 | 0.0 | 36.1 | 4.4 | 48_ABT-888 | PARP | 100.0 | 0.0 | 82.2 | 5.4 | 100.0 | 0.0 | 112.3 | 8.7 |
| 13_BYL719 | PI3K | 100.0 | 0.0 | 13.0 | 4.2 | 100.0 | 0.0 | 49.0 | 9.1 | 49_AUY922 | HSP (e.g. HSP90) | 27.8 | 5.5 | 8.5 | 2.7 | 63.8 | 12.4 | 80.2 | 11.7 |
| 14_XL147 | PI3K | 65.1 | 11.1 | 7.7 | 2.1 | 38.0 | 8.6 | 28.0 | 3.8 | 50_Dabrafenib | BRAFV600 | 29.3 | 11.0 | 47.0 | 10.1 | 100.0 | 0.0 | 95.1 | 6.8 |
| 15_Everolimus | mTOR | 100.0 | 0.0 | 57.5 | 4.0 | 67.6 | 16.3 | 86.6 | 12.1 | 51_Ibrutinib | Btk, modestly potent to Bmx, CSK, FGR, BRK, HCK | 12.4 | 5.6 | 74.5 | 9.4 | 35.0 | 2.8 | 94.5 | 16.8 |
| 16_AZD2014 | mTOR | 73.1 | 9.6 | 6.9 | 2.1 | 100.0 | 0.0 | 33.4 | 7.6 | 52_LDE225 | Smoothened | 100.0 | 0.0 | 90.3 | 12.1 | 100.0 | 0.0 | 94.3 | 15.9 |
| 17_PF-05212384 | P3k/mTOR | 100.0 | 0.0 | 1.6 | 0.6 | 100.0 | 0.0 | 34.9 | 5.9 | 53_LDK378 | ALK | 100.0 | 0.0 | 0.0 | 0.0 | 64.2 | 19.9 | 0.0 | 0.0 |
| 18_XL765 | P3k/mTOR | 100.0 | 0.0 | 42.5 | 3.8 | 100.0 | 0.0 | 68.7 | 3.0 | 54_LGK-974 | PORCN | 100.0 | 0.0 | 72.8 | 4.5 | 100.0 | 0.0 | 81.9 | 7.3 |
| 19_BEZ235 | P3k/mTOR | 100.0 | 0.0 | 10.0 | 3.4 | 63.8 | 4.8 | 58.3 | 7.7 | 55_Olaparib | PARP1/2 | 100.0 | 0.0 | 59.2 | 5.3 | 67.8 | 18.0 | 74.5 | 17.3 |
| 20_AZD5363 | Akt1/2/3 | 68.7 | 7.5 | 22.4 | 8.0 | 69.2 | 5.5 | 77.6 | 9.0 | 56_Panobinostat | HDAC | 62.8 | 8.6 | 0.1 | 0.1 | 76.4 | 2.5 | 4.2 | 1.3 |
| 21_Axitinib | VEGFR1/2/3, PDGFRβ and c-Kit | 100.0 | 0.0 | 34.4 | 12.0 | 100.0 | 0.0 | 119.0 | 9.6 | 57_PF-04449913 | HSP90 | 100.0 | 0.0 | 80.6 | 15.1 | 100.0 | 9.2 | 87.8 | 15.2 |
| 22_Cediranib | VEGFR, Flt | 100.0 | 0.0 | 0.0 | 0.0 | 72.9 | 14.9 | 32.7 | 44.7 | 58_Ruxolitinib | JAK1/2 | 29.9 | 12.2 | 6.5 | 2.3 | 100.0 | 0.0 | 67.8 | 8.1 |
| 23_Imatinib | v-Abl, c-Kit and PDGFR | 45.3 | 9.3 | 86.4 | 3.7 | 49.9 | 10.9 | 87.9 | 7.8 | 59_Sotrastaurin | PKC | 100.0 | 0.0 | 30.0 | 8.3 | 100.0 | 0.0 | 51.0 | 14.9 |
| 24_Pazopanib HCl | VEGFR1/2/3, PDGFR, FGFR, c-Kit | 89.0 | 14.2 | 46.2 | 11.5 | 60.4 | 23.1 | 95.8 | 8.5 | 60_Vemurafenib | B-RafV600E | 34.8 | 5.5 | 44.5 | 2.8 | 97.1 | 5.8 | 70.7 | 24.4 |
| 25_Sunitinib | VEGFR2 and PDGFRβ | 100.0 | 0.0 | 0.0 | 0.0 | 100.0 | 0.0 | 39.3 | 43.6 | 61_Vismodegib | Hedgehog/smothen | 100.0 | 0.0 | 97.2 | 9.7 | 100.0 | 0.0 | 111.9 | 5.6 |
| 26_Tandutinib | FLT3, PDGFR, and KIT | 100.0 | 0.0 | 8.0 | 5.0 | 100.0 | 0.0 | 23.6 | 4.2 | 62_PHA-665752 | c-Met inhibitor | 100.0 | 0.0 | 102.0 | 11.4 | 100.0 | 0.0 | 130.0 | 13.2 |
| 27_Tivozanib | VEGFR, c-Kit, PDGFR | 19.8 | 4.6 | 3.2 | 2.2 | 64.0 | 22.2 | 56.7 | 5.9 | 63_TMZ | alkylating agent | 88.5 | 6.3 | 80.7 | 7.8 | 49.6 | 21.1 | 97.3 | 13.2 |
| 28_Regorafenib | VEGFR1/2/3, PDGFRβ, Kit, RET and Raf-1 | 20.7 | 4.8 | 0.6 | 0.7 | 100.0 | 0.0 | 6.3 | 1.1 | 64_Amoral | morpholine antifungal drug | 87.9 | 16.6 | 86.0 | 9.5 | 51.9 | 22.1 | 99.9 | 9.5 |
| 29_Vandetanib | VEGFR2 | 53.3 | 4.7 | 0.7 | 1.1 | 47.0 | 23.6 | 96.0 | 14.6 | 65_Mevas | HMG-CoA reductase inhibitor | 100.0 | 0.0 | 88.9 | 7.2 | 84.0 | 11.4 | 107.4 | 8.1 |
| 30_Cabozantinib | VEGFR2,c-Met, Ret, Kit, Flt-1/3/4, Tie2, and AXL | 28.7 | 7.0 | 8.0 | 5.1 | 29.3 | 13.3 | 83.0 | 16.8 | 66_Amio | antiarrhythmic medication | 75.2 | 27.9 | 96.6 | 3.9 | 73.1 | 10.7 | 106.5 | 2.5 |
| 31_Foretinib | HGFR and VEGFR, mostly for Met and KDR | 26.8 | 6.5 | 5.4 | 3.3 | 37.4 | 3.3 | 69.1 | 19.1 | 67_Flu | Anticholesterol agent. HMG-CoA inhibitor | 100.0 | 0.0 | 87.4 | 15.2 | 100.0 | 0.0 | 105.7 | 5.3 |
| 32_Crizotinib | Met, ALK | 70.4 | 4.4 | 0.0 | 0.0 | 78.1 | 23.7 | 0.2 | 0.2 | 68_Myco_acid | Inosine-5’-monophosphate dehydrogenase inhibitor | 100.0 | 0.0 | 33.5 | 8.7 | 100.0 | 0.0 | 100.3 | 4.8 |
| 33_INCB28060 | Met | 62.5 | 13.5 | 88.9 | 7.4 | 52.2 | 10.6 | 115.4 | 12.9 | 69_Raloxi | Estrogen receptor inhibitor | 100.0 | 0.0 | 95.1 | 4.9 | 100.0 | 0.0 | 103.0 | 13.4 |
| 34_LEE011 | CDK4/6 | 100.0 | 0.0 | 64.2 | 9.0 | 100.0 | 0.0 | 86.0 | 5.7 | 70_Astemi | Histamine receptor ligand | 100.0 | 0.0 | 78.6 | 8.5 | 75.4 | 14.9 | 93.5 | 11.4 |
| 35_PD 0332991 | CDK4/6 | 100.0 | 0.0 | 1.2 | 0.9 | 100.0 | 0.0 | 85.7 | 10.1 | 71_Ferre | Retinoic acid receptor ligand | 100.0 | 0.0 | 71.0 | 7.4 | 100.0 | 0.0 | 111.9 | 16.6 |
| 36_LY2835219 | CDK4/6 | 66.6 | 8.5 | 0.0 | 0.0 | 75.1 | 25.9 | 82.1 | 11.2 | - | - | - | - | - | - | - | - | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, J.W.; Lee, S.-Y.; Lee, D.W. A Cancer Spheroid Array Chip for Selecting Effective Drug. Micromachines 2019, 10, 688. https://doi.org/10.3390/mi10100688
Choi JW, Lee S-Y, Lee DW. A Cancer Spheroid Array Chip for Selecting Effective Drug. Micromachines. 2019; 10(10):688. https://doi.org/10.3390/mi10100688
Chicago/Turabian StyleChoi, Jae Won, Sang-Yun Lee, and Dong Woo Lee. 2019. "A Cancer Spheroid Array Chip for Selecting Effective Drug" Micromachines 10, no. 10: 688. https://doi.org/10.3390/mi10100688
APA StyleChoi, J. W., Lee, S.-Y., & Lee, D. W. (2019). A Cancer Spheroid Array Chip for Selecting Effective Drug. Micromachines, 10(10), 688. https://doi.org/10.3390/mi10100688





