Probing Bacterial Interactions with the Schistosoma mansoni-Killing Toxin Biomphalysin via Atomic Force Microscopy and Single Molecule Force Spectroscopy
Abstract
1. Introduction
2. Results and Discussion
2.1. Bacterial Aggregation Mediated by B. glabrata Plasma Components, Including Biomphalysin
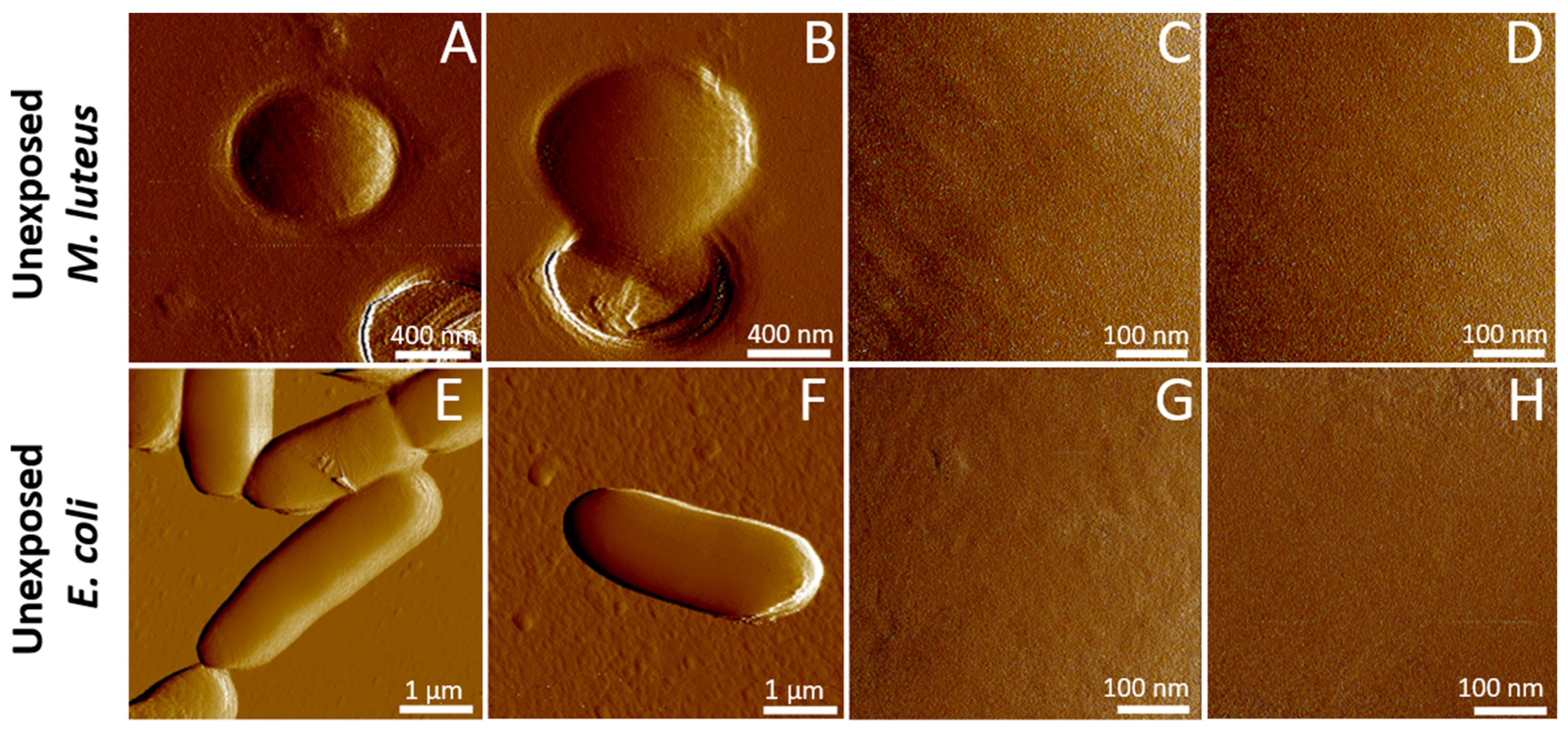
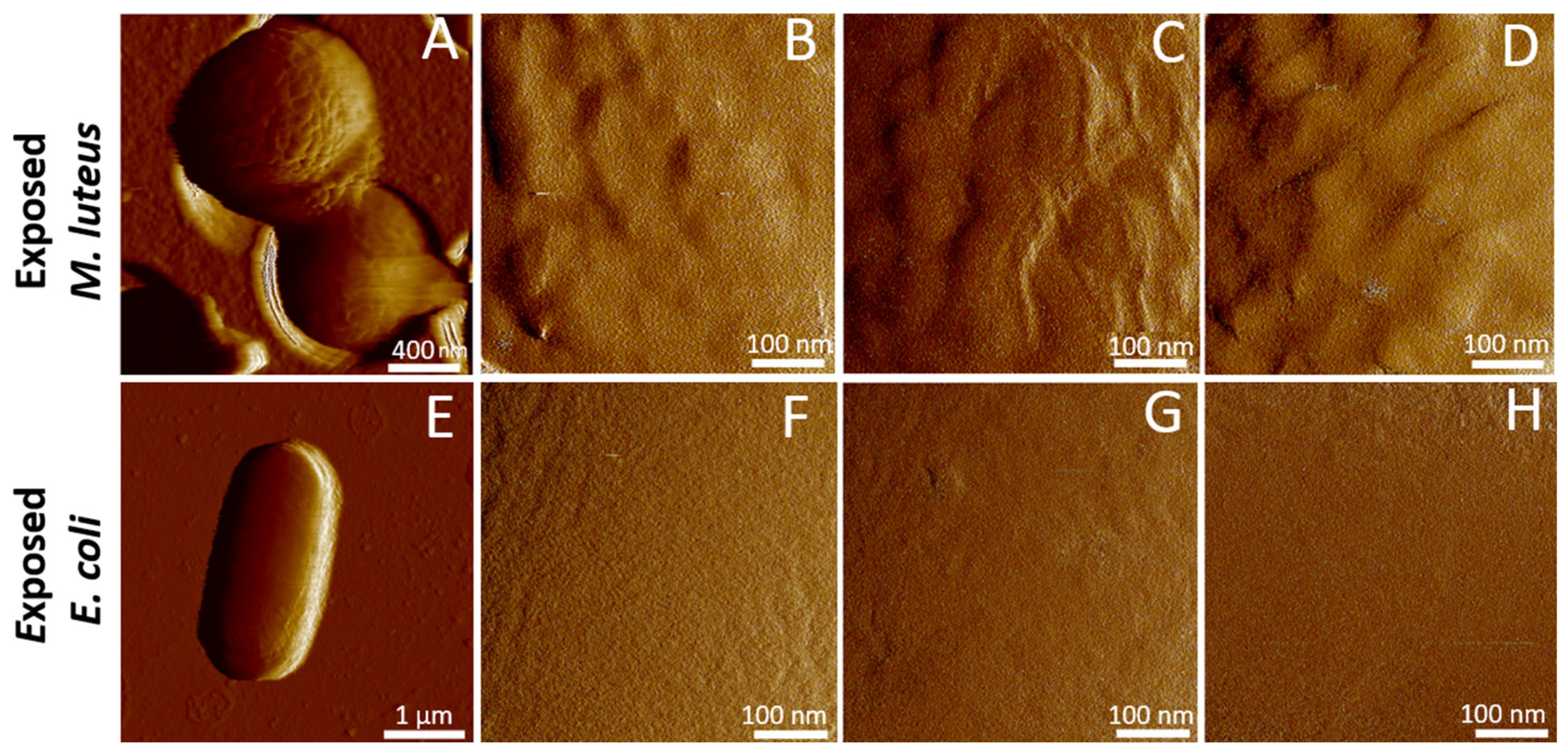
2.2. Cell Surface Roughness Measurements by AFM Imaging
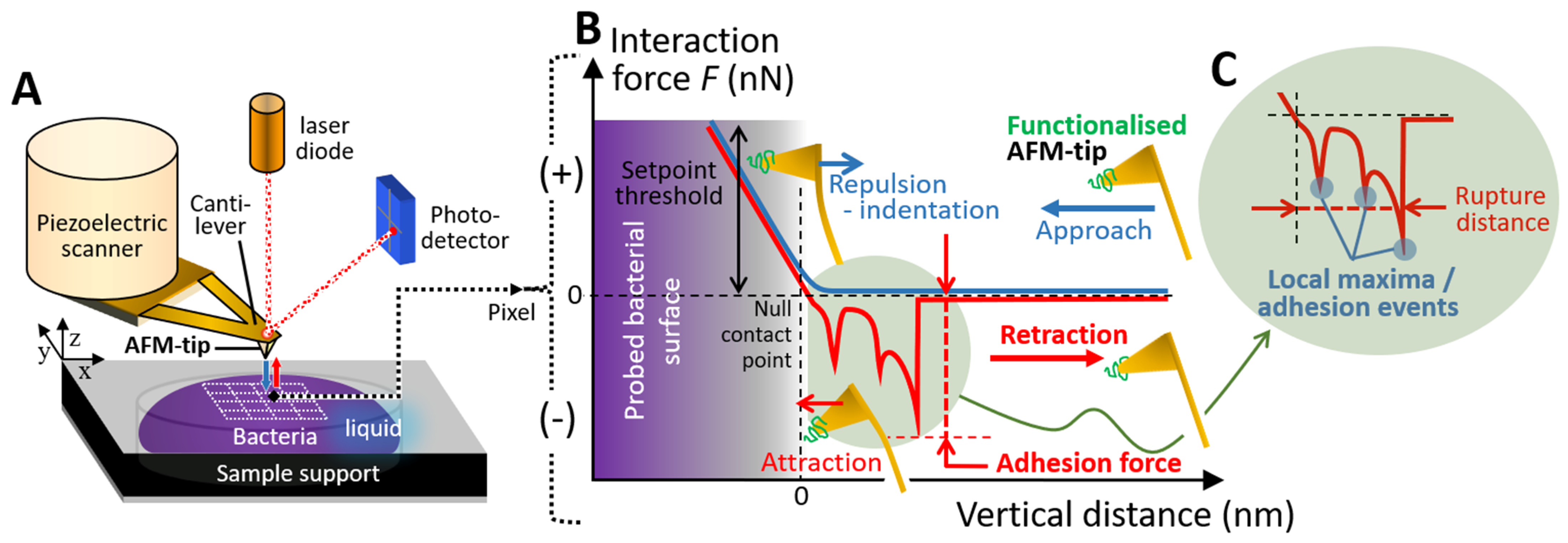
2.3. SMFS Interaction Force Measurements
3. Materials and Methods
3.1. Preparation of Biomphalysin-Containing Plasma
3.2. Bacterial Cell Culture
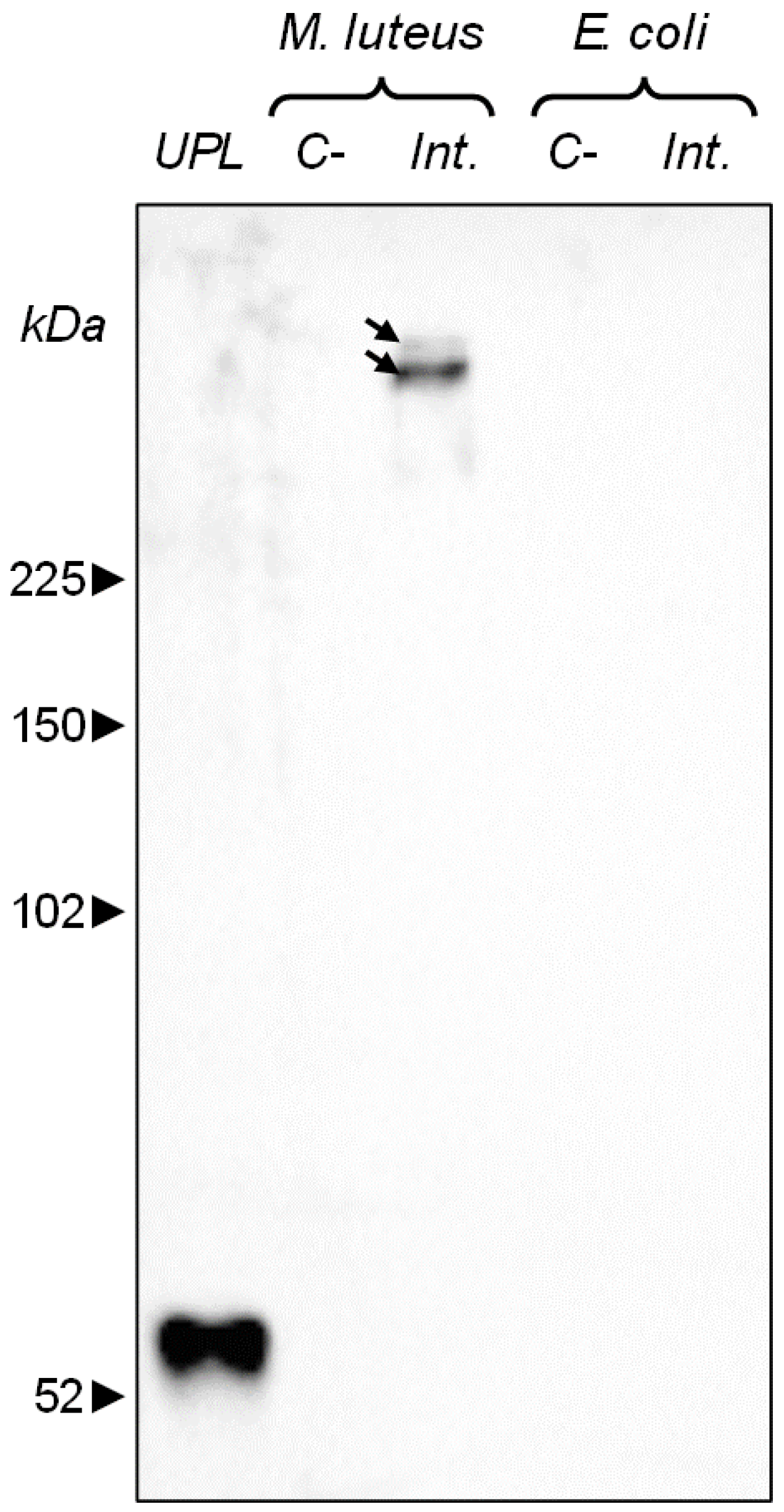
3.3. Cell Plasma-Exposure Conditions, SDS-PAGE and Western Blot Experiments
3.4. Cell Aggregation Assay
3.5. Cell Immobilization for Atomic Force Microscopy (AFM) Imaging and Single Molecule Force Spectroscopy (SMFS) Measurements
3.6. Cell Imaging and Surface Roughness Evaluation by AFM, and SMFS Experiments
3.7. AFM Tip Functionalization with Anti-Biomphalysin 1/2 Antibody
3.8. Processing of SMFS Interaction Measurements
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Report on Neglected Tropical Diseases 2024; World Health Organization: Geneva, Switzerland, 2024; Available online: https://www.who.int/teams/control-of-neglected-tropical-diseases/global-report-on-neglected-tropical-diseases-2024 (accessed on 3 January 2025).
- Galinier, R.; Roger, E.; Moné, Y.; Duval, D.; Portet, A.; Pinaud, S.; Chaparro, C.; Grunau, C.; Genthon, C.; Dubois, E.; et al. A multistrain approach to studying the mechanisms underlying compatibility in the interaction between Biomphalaria glabrata and Schistosoma mansoni. PLoS Negl. Trop. Dis. 2017, 11, e0005398. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hambrook, J.R.; Pila, E.A.; Gharamah, A.A.; Fang, J.; Wu, X.; Hanington, P. Coordination of humoral immune factors dictates compatibility between Schistosoma mansoni and Biomphalaria glabrata. eLife 2020, 9, e51708. [Google Scholar] [CrossRef] [PubMed]
- Mitta, G.; Adema, C.M.; Gourbal, B.; Loker, E.S.; Theron, A. Compatibility polymorphism in snail/schistosome interactions: From field to theory to molecular mechanisms. Dev. Comp. Immunol. 2012, 37, 1–8. [Google Scholar] [CrossRef]
- Bouchut, A.; Sautiere, P.E.; Coustau, C.; Mitta, G. Compatibility in the Biomphalaria glabrata/Echinostoma caproni model: Potential involvement of proteins from hemocytes revealed by a proteomic approach. Acta Trop. 2006, 98, 234–246. [Google Scholar] [CrossRef]
- Loker, E.S.; Cimino, D.F.; Stryker, G.A.; Hertel, L.A. The effect of size of M line Biomphalaria glabrata on the course of development of Echinostoma paraensei. J. Parasitol. 1987, 73, 1090–1098. [Google Scholar] [CrossRef]
- Nassi, H. Sur quatre furcocercaires émises par Biomphalaria glabrata en Guadeloupe. Ann. Parasitol. Hum. Comp. 1987, 62, 17–35. [Google Scholar] [CrossRef]
- Deleury, E.; Dubreuil, G.; Elangovan, N.; Wajnberg, E.; Reichhart, J.-M.; Gourbal, B.; Duval, D.; Baron, O.L.; Gouzy, J.; Coustau, C. Specific versus non-specific immune responses in an invertebrate species evidenced by a comparative de novo sequencing study. PLoS ONE 2012, 7, e32512. [Google Scholar] [CrossRef]
- Duval, D.; Galinier, R.; Mouahid, G.; Toulza, E.; Allienne, J.F.; Portela, J.; Calvayrac, C.; Rognon, A.; Arancibia, N.; Mitta, G.; et al. A novel bacterial pathogen of Biomphalaria glabrata: A potential weapon for Schistosomiasis control. PLoS Negl. Trop. Dis. 2015, 9, e0003815. [Google Scholar] [CrossRef]
- Granath, W.O. Identification of hemeproteins in the hemolymph of Biomphalaria glabrata (Gastropoda) separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Invertebr. Pathol. 1988, 52, 43–48. [Google Scholar] [CrossRef]
- Zelck, U.E.; Becker, W.; Bayne, C.J. The plasma proteins of Biomphalaria glabrata in the presence and absence of Schistosoma mansoni. Dev. Comp. Immunol. 1995, 19, 181–194. [Google Scholar] [CrossRef]
- Bayne, C.J. Successful parasitism of vector snail Biomphalaria glabrata by the human blood fluke (trematode) Schistosoma mansoni: A 2009 assessment. Mol. Biochem. Parasitol. 2009, 165, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Dheilly, N.M.; Duval, D.; Mouahid, G.; Emans, R.; Allienne, J.F.; Galinier, R.; Genthon, C.; Dubois, E.; Du Pasquier, L.; Adema, C.M.; et al. A family of variable immunoglobulin and lectin domain containing molecules in the snail Biomphalaria glabrata. Dev. Comp. Immunol. 2015, 48, 234–243. [Google Scholar] [CrossRef]
- Lu, L.; Loker, E.S.; Adema, C.M.; Zhang, S.-M.; Bu, L. Genomic and transcriptional analysis of genes containing fibrinogen and IgSF domains in the schistosome vector Biomphalaria glabrata, with emphasis on the differential responses of snails susceptible or resistant to Schistosoma mansoni. PLoS Negl. Trop. Dis. 2020, 14, e0008780. [Google Scholar] [CrossRef]
- Bender, R.C.; Bayne, C.J. Purification and characterization of a tetrameric alpha-macroglobulin proteinase inhibitor from the gastropod mollusc Biomphalaria glabrata. Biochem. J. 1996, 316, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Duval, D.; Pichon, R.; Lassalle, D.; Laffitte, M.; Gourbal, B.; Galinier, R. A new assessment of thioester-containing proteins diversity of the freshwater snail Biomphalaria glabrata. Genes 2020, 11, 69. [Google Scholar] [CrossRef]
- Portet, A.; Galinier, R.; Pinaud, S.; Portela, J.; Nowacki, F.; Gourbal, B.; Duval, D. BgTEP: An antiprotease involved in innate immune sensing in Biomphalaria glabrata. Front. Immunol. 2018, 9, 1206. [Google Scholar] [CrossRef]
- Galinier, R.; Portela, J.; Moné, Y.; Allienne, J.F.; Henri, H.; Delbecq, S.; Mitta, G.; Gourbal, B.; Duval, D. Biomphalysin, a new β pore-forming toxin involved in Biomphalaria glabrata immune defense against Schistosoma mansoni. PLoS Pathog. 2013, 9, e1003216. [Google Scholar] [CrossRef]
- Pinaud, S.; Tetreau, G.; Poteaux, P.; Galinier, R.; Chaparro, C.; Lassalle, D.; Portet, A.; Simphor, E.; Gourbal, B.; Duval, D. New insights into biomphalysin gene family diversification in the vector snail Biomphalaria glabrata. Front. Immunol. 2021, 12, 635131. [Google Scholar] [CrossRef] [PubMed]
- Gordy, M.A.; Pila, E.A.; Hanington, P.C. The role of fibrinogen-related proteins in the gastropod immune response. Fish Shellfish. Immunol. 2015, 46, 39–49. [Google Scholar] [CrossRef]
- Hambrook, J.R.; Hanington, P.C. Immune evasion strategies of schistosomes. Front. Immunol. 2021, 11, 624178. [Google Scholar] [CrossRef]
- Hanington, P.C.; Forys, M.A.; Dragoo, J.W.; Zhang, S.-M.; Adema, C.M.; Loker, E.S. Role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proc. Natl. Acad. Sci. USA 2010, 107, 21087–21092. [Google Scholar] [CrossRef] [PubMed]
- Abrami, L.; Velluz, M.C.; Hong, Y.; Ohishi, K.; Mehlert, A.; Ferguson, M.; Kinoshita, T.; van der Goot, F.G. The glycan core of GPI-anchored proteins modulates aerolysin binding but is not sufficient: The polypeptide moiety is required for the toxin–receptor interaction. FEBS Lett. 2002, 512, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Ohishi, K.; Inoue, N.; Kang, J.Y.; Shime, H.; Horiguchi, Y.; van der Goot, F.G.; Sugimoto, N.; Kinoshita, T. Requirement of N-glycan on GPI-anchored proteins for efficient binding of aerolysin but not Clostridium septicum α-toxin. EMBO J. 2002, 21, 5047–5056. [Google Scholar] [CrossRef]
- Iacovache, I.; De Carlo, S.; Cirauqui, N.; Dal Peraro, M.; van der Goot, F.G.; Zuber, B. Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process. Nat. Comm. 2016, 7, 12062. [Google Scholar] [CrossRef] [PubMed]
- Lima, C.; Disner, G.R.; Falcão, M.A.P.; Seni-Silva, A.C.; Maleski, A.L.A.; Souza, M.M.; Reis Tonello, M.C.; Lopes-Ferreira, M. The natterin proteins diversity: A review on phylogeny, structure, and immune function. Toxins 2021, 13, 538. [Google Scholar] [CrossRef]
- Moran, Y.; Fredman, D.; Szczesny, P.; Grynberg, M.; Technau, U. Recurrent horizontal transfer of bacterial toxin genes to eukaryotes. Mol. Biol. Evol. 2012, 29, 2223–2230. [Google Scholar] [CrossRef]
- Szczesny, P.; Iacovache, I.; Muszewska, A.; Ginalski, K.; van der Goot, F.G.; Grynberg, M. Extending the aerolysin family: From bacteria to vertebrates. PLoS ONE 2011, 6, e20349. [Google Scholar] [CrossRef]
- Knapp, O.; Stiles, B.; Popoff, M.R. The aerolysin-like toxin family of cytolytic, pore-forming toxins. Open Toxinology J. 2010, 3, 53–68. [Google Scholar] [CrossRef]
- Chen, L.L.; Xie, J.; Cao, D.D.; Jia, N.; Li, Y.J.; Sun, H.; Li, W.F.; Hu, B.; Chen, Y.; Zhou, C.Z. The pore-forming protein Aep1 is an innate immune molecule that prevents zebrafish from bacterial infection. Dev. Comp. Immunol. 2018, 82, 49–54. [Google Scholar] [CrossRef]
- Bruhn, H.; Winkelmann, J.; Andersen, C.; Andrä, J.; Leippe, M. Dissection of the mechanisms of cytolytic and antibacterial activity of lysenin, a defence protein of the annelid Eisenia fetida. Dev. Comp. Immunol. 2006, 30, 597–606. [Google Scholar] [CrossRef]
- Tetreau, G.; Pinaud, S.; Portet, A.; Galinier, R.; Gourbal, B.; Duval, D. Specific pathogen recognition by multiple innate immune sensors in an invertebrate. Front. Immunol. 2017, 8, 1249. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Balta, V.A.; West, R.; Newlin, K.N.; Miljanić, O.Š.; Sullivan, D.J.; Vekilov, P.G.; Rimer, J.D. A second mechanism employed by artemisinins to suppress Plasmodium falciparum hinges on inhibition of hematin crystallization. J. Biol. Chem. 2020, 296, 100123. [Google Scholar] [CrossRef]
- Bastounis, E.E.; Radhakrishnan, P.; Prinz, C.K.; Theriot, J.A. Mechanical forces govern interactions of host cells with intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 2022, 86, e0009420. [Google Scholar] [CrossRef]
- Chattopadhyay, P.K.; Roederer, M.; Bolton, D.L. A deadly dance: The choreography of host-pathogen interactions, as revealed by single-cell technologies. Nat. Comm. 2018, 9, 4638. [Google Scholar] [CrossRef]
- Beaussart, A.; Abellán-Flos, M.; El-Kirat-Chatel, S.; Vincent, S.P.; Dufrêne, Y.F. Force nanoscopy as a versatile platform for quantifying the activity of antiadhesion compounds targeting bacterial pathogens. Nano Lett. 2016, 16, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.F. Atomic Force Microscopy in microbiology: New structural and functional insights into the microbial cell surface. mBio 2014, 5, e01363-14. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Beaussart, A.; El-Kirat-Chatel, S.; Sullan, R.M.A.; Dufrêne, Y.F. Atomic Force Microscopy: A new look at pathogens. PLoS Pathog. 2013, 9, e1003516. [Google Scholar] [CrossRef]
- Vogel, V.; Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell. Biol. 2006, 7, 265–275. [Google Scholar] [CrossRef]
- Sumbul, F.; Rico, F. Single-molecule force spectroscopy: Experiments, analysis, and simulations. Methods Mol. Biol. 2019, 1886, 163–189. [Google Scholar] [CrossRef]
- Dufrêne, Y.F. Towards nanomicrobiology using atomic force microscopy. Nat. Rev. Microbiol. 2008, 6, 674–680. [Google Scholar] [CrossRef]
- Sbarigia, C.; Tacconi, S.; Mura, F.; Rossi, M.; Dinarelli, S.; Dini, L. High-resolution atomic force microscopy as a tool for topographical mapping of surface budding. Front. Cell Dev. Biol. 2022, 10, 975919. [Google Scholar] [CrossRef] [PubMed]
- Dorobantu, L.S.; Goss, G.G.; Burrell, R.E. Atomic Force Microscopy: A nanoscopic view of microbial cell surfaces. Micron 2012, 43, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Dudko, O.K.; Hummer, G.; Szabo, A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. USA 2008, 105, 15755–15760. [Google Scholar] [CrossRef]
- Jacquot, A.; Sakamoto, C.; Razafitianamaharavo, A.; Caillet, C.; Merlin, J.; Fahs, A.; Ghigo, J.M.; Duval, J.F.L.; Beloin, C.; Francius, G. The dynamics and pH-dependence of Ag43 adhesins’ self-association probed by atomic force spectroscopy. Nanoscale 2014, 6, 12665–12681. [Google Scholar] [CrossRef] [PubMed]
- Francius, G.; Petit, F.; Clement, E.; Chekli, Y.; Ghigo, J.-M.; Beloin, C.; Duval, J.F.L. On the strong connection between nanoscale adhesion of Yad fimbriae and macroscale attachment of Yad-decorated bacteria to glycosylated, hydrophobic and hydrophilic surfaces. Nanoscale 2021, 13, 1257–1272. [Google Scholar] [CrossRef]
- Jacquot, A.; Sakamoto, C.; Razafitianamaharavo, A.; Caillet, C.; Merlin, J.; Fahs, A.; Ghigo, J.M.; Beloin, C.; Duval, J.F.L.; Francius, G. Dynamic modulation of fimbrial extension and FimH-mannose binding force on live bacteria under pH changes: A molecular atomic force microscopy analysis. J. Biomed. Nanotechnol. 2014, 10, 3361–3372. [Google Scholar] [CrossRef][Green Version]
- Beaussart, A.; El-Kirat-Chatel, S. Microbial adhesion and ultrastructure from the single-molecule to the single-cell levels by atomic force microscopy. Cell. Surf. 2019, 5, 100031. [Google Scholar] [CrossRef]
- Benoit, M.; Gabriel, D.; Gerisch, G.; Gaub, H.E. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat. Cell. Biol. 2000, 2, 313–317. [Google Scholar] [CrossRef]
- El-Kirat-Chatel, S.; Beaussart, A.; Boyd, C.D.; O’Toole, G.A.; Dufrêne, Y.F. Single-cell and single-molecule analysis deciphers the localization, adhesion, and mechanics of the biofilm adhesin LapA. ACS Chem. Biol. 2014, 9, 485–494. [Google Scholar] [CrossRef]
- Alsteens, D.; Garcia, M.C.; Lipke, P.N.; Dufrêne, Y.F. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. USA 2010, 107, 20744–20749. [Google Scholar] [CrossRef]
- Mathelié-Guinlet, M.; Viela, F.; Viljoen, A.; Dehullu, J.; Dufrêne, Y.F. Single-molecule atomic force microscopy studies of microbial pathogens. Curr. Opin. Biomed. Eng. 2019, 12, 1–7. [Google Scholar] [CrossRef]
- Herman, P.; El-Kirat-Chatel, S.; Beaussart, A.; Geoghegan, J.A.; Foster, T.J.; Dufrêne, Y.F. The binding force of the staphylococcal adhesin SdrG is remarkably strong. Mol. Microbiol. 2014, 93, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Lower, B.H.; Yongsunthon, R.; Shi, L.; Wildling, L.; Gruber, H.J.; Wigginton, N.S.; Reardon, C.L.; Pinchuk, G.E.; Droubay, T.C.; Boily, J.-F.; et al. Antibody recognition force microscopy shows that outer membrane cytochromes OmcA and MtrC are expressed on the exterior surface of shewanella oneidensis MR-1. Appl. Environ. Microbiol. 2009, 75, 2931–2935. [Google Scholar] [CrossRef] [PubMed]
- Poteaux, P.; Parpinel, A.; Ripoll, C.; Sarrazin, A.; Galinier, R.; Brugière, S.; Couté, Y.; Mourey, L.; Hanington, P.C.; Gourbal, B.; et al. The structural features and immunological role of biomphalysins in the snail Biomphalaria glabrata. PLoS Pathogens, 2025; in press. [Google Scholar] [CrossRef]
- Zhang, S.-M.; Zeng, Y.; Loker, E.S. Expression profiling and binding properties of fibrinogen-related proteins (FREPs), plasma proteins from the schistosome snail host Biomphalaria glabrata. Innate Immun. 2008, 14, 175–189. [Google Scholar] [CrossRef]
- Hanington, P.C.; Forys, M.A.; Loker, E.S. A somatically diversified defense factor, FREP3, is a determinant of snail resistance to Schistosome infection. PLoS Negl. Trop. Dis. 2012, 6, e1591. [Google Scholar] [CrossRef]
- Gorbushin, A.M. Derivatives of the lectin complement pathway in Lophotrochozoa. Dev. Comp. Immunol. 2019, 94, 35–58. [Google Scholar] [CrossRef]
- Abrami, L.; Goot, F.G.V.D. Plasma membrane microdomains act as concentration platforms to facilitate intoxication by aerolysin. J. Cell. 1999, 147, 175–184. [Google Scholar] [CrossRef]
- López-Diaz, J.A.; Cantón, P.E.; Gill, S.S.; Soberón, M.; Bravo, A. Oligomerization is a key step in Cyt1Aa membrane insertion and toxicity but not necessary to synergize Cry11Aa toxicity in Aedes aegypti larvae. Environ. Microbiol. 2013, 15, 3030–3039. [Google Scholar] [CrossRef]
- Khorramnejad, A.; Domínguez-Arrizabalaga, M.; Caballero, P.; Escriche, B.; Bel, Y. Study of the Bacillus thuringiensis Cry1Ia protein oligomerization promoted by midgut brush border membrane vesicles of Lepidopteran and Coleopteran insects, or cultured insect cells. Toxins 2020, 12, 133. [Google Scholar] [CrossRef]
- Okumura, S.; Saitoh, H.; Ishikawa, T.; Inouye, K.; Mizuki, E. Mode of action of parasporin-4, a cytocidal protein from Bacillus thuringiensis. Biochim. Biophys. Acta (BBA)-Biomembr. 2011, 1808, 1476–1482. [Google Scholar] [CrossRef]
- Lesieur, C.; Frutiger, S.; Hughes, G.; Kellner, R.; Pattus, F.; van der Goot, F.G. Increased stability upon heptamerization of the pore-forming toxin aerolysin. J. Biol. Chem. 1999, 274, 36722–36728. [Google Scholar] [CrossRef]
- Petit, L.; Gibert, M.; Gillet, D.; Laurent-Winter, C.; Boquet, P.; Popoff, M.R. Clostridium perfringens epsilon-toxin acts on MDCK cells by forming a large membrane complex. J. Bacteriol. 1997, 179, 6480–6487. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Shimada, H.; Kitada, S. Raft-targeting and oligomerization of parasporin-2, a Bacillus thuringiensis crystal protein with anti-tumour activity. J. Biochem. 2008, 143, 269–275. [Google Scholar] [CrossRef]
- Degiacomi, M.T.; Iacovache, I.; Pernot, L.; Chami, M.; Kudryashev, M.; Stahlberg, H.; van der Goot, F.G.; Dal Peraro, M. Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism. Nat. Chem. Biol. 2013, 9, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Moné, Y.; Gourbal, B.; Duval, D.; Du Pasquier, L.; Kieffer-Jaquinod, S.; Mitta, G. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PLoS Negl. Trop. Dis. 2010, 4, e813. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Patel, N.; Fukuda, H.; Nagata, T.; Mitarai, S.; Azam, F. Bacterial surface roughness regulates nanoparticle scavenging in seawater. Limnol. Oceanogr. 2023, 68, 780–789. [Google Scholar] [CrossRef]
- Pagnout, C.; Sohm, B.; Razafitianamaharavo, A.; Caillet, C.; Offroy, M.; Leduc, M.; Gendre, H.; Jomini, S.; Beaussart, A.; Bauda, P.; et al. Pleiotropic effects of rfa-gene mutations on Escherichia coli envelope properties. Sci. Rep. 2019, 9, 9696. [Google Scholar] [CrossRef]
- Pagnout, C.; Razafitianamaharavo, A.; Sohm, B.; Caillet, C.; Beaussart, A.; Delatour, E.; Bihannic, I.; Offroy, M.; Duval, J.F.L. Osmotic stress and vesiculation as key mechanisms controlling bacterial sensitivity and resistance to TiO2 nanoparticles. Comm. Biol. 2021, 4, 678. [Google Scholar] [CrossRef]
- Meincken, M.; Holroyd, D.L.; Rautenbach, M. Atomic force microscopy study of the effect of antimicrobial peptides on the cell envelope of Escherichia coli. Antimicrob. Agents Chemother. 2005, 49, 4085–4092. [Google Scholar] [CrossRef]
- Snoussi, M.; Talledo, J.P.; Del Rosario, N.A.; Mohammadi, S.; Ha, B.Y.; Košmrlj, A.; Taheri-Araghi, S. Heterogeneous absorption of antimicrobial peptide LL37 in Escherichia coli cells enhances population survivability. eLife 2018, 7, e38174. [Google Scholar] [CrossRef]
- Miller, M.A.; Zachary, J.F. Chapter 1-Mechanisms and Morphology of Cellular Injury, Adaptation, and Death. In Pathologic Basis of Veterinary Disease, 6th ed.; Zachary, J.F., Ed.; Mosby: Maryland Heights, MO, USA, 2017; p. 2-43.e19. ISBN 9780323357753. [Google Scholar] [CrossRef]
- Hu, H.; Liu, M.; Sun, S. Pore-forming toxins during bacterial infection: Molecular mechanisms and potential therapeutic targets. Drug Des. Devel. Ther. 2021, 15, 3773–3781. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.K.; Chattopadhyay, K. Taking toll on membranes: Curious cases of bacterial β-barrel pore-forming toxins. Biochem. 2020, 59, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Dang, D.; Liu, L.; Xi, N.; Wang, Y. Imaging and force recognition of single molecular behaviors using atomic force microscopy. Sensors 2017, 17, 200. [Google Scholar] [CrossRef]
- Li, H.; Oberhauser, A.F.; Fowler, S.B.; Clarke, J.; Fernandez, J.M. Atomic force microscopy reveals the mechanical design of a modular protein. Proc. Natl. Acad. Sci. USA 2000, 97, 6527–6531. [Google Scholar] [CrossRef]
- Butt, H.-J.; Cappella, B.; Kappl, M. Force measurements with the atomic force microscope: Technique, interpretation and applications. Surf. Sci. Rep. 2005, 59, 1–152. [Google Scholar] [CrossRef]
- Touhami, A.; Jericho, M.H.; Boyd, J.M.; Beveridge, T.J. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J. Bacteriol. 2006, 188, 370–377. [Google Scholar] [CrossRef]
- Cremin, K.; Jones, B.A.; Teahan, J.; Meloni, G.N.; Perry, D.; Zerfass, C.; Asally, M.; Soyer, O.S.; Unwin, P.R. Scanning ion conductance microscopy reveals differences in the ionic environments of Gram-positive and negative bacteria. Anal. Chem. 2020, 92, 16024–16032. [Google Scholar] [CrossRef]
- Müller, D.J.; Dumitru, A.C.; Lo Giudice, C.; Gaub, H.E.; Hinterdorfer, P.; Hummer, G.; De Yoreo, J.J.; Dufrêne, Y.F.; Alsteens, D. Atomic force microscopy-based force spectroscopy and multiparametric imaging of biomolecular and cellular systems. Chem. Rev. 2021, 121, 11701–11725. [Google Scholar] [CrossRef]
- Berquand, A.; Xia, N.; Castner, D.G.; Clare, B.H.; Abbott, N.L.; Dupres, V.; Adriaensen, Y.; Dufrêne, Y.F. Antigen binding forces of single antilysozyme Fv fragments explored by atomic force microscopy. Langmuir 2005, 21, 5517–5523. [Google Scholar] [CrossRef]
- Lostao, A.; Lim, K.; Pallarés, M.C.; Ptak, A.; Marcuello, C. Recent advances in sensing the inter-biomolecular interactions at the nanoscale—A comprehensive review of AFM-based force spectroscopy. Int. J. Biol. Macromol. 2023, 238, 124089. [Google Scholar] [CrossRef]
- Mechaly, A.E.; Bellomio, A.; Gil-Cartón, D.; Morante, K.; Valle, M.; González-Mañas, J.M.; Guérin, D.M.A. Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins. Structure 2011, 19, 181–191. [Google Scholar] [CrossRef]
- Opper, B.; Bognár, A.; Heidt, D.; Németh, P.; Engelmann, P. Revising lysenin expression of earthworm coelomocytes. Dev. Comp. Immunol. 2013, 39, 214–218. [Google Scholar] [CrossRef]
- Xiang, Y.; Yan, C.; Guo, X.; Zhou, K.; Li, S.; Gao, Q.; Wang, X.; Zhao, F.; Liu, J.; Lee, W.; et al. Host-derived, pore-forming toxin–like protein and trefoil factor complex protects the host against microbial infection, Proc. Natl. Acad. Sci. USA 2014, 111, 6702–6707. [Google Scholar] [CrossRef] [PubMed]
- Marszalek, P.E.; Oberhauser, A.F.; Pang, Y.P.; Fernandez, J.M. Polysaccharide elasticity governed by chair-boat transitions of the glucopyranose ring. Nature 1998, 396, 661–664. [Google Scholar] [CrossRef]
- Chernin, E. Observations on hearts explanted in vitro from the snail Australorbis glabratus. J. Parasitol. 1963, 49, 353–364. [Google Scholar] [CrossRef]
- Thornton, D.J.; Sheehan, J.K.; Carlstedt, I. Identification of glycoproteins on nitrocellulose membranes and gels. In Basic Protein and Peptide Protocols, 1st ed.; In Methods in Molecular BiologyTM Series; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 1994; Volume 32, pp. 119–128. [Google Scholar] [CrossRef]
- Butt, H.-J.; Jascke, M. Calculation of thermal noise in atomic force microscopy. Nanotechnology 1995, 6, 1–7. [Google Scholar] [CrossRef]
- Greif, D.; Wesner, D.; Regtmeier, J.; Anselmetti, D. High resolution imaging of surface patterns of single bacterial cells. Ultramicroscopy 2010, 110, 1290–1296. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xi, N.; Liu, L. Peak force tapping atomic force microscopy for advancing cell and molecular biology. Nanoscale 2021, 13, 8358–8375. [Google Scholar] [CrossRef]
- Offroy, M.; Razafitianamaharavo, A.; Beaussart, A.; Pagnout, C.; Duval, J.F.L. Fast automated processing of AFM PeakForce curves to evaluate spatially-resolved Young modulus and stiffness of turgescent cells. RSC Adv. 2020, 10, 19258–19275. [Google Scholar] [CrossRef]
- Sehgal, D.; Vijay, I.K. A method for the high efficiency of water-soluble carbodiimide-mediated amidation. Anal. Biochem. 1994, 218, 87–91. [Google Scholar] [CrossRef]
- Becke, T.D.; Ness, S.; Sudhop, S.; Gaub, H.E.; Hilleringmann, M.; Schilling, A.F.; Clausen-Schaumann, H. Covalent immobilization of proteins for the single molecule force spectroscopy. J. Vis. Exp. 2018, 138, 58167. [Google Scholar] [CrossRef]



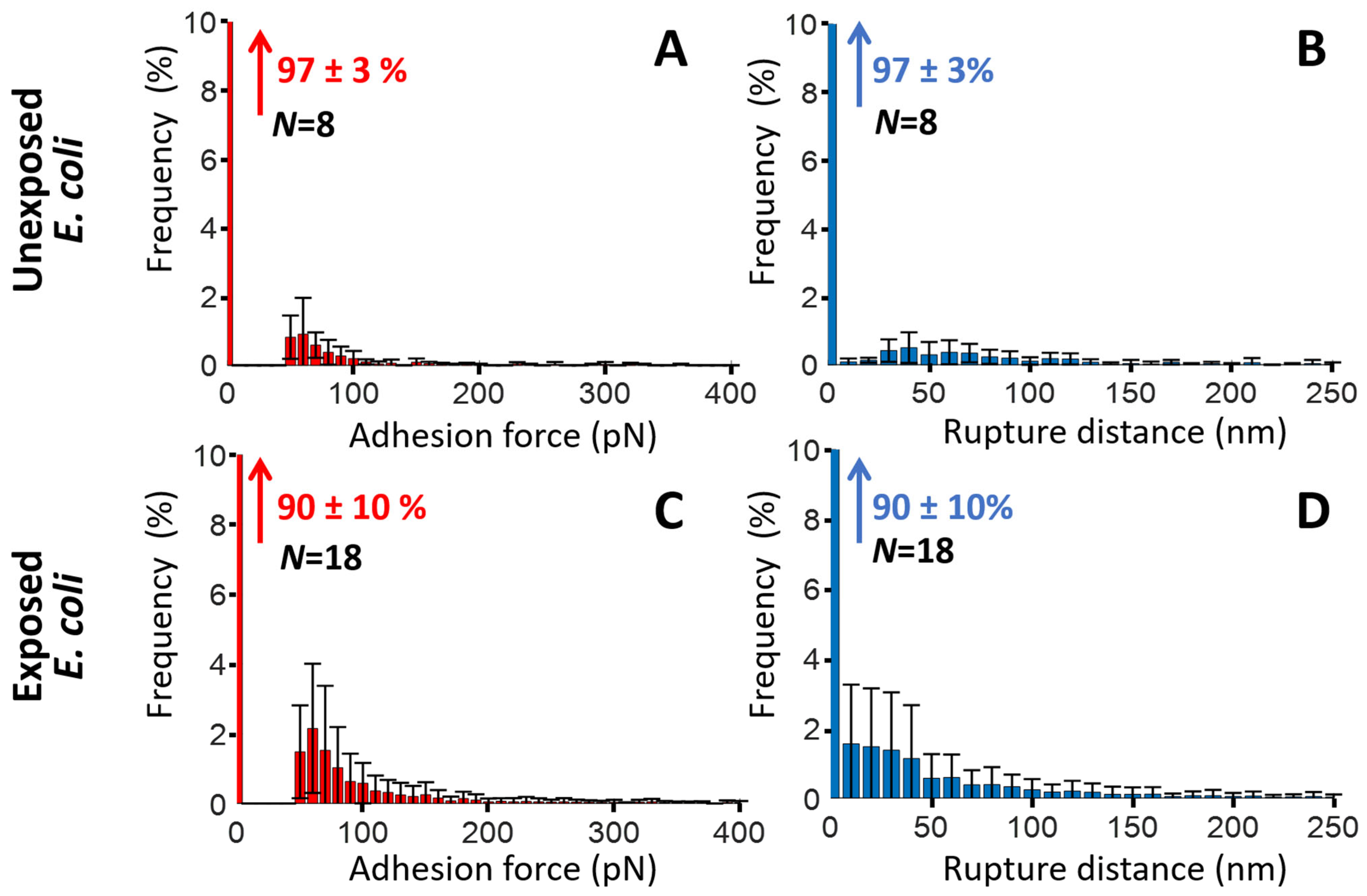


| E. coli | M. luteus | |||
|---|---|---|---|---|
| Before Exposure | After Exposure | Before Exposure | After Exposure | |
| Surface Roughness (nm) | 3.0 ± 0.9 nm | 2.9 ± 1.8 nm | 2.1 ± 0.8 nm | 4.3 ± 2.4 nm |
| % of SMFS force curves (or cell pixels) free of BM adhesion events | 97 ± 3% | 90 ± 10% | 88 ± 10% | 70 ± 19% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zouaoui, J.; Poteaux, P.; Beaussart, A.; Lesniewska, N.; Duval, D.; Duval, J.F.L. Probing Bacterial Interactions with the Schistosoma mansoni-Killing Toxin Biomphalysin via Atomic Force Microscopy and Single Molecule Force Spectroscopy. Toxins 2025, 17, 269. https://doi.org/10.3390/toxins17060269
Zouaoui J, Poteaux P, Beaussart A, Lesniewska N, Duval D, Duval JFL. Probing Bacterial Interactions with the Schistosoma mansoni-Killing Toxin Biomphalysin via Atomic Force Microscopy and Single Molecule Force Spectroscopy. Toxins. 2025; 17(6):269. https://doi.org/10.3390/toxins17060269
Chicago/Turabian StyleZouaoui, Jihen, Pierre Poteaux, Audrey Beaussart, Nicolas Lesniewska, David Duval, and Jérôme F. L. Duval. 2025. "Probing Bacterial Interactions with the Schistosoma mansoni-Killing Toxin Biomphalysin via Atomic Force Microscopy and Single Molecule Force Spectroscopy" Toxins 17, no. 6: 269. https://doi.org/10.3390/toxins17060269
APA StyleZouaoui, J., Poteaux, P., Beaussart, A., Lesniewska, N., Duval, D., & Duval, J. F. L. (2025). Probing Bacterial Interactions with the Schistosoma mansoni-Killing Toxin Biomphalysin via Atomic Force Microscopy and Single Molecule Force Spectroscopy. Toxins, 17(6), 269. https://doi.org/10.3390/toxins17060269






