Recombinant Oxidase from Armillaria tabescens as a Potential Tool for Aflatoxin B1 Degradation in Contaminated Cereal Grain
Abstract
:1. Introduction
2. Results
2.1. Recombinant Aflatoxin B1 Oxidase Obtained Using a Pichia Pastoris Expression System
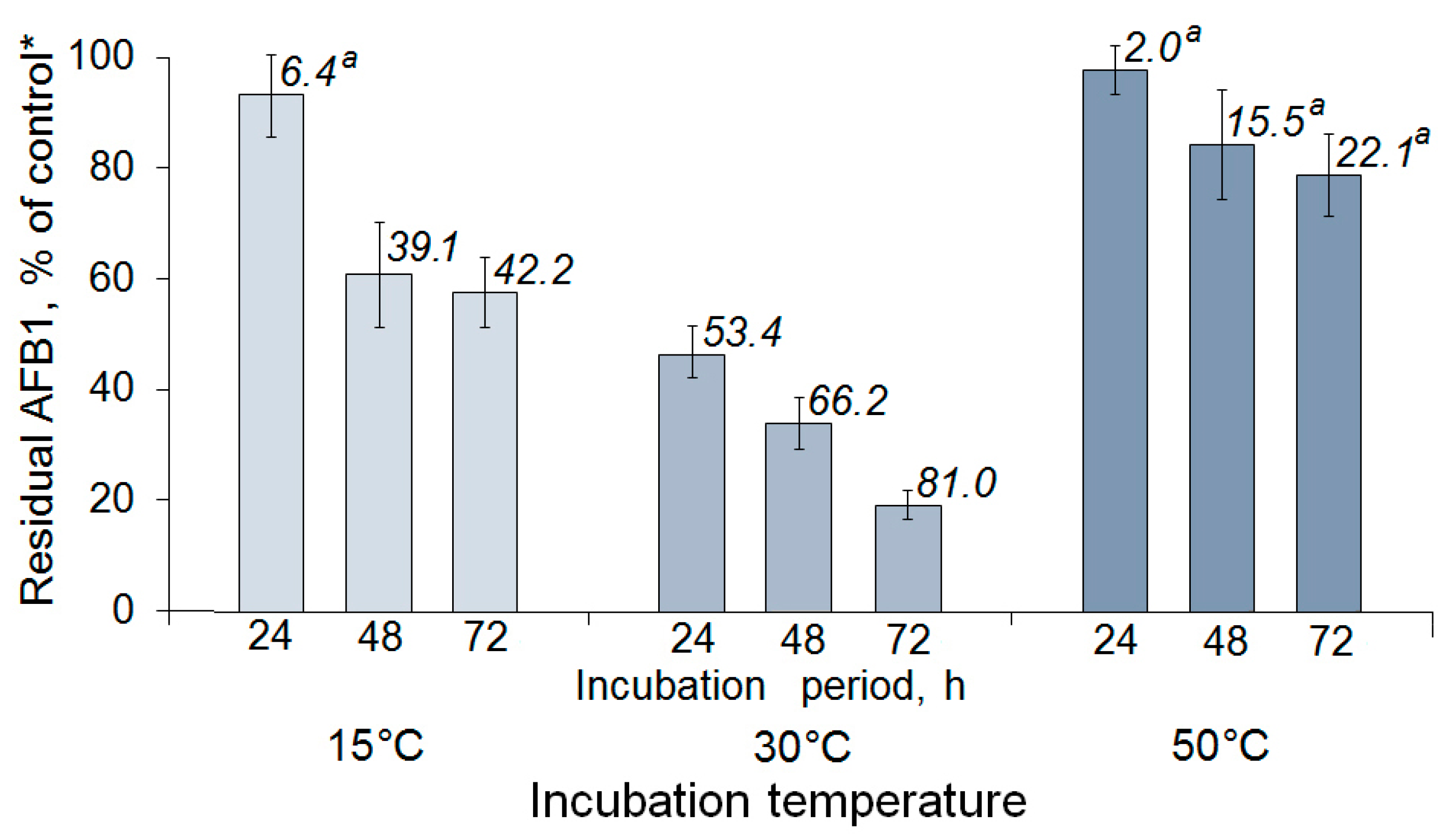
2.2. Enzymatic AFB1 Degradation in Model Solutions under Different Conditions
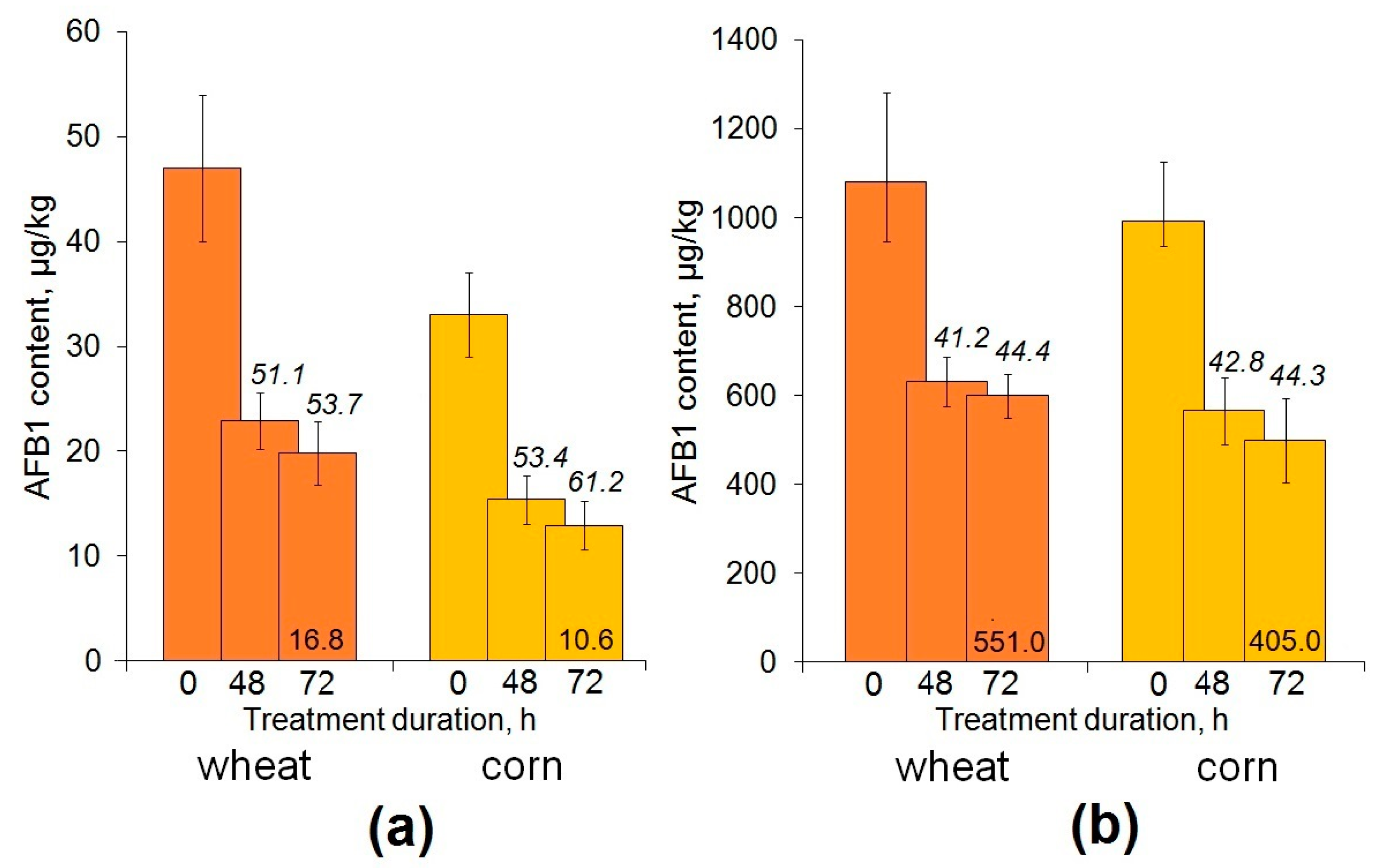
2.3. Enzymatic Decontamination of Cereal Grain Inoculated with a Toxigenic Aspergillus flaus
3. Discussion
4. Materials and Methods
4.1. Generation of a Yeast Strain Producing Recombinant AFO
4.2. Recombinant AFO Production in Yeast Cells
4.3. AFO Purification Procedure
4.4. AFB1 Degradation by AFO in Buffer Solutions
4.5. AFB1 Degradation by AFO in Artificially Inoculated Grain
4.5.1. Grain Inoculation with Aspergillus flavus
4.5.2. Treatment of Inoculated Grain with AFO
4.6. Isolation of Residual AFB1 from Cereal Grain after Enzymatic Decontamination
4.7. AFB1 Quantification in Buffer Solutions and AFO-Exposed Grain
4.8. Statistical Treatment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dzhavakhiya, V.G.; Voinova, T.M.; Popletaeva, S.B.; Statsyuk, N.V.; Limantseva, L.A.; Shcherbakova, L.A. Effect of various compounds blocking the colony pigmentation on the aflatoxin B1 production by Aspergillus flavus. Toxins 2016, 8, 313. [Google Scholar] [CrossRef] [PubMed]
- Sipos, P.; Peles, F.; Brassó, D.L.; Béri, B.; Pusztahelyi, T.; Pócsi, I.; Győri, Z. Physical and chemical methods for reduction in aflatoxin content of feed and food. Toxins 2021, 13, 204. [Google Scholar] [CrossRef] [PubMed]
- Umaya, S.R.; Vijayalakshmi, Y.C.; Sejian, V. Exploration of plant products and phytochemicals against aflatoxin toxicity in broiler chicken production: Present status. Toxicon 2021, 200, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Salazar, J.A.; Ruiz-Hernández, K.; Martínez-Miranda, M.M.; Castro-Ríos, K. Postharvest strategies for decontamination of aflatoxins in cereals. Food Rev. Int. 2023, 39, 3635–3662. [Google Scholar] [CrossRef]
- Marshall, H.; Meneely, J.P.; Quinn, B.; Zhao, Y.; Bourke, P.; Gilmore, B.F.; Zhang, G.; Elliott, C.T. Novel decontamination approaches and their potential application for post-harvest aflatoxin control. Trends Food Sci. Technol. 2020, 106, 489–496. [Google Scholar] [CrossRef]
- Rajasekaran, K.; Sayler, R.J.; Majumdar, R.; Sickler, C.M.; Cary, J.W. Inhibition of Aspergillus flavus growth and aflatoxin production in transgenic maize expressing the α-amylase inhibitor from Lablab purpureus L. J. Vis. Exp. 2019, 144, e59169. [Google Scholar] [CrossRef]
- Popescu, R.G.; Rădulescu, A.L.; Georgescu, S.E.; Dinischiotu, A. Aflatoxins in feed: Types, metabolism, health consequences in swine and mitigation strategies. Toxins 2022, 14, 853. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Pathak, H.; Bhadauria, S.; Sudan, J. Aflatoxin contamination in food crops: Causes, detection, and management: A review. Food Prod. Process. Nutr. 2021, 3, 17. [Google Scholar] [CrossRef]
- Mahato, D.K.; Lee, K.E.; Kamle, M.; Devi, S.; Dewangan, K.N.; Kumar, P.; Kang, S.G. Aflatoxins in food and feed: An overview on prevalence, detection and control strategies. Front. Microbiol. 2019, 10, 2266. [Google Scholar] [CrossRef]
- Kumar, P.; Gupta, A.; Mahato, D.K.; Pandhi, S.; Pandey, A.K.; Kargwal, R.; Mishra, S.; Suhag, R.; Sharma, N.; Saurabh, V.; et al. Aflatoxins in cereals and cereal-based products: Occurrence, toxicity, impact on human health, and their detoxification and management strategies. Toxins 2022, 14, 687. [Google Scholar] [CrossRef]
- Tadele, F.; Demissie, B.; Amsalu, A.; Demelash, H.; Mengist, Z.; Ambelu, A.; Yenew, C. Aflatoxin contamination of animal feeds and its predictors among dairy farms in northwest Ethiopia: One health approach implications. Front. Vet. Sci. 2023, 10, 1123573. [Google Scholar] [CrossRef] [PubMed]
- Putman, B.; Thoma, G.; Burek, J.; Matlock, M. A retrospective analysis of the United States poultry industry: 1965 compared with 2010. Agric. Syst. 2017, 157, 107–117. [Google Scholar] [CrossRef]
- Lee, J.; Nam, D.S.; Kong, C. Variability in nutrient composition of cereal grains from different origins. SpringerPlus 2016, 5, 419. [Google Scholar] [CrossRef]
- Filazi, A.; Sireli, U.T.; Filazi, A.; Sireli, U.T. Occurrence of aflatoxins in food. In Aflatoxins–Recent Advances and Future Prospects; Razzaghi-Abyaneh, M., Ed.; IntechOpen: Rijeka, Croatia, 2013; pp. 143–171. [Google Scholar]
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajšlová, J.; Mayar, S.; Krska, R. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited “FAO estimate” of 25%. Crit. Rev. Food Sci. Nutr. 2020, 60, 2773–2789. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S.; Coulombe, R.A.; Reed, K.M. Aflatoxicosis: Lessons from toxicity and responses to aflatoxin B1 in poultry. Agriculture 2015, 5, 742–777. [Google Scholar] [CrossRef]
- Rawal, S.; Kim, J.E.; Coulombe, R. Aflatoxin B1 in poultry: Toxicology, metabolism and prevention. Res. Vet. Sci. 2010, 89, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Loi, M.; Paciolla, C.; Logrieco, A.F.; Mulè, G. Plant bioactive compounds in pre- and postharvest management for aflatoxins reduction. Front. Microbiol. 2020, 11, 243. [Google Scholar] [CrossRef]
- Wahyono, N.D.; Utami, M.D. A review of the poultry meat production industry for food safety in Indonesia. J. Phys. Conf. Ser. 2018, 953, 012125. [Google Scholar] [CrossRef]
- Alberts, J.F.; Gelderblom, W.C.A.; Botha, A.; van Zyl, W.H. Degradation of aflatoxin B(1) by fungal laccase enzymes. Int. J. Food Microbiol. 2009, 135, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.C.; Jackson, C.J.; Tattersall, D.B.; French, N.; Peat, T.S.; Newman, J.; Briggs, L.J.; Lapalikar, G.V.; Campbell, P.M.; Scott, C.; et al. Identification and characterization of two families of F420 H2-dependent reductases from mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 2010, 78, 561–575. [Google Scholar] [CrossRef]
- Zhao, L.H.; Guan, S.; Gao, X.; Ma, Q.G.; Lei, Y.P.; Bai, X.M.; Ji, C. Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 2011, 110, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qin, X.; Tang, Y.; Ma, Q.; Zhang, J.; Zhao, L. CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chem. 2020, 325, 126877. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Fan, Y.; Zhao, L. Review on biological degradation of mycotoxins. Anim. Nutr. 2016, 2, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Adebo, O.A.; Njobeh, P.B.; Gbashi, S.; Nwinyi, O.C.; Mavumengwana, V. Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. Nutr. 2017, 57, 3208–3217. [Google Scholar] [CrossRef] [PubMed]
- Lyagin, I.; Efremenko, E. Enzymes for detoxification of various mycotoxins: Origins and mechanisms of catalytic action. Molecules 2019, 24, 2362. [Google Scholar] [CrossRef]
- Guan, Y.; Chen, J.; Nepovimova, E.; Long, M.; Wu, W.; Kuca, K. Aflatoxin detoxification using microorganisms and enzymes. Toxins 2021, 13, 46. [Google Scholar] [CrossRef] [PubMed]
- Verheecke, C.; Liboz, T.; Mathieu, F. Microbial degradation of aflatoxin B1: Current status and future advances. Int. J. Food Microbiol. 2016, 237, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, L.; Sun, J.; Wang, L.; Guo, H.; Ye, Y.; Sun, X. Microbial detoxification of mycotoxins in food and feed. Crit. Rev. Food Sci. Nutr. 2022, 62, 4951–4969. [Google Scholar] [CrossRef]
- Smaoui, S.; D’Amore, T.; Tarapoulouzi, M.; Agriopoulou, S.; Varzakas, T. Aflatoxins contamination in feed commodities: From occurrence and toxicity to recent advances in analytical methods and detoxification. Microorganisms 2023, 11, 2614. [Google Scholar] [CrossRef]
- Kolosova, A.; Stroka, J. Substances for reduction of the contamination of feed by mycotoxins: A review. World Mycotoxin J. 2011, 4, 225–256. [Google Scholar] [CrossRef]
- Loi, M.; Fanelli, F.; Zucca, P.; Liuzzi, V.C.; Quintieri, L.; Cimmarusti, M.T.; Monaci, L.; Haidukowski, M.; Logrieco, A.F.; Sanjust, E.; et al. Aflatoxin B1 and M1 degradation by Lac2 from Pleurotus pulmonarius and redox mediators. Toxins 2016, 8, 245. [Google Scholar] [CrossRef]
- Branà, M.T.; Sergio, L.; Haidukowski, M.; Logrieco, A.F.; Altomare, C. Degradation of Aflatoxin B1 by a sustainable enzymatic extract from spent mushroom substrate of Pleurotus eryngii. Toxins 2020, 12, 49. [Google Scholar] [CrossRef]
- Liu, D.L.; Yao, D.S.; Liang, Y.Q.; Zhou, T.H.; Song, Y.P.; Zhao, L.; Ma, L. Production, purification, and characterization of an intracellular aflatoxin-detoxifizyme from Armillariella tabescens (E-20). Food Chem. Toxicol. 2001, 39, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ogata, M.; Hirai, H.; Kawagishi, H. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 2011, 314, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mao, H.; Hu, C.; Tron, T.; Lin, J.; Wang, J.; Sun, B. Molecular docking studies and in vitro degradation of four aflatoxins (AFB1, AFB2, AFG1, and AFG2) by a recombinant laccase from Saccharomyces cerevisiae. J. Food Sci. 2020, 85, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Ohkusu, K.; Okuda, M.; Imataki, O.; Ishii, T.; Negayama, K.; Tadokoro, A.; Kita, N.; Takagi, T.; Kanaji, N.; et al. Phanerochaete sordida as a cause of pulmonary nodule in an immunocompromised patient: A case report. BMC Infect. Dis. 2017, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Brugnari, T.; Braga, D.M.; dos Santos, C.S.A.; Torres, B.H.C.; Modkovski, T.A.; Haminiuk, C.W.I.; Maciel, G.M. Laccases as green and versatile biocatalysts: From lab to enzyme market—An overview. Bioresour. Bioprocess. 2021, 8, 131. [Google Scholar] [CrossRef]
- Rodríguez-Escribano, D.; Pliego-Magán, R.; de Salas, F.; Aza, P.; Gentili, P.; Ihalainen, P.; Levée, T.; Meyer, V.; Petit-Conil, M.; Tapin-Lingua, S.; et al. Tailor-made alkaliphilic and thermostable fungal laccases for industrial wood processing. Biotechnol. Biofuels 2022, 15, 149. [Google Scholar] [CrossRef]
- Bao, C.; Liu, Y.; Li, F.; Cao, H.; Dong, B.; Cao, Y. Expression and characterization of laccase Lac1 from Coriolopsis trogii strain Mafic-2001 in Pichia pastoris and its degradation of lignin. Appl. Biochem. Biotechnol. 2023, 195, 6150–6167. [Google Scholar] [CrossRef]
- Cao, H.; Liu, D.; Mo, X.; Xie, C.; Yao, D. A fungal enzyme with the ability of aflatoxin B1 conversion: Purification and ESI-MS/MS identification. Microbiol. Res. 2011, 166, 475–483. [Google Scholar] [CrossRef]
- Schuda, P.F. Aflatoxin chemistry and syntheses. In Syntheses of Natural Products. Topics in Current Chemistry; Springer: Berlin/Heidelberg, Germany, 1980; Volume 91, pp. 75–111. [Google Scholar] [CrossRef]
- Wu, Y.Z.; Lu, F.P.; Jiang, H.L.; Tan, C.P.; Yao, D.S.; Xie, C.F.; Liu, D.L. The furofuran-ring selectivity, hydrogen peroxide-production and low km value are the three elements for highly effective detoxification of aflatoxin oxidase. Food Chem. Toxicol. 2015, 76, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, H.W.; Kelton, W.H. Production of sterigmatocystin by some species of the genus Aspergillus and its toxicity to chicken embryos. Appl. Microbiol. 1975, 30, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mei, M.; Wang, J.; Huang, J.; Zong, X.; Wang, X. Expression and Application of Aflatoxin Degrading Enzyme Gene in Pichia pastoris. Biotechnol. J. 2023, 2300167. [Google Scholar] [CrossRef]
- Chen, J.; Liu, D.; Li, S.; Yao, D. Development of an amperometric enzyme electrode biosensor for sterigmatocystin detection. Enz. Microb. Technol. 2010, 47, 119–126. [Google Scholar] [CrossRef]
- Wen, S.; Guan, M.; Zhou, T.; Cao, H.; Xie, C.; Liu, D.; Yao, D. Cloning, expression, purification and characterization of an aflatoxin-converting enzyme from Armillaria tabescens. Wei Sheng Wu Xue Bao 2011, 51, 1212–1221. [Google Scholar] [PubMed]
- Sinelnikov, I.G.; Zorov, I.N.; Denisenko, Y.A.; Mikityuk, O.O.; Sinitsyn, A.P.; Shcherbakova, L.A. A new producer of a recombinant aflatoxin-degrading enzyme obtained via heterologous expression in Pichia pastoris. Agric. Biol. 2022, 57, 1166–1177. (In Russian) [Google Scholar] [CrossRef]
- Yang, P.; Xiao, W.; Lu, S.; Jiang, S.; Zheng, Z.; Zhang, D.; Zhang, M.; Jiang, S.; Jiang, S. Recombinant expression of Trametes versicolor aflatoxin B1-degrading enzyme (TV-AFB1D) in engineering Pichia pastoris GS115 and application in AFB1 degradation in AFB1-contaminated peanuts. Toxins 2021, 13, 349. [Google Scholar] [CrossRef]
- Li, C.H.; Li, W.Y.; Hsu, I.N.; Liao, Y.Y.; Yang, C.Y.; Taylor, M.C.; Liu, Y.F.; Huang, W.H.; Chang, H.H.; Huang, H.L.; et al. Recombinant aflatoxin-degrading F420H2-dependent reductase from Mycobacterium smegmatis protects mammalian cells from aflatoxin toxicity. Toxins 2019, 11, 259. [Google Scholar] [CrossRef]
- Shcherbakova, L.; Rozhkova, A.; Osipov, D.; Zorov, I.; Mikityuk, O.; Statsyuk, N.; Sinitsyna, O.; Dzhavakhiya, V.; Sinitsyn, A. Effective zearalenone degradation in model solutions and infected wheat grain using a novel heterologous lactonohydrolase secreted by recombinant Penicillium canescens. Toxins 2020, 12, 475. [Google Scholar] [CrossRef]
- Michlmayr, H.; Malachová, A.; Varga, E.; Kleinová, J.; Lemmens, M.; Newmister, S.; Rayment, I.; Berthiller, F.; Adam, G. Biochemical characterization of a recombinant UDP-glucosyltransferase from rice and enzymatic production of deoxynivalenol-3-O-β-D-glucoside. Toxins 2015, 7, 2685–2700. [Google Scholar] [CrossRef]
- Xu, T.; Xie, C.; Yao, D.; Zhou, C.Z.; Liu, J. Crystal structures of aflatoxin-oxidase from Armillariella tabescens reveal a dual activity enzyme. Biochem. Biophys. Res. Commun. 2017, 494, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, I.R.; Mikhalev, A.V.; Satyukova, L.P.; Borisova, V.S. Current state of feed quality and safety in Russia. Veterinariya 2009, 2, 3–7. (In Russian) [Google Scholar]
- Borisova, T. Screening analysis of mycotoxins in grain and food products. Analytics 2017, 2, 98–103. (In Russian) [Google Scholar] [CrossRef]
- Loi, M.; Logrieco, A.F.; Pusztahelyi, T.; Leiter, É.; Hornok, L.; Pócsi, I. Advanced mycotoxin control and decontamination techniques in view of an increased aflatoxin risk in Europe due to climate change. Front. Microbiol. 2023, 13, 1085891. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, A.; Soldatenko, N.; Fetisov, L.; Sukhikh, E. Mycotoxicological monitoring of feed in the North Caucasus region. Kombikorma 2011, 3, 98–99. (In Russian) [Google Scholar]
- Moretti, A.; Pascale, M.; Logrieco, A.F. Mycotoxin risks under a climate change scenario in Europe. Trends Food Sci. Technol. 2019, 84, 38–40. [Google Scholar] [CrossRef]
- Hawkins, L.K.; Windham, G.L.; Williams, W.P. Effect of different postharvest drying temperatures on Aspergillus flavus survival and aflatoxin content in five maize hybrids. J. Food Prot. 2005, 68, 1521–1524. [Google Scholar] [CrossRef]
- Siciliano, I.; Berta, F.; Bosio, P.; Gullino, M.L.; Garibaldi, A. Effect of different temperatures and CO2 levels on Alternaria toxins produced on cultivated rocket, cabbage and cauliflower. World Mycotoxin J. 2017, 10, 63–71. [Google Scholar] [CrossRef]
- Rushing, B.R.; Selim, M.I. Aflatoxin B1: A review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem. Toxicol. 2019, 124, 81–100. [Google Scholar] [CrossRef]
- Toteja, G.S.; Mukherjee, A.; Diwakar, S.; Singh, P.; Saxena, B.N.; Sinha, K.K.; Sinha, A.K.; Kumar, N.; Nagaraja, K.V.; Bai, G.; et al. Aflatoxin B1 contamination in wheat grain samples collected from different geographical regions of India: A multicenter study. J. Food Prot. 2006, 69, 1463–1467. [Google Scholar] [CrossRef]
- Summary of Food and Agricultural Statistics 2004. Available online: https://www.fao.org/3/ae881e/ae881e00.htm (accessed on 29 October 2023).
- Dobolyi, C.; Sebők, F.; Varga, J.; Kocsubé, S.; Szigeti, G.; Baranyi, N.; Szécsi, Á.; Tóth, B.; Varga, M.; Kriszt, B.; et al. Occurrence of aflatoxin producing Aspergillus flavus isolates in maize kernel in Hungary. Acta Aliment. 2013, 42, 451–459. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R. Simplified protocol for faster transformation of (a large number of) Pichia pastoris strains. Yeast 2019, 36, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques 2011, 50, 325–328. [Google Scholar] [CrossRef]
- Ghosalkar, A.; Sahai, V.; Srivastava, A. Optimization of chemically defined medium for recombinant Pichia pastoris for biomass production. Bioresour. Technol. 2008, 99, 7906–7910. [Google Scholar] [CrossRef]
- Bernáldez, V.; Córdoba, J.J.; Magan, N.; Peromingo, B.; Rodríguez, A. The influence of ecophysiological factors on growth, aflR gene expression and aflatoxin B1 production by a type strain of Aspergillus flavus. LWT-Food Sci. Technol. 2017, 83, 283–291. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinelnikov, I.; Mikityuk, O.; Shcherbakova, L.; Nazarova, T.; Denisenko, Y.; Rozhkova, A.; Statsyuk, N.; Zorov, I. Recombinant Oxidase from Armillaria tabescens as a Potential Tool for Aflatoxin B1 Degradation in Contaminated Cereal Grain. Toxins 2023, 15, 678. https://doi.org/10.3390/toxins15120678
Sinelnikov I, Mikityuk O, Shcherbakova L, Nazarova T, Denisenko Y, Rozhkova A, Statsyuk N, Zorov I. Recombinant Oxidase from Armillaria tabescens as a Potential Tool for Aflatoxin B1 Degradation in Contaminated Cereal Grain. Toxins. 2023; 15(12):678. https://doi.org/10.3390/toxins15120678
Chicago/Turabian StyleSinelnikov, Igor, Oleg Mikityuk, Larisa Shcherbakova, Tatyana Nazarova, Yury Denisenko, Alexandra Rozhkova, Natalia Statsyuk, and Ivan Zorov. 2023. "Recombinant Oxidase from Armillaria tabescens as a Potential Tool for Aflatoxin B1 Degradation in Contaminated Cereal Grain" Toxins 15, no. 12: 678. https://doi.org/10.3390/toxins15120678
APA StyleSinelnikov, I., Mikityuk, O., Shcherbakova, L., Nazarova, T., Denisenko, Y., Rozhkova, A., Statsyuk, N., & Zorov, I. (2023). Recombinant Oxidase from Armillaria tabescens as a Potential Tool for Aflatoxin B1 Degradation in Contaminated Cereal Grain. Toxins, 15(12), 678. https://doi.org/10.3390/toxins15120678




