The Origin of Teratogenic Retinoids in Cyanobacteria
Abstract
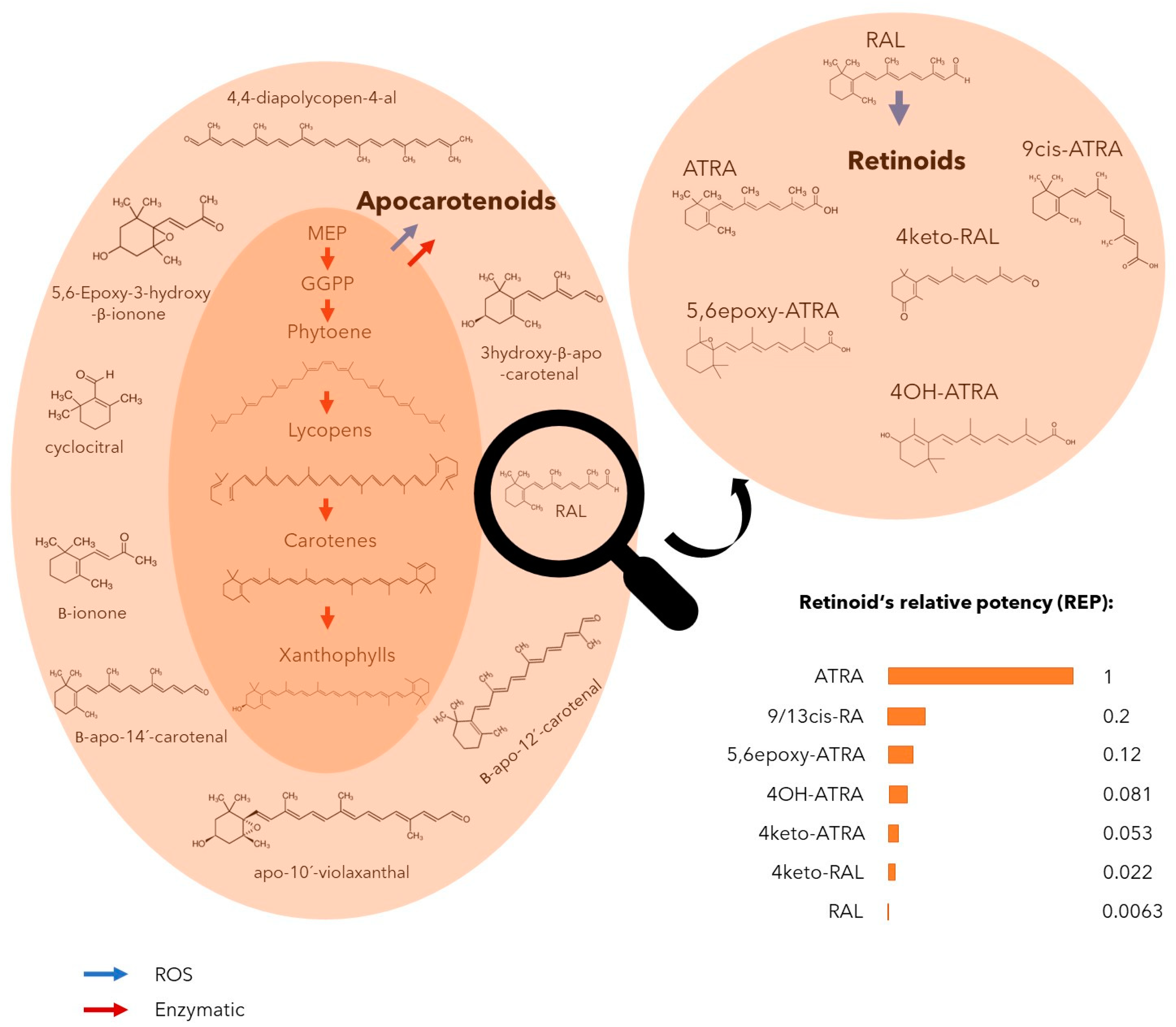
:1. Introduction
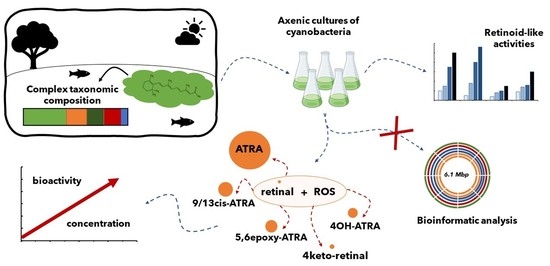
2. Results and Discussion
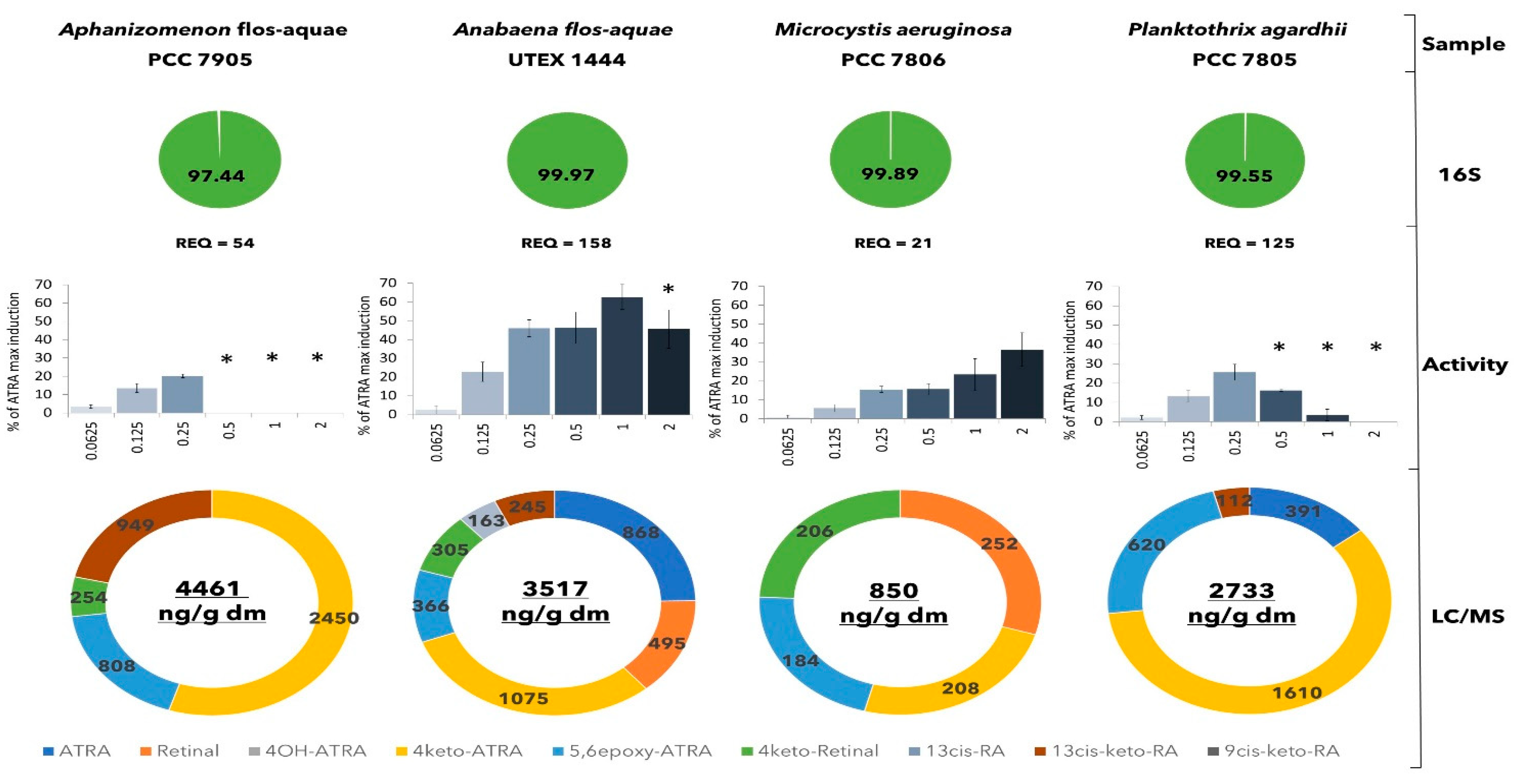
2.1. Lab Cultures of Cyanobacteria and Their Retinoids Production
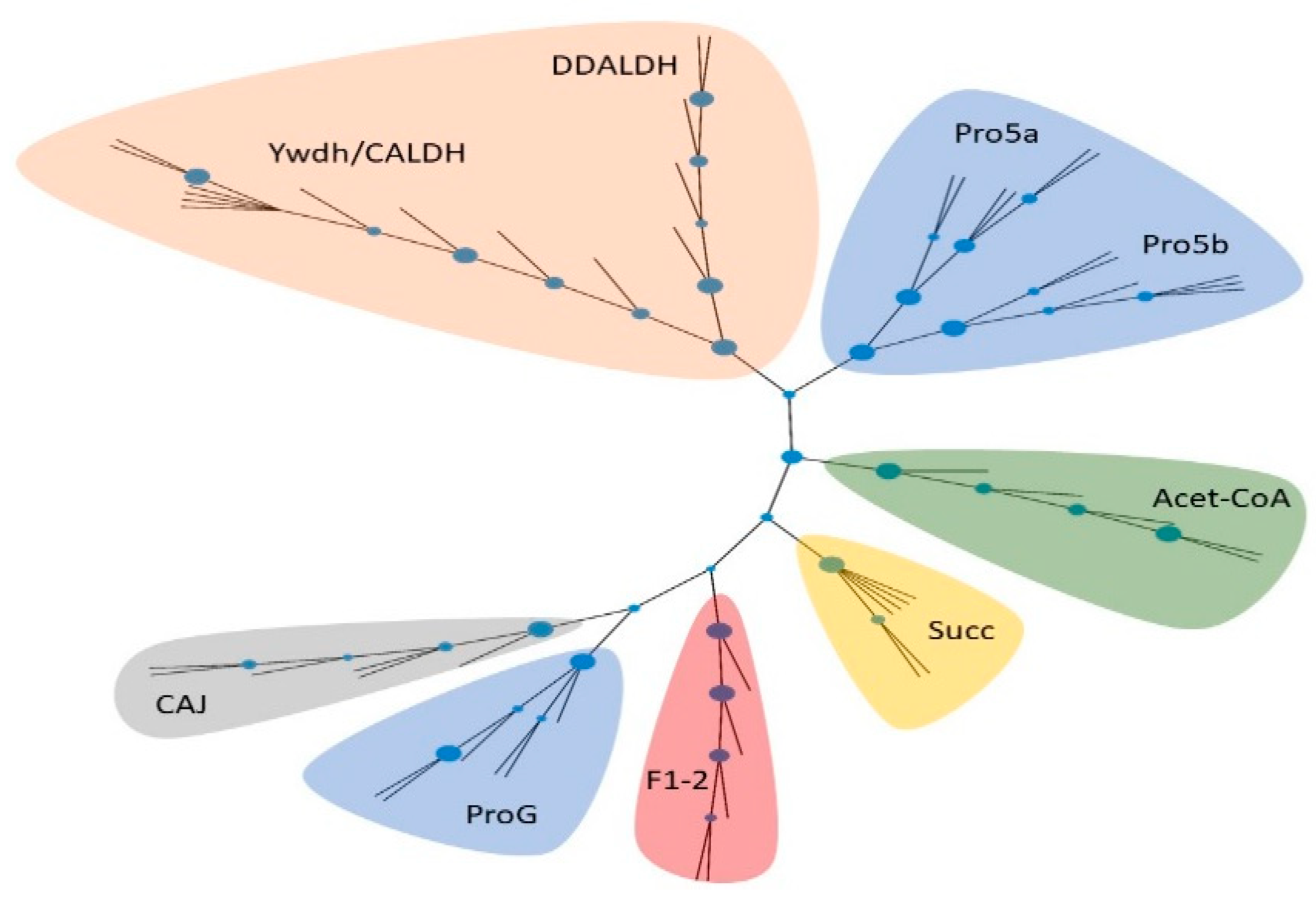
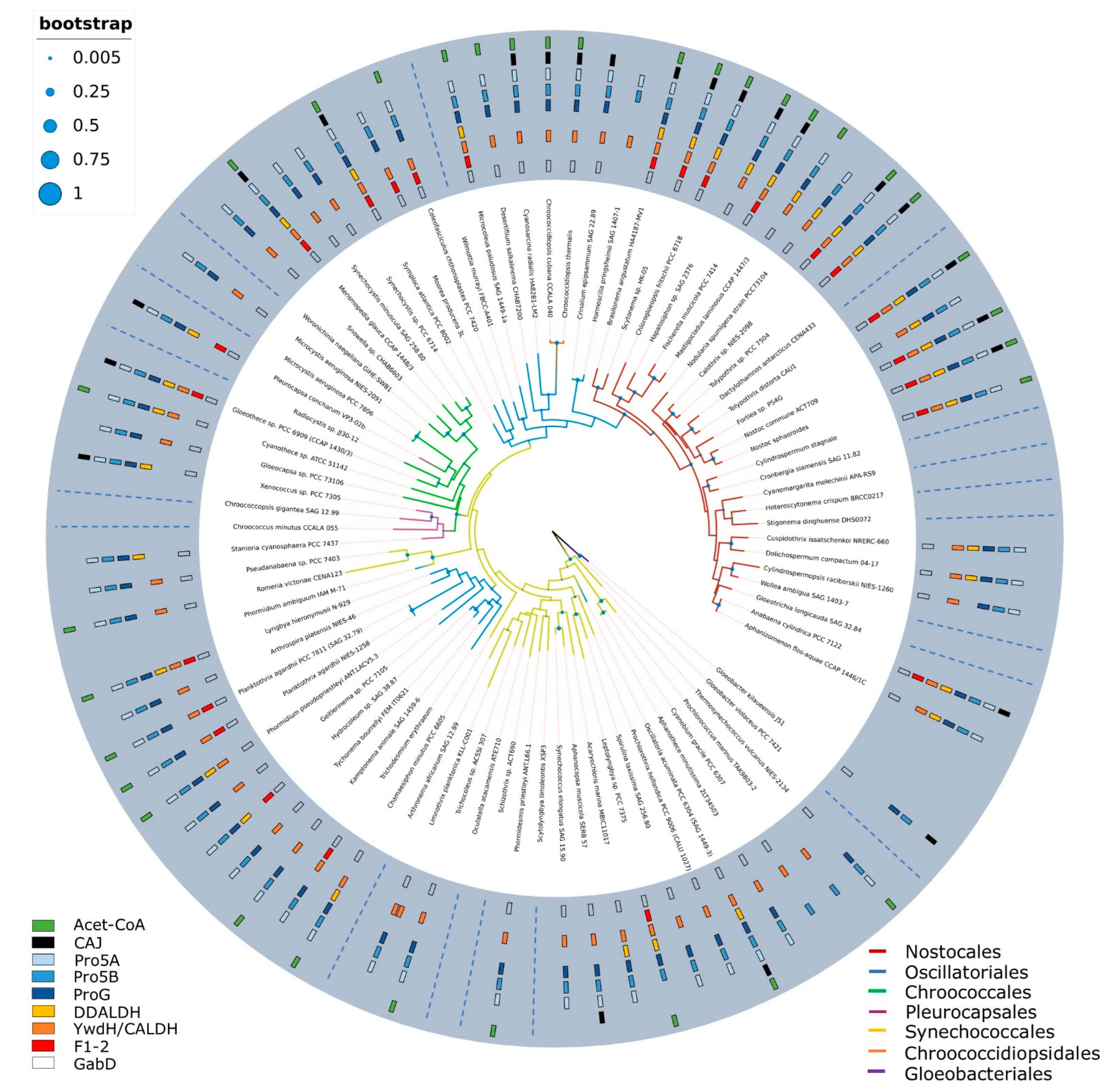
2.2. Absence of Genetic Basis for ATRA Biosynthesis across Cyanobacterial Genomes
2.3. ROS-Mediated Oxidative Conversion of Retinal to More Bioactive Retinoids
2.4. Production of Retinoids and Biological Function
3. Conclusions
4. Materials and Methods
4.1. Cultivation of Axenic Cyanobacterial Cultures
4.2. 16S Amplicon Sequencing
4.2.1. DNA Extraction
4.2.2. Illumina Library Preparation
4.2.3. Bioinformatic Processing of 16S rRNA Data
4.3. Search for Enzyme Converting RAL to ATRA
4.4. Distribution of Aldh across Cyanobacterial Genomes
4.5. Phylogenetic Analysis of Aldh
4.6. Biomass Extraction
4.7. Fenton Reaction
4.7.1. Experimental Design
4.7.2. Experimental Set-Up
4.8. Retinoid-like Activity Analysis
4.9. LC-MS/MS Analysis of Retinoids
4.10. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
- Illumina library preparation
- Bioinformatic processing of 16S rRNA data
- List of chemicals and reagents
- LC-MS/MS analysis of retinoids
References
- Gutierrez-Mazariegos, J.; Nadendla, E.K.; Lima, D.; Pierzchalski, K.; Jones, J.W.; Kane, M.; Nishikawa, J.-I.; Hiromori, Y.; Nakanishi, T.; Santos, M.M.; et al. A Mollusk Retinoic Acid Receptor (RAR) Ortholog Sheds Light on the Evolution of Ligand Binding. Endocrinology 2014, 155, 4275–4286. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Gibert, Y. Retinoids in Embryonic Development. Biomolecules 2020, 10, 1278. [Google Scholar] [CrossRef] [PubMed]
- Chambon, P. A Decade of Molecular Biology of Retinoic Acid Receptors. FASEB J. 1996, 10, 940–954. [Google Scholar] [CrossRef] [PubMed]
- Kam, R.K.T.; Deng, Y.; Chen, Y.; Zhao, H. Retinoic Acid Synthesis and Functions in Early Embryonic Development. Cell Biosci. 2012, 2, 11. [Google Scholar] [CrossRef]
- Das, B.C.; Thapa, P.; Karki, R.; Das, S.; Mahapatra, S.; Liu, T.-C.; Torregroza, I.; Wallace, D.P.; Kambhampati, S.; Van Veldhuizen, P.; et al. Retinoic Acid Signaling Pathways in Development and Diseases. Bioorg. Med. Chem. 2014, 22, 673–683. [Google Scholar] [CrossRef]
- Eroglu, A.; Hruszkewycz, D.P.; Dela Sena, C.; Narayanasamy, S.; Riedl, K.M.; Kopec, R.E.; Schwartz, S.J.; Curley, R.W.; Harrison, E.H. Naturally Occurring Eccentric Cleavage Products of Provitamin A β-Carotene Function as Antagonists of Retinoic Acid Receptors. J. Biol. Chem. 2012, 287, 15886–15895. [Google Scholar] [CrossRef]
- Morriss-Kay, G.M.; Wardt, S.J. Retinoids and Mammalian Development. Int. Rev. Cytol. 1999, 188, 73–131. [Google Scholar] [CrossRef]
- Javůrek, J.; Sychrová, E.; Smutná, M.; Bittner, M.; Kohoutek, J.; Adamovský, O.; Nováková, K.; Smetanová, S.; Hilscherová, K. Retinoid Compounds Associated with Water Blooms Dominated by Microcystis Species. Harmful Algae 2015, 47, 116–125. [Google Scholar] [CrossRef]
- Wu, X.; Jiang, J.; Hu, J. Determination and Occurrence of Retinoids in a Eutrophic Lake (Taihu Lake, China): Cyanobacteria Blooms Produce Teratogenic Retinal. Environ. Sci. Technol. 2013, 47, 807–814. [Google Scholar] [CrossRef]
- Wu, X.; Jiang, J.; Wan, Y.; Giesy, J.P.; Hu, J. Cyanobacteria Blooms Produce Teratogenic Retinoic Acids. Proc. Natl. Acad. Sci. USA 2012, 109, 9477–9482. [Google Scholar] [CrossRef] [Green Version]
- Sehnal, L.; Procházková, T.; Smutná, M.; Kohoutek, J.; Lepšová-Skácelová, O.; Hilscherová, K. Widespread Occurrence of Retinoids in Water Bodies Associated with Cyanobacterial Blooms Dominated by Diverse Species. Water Res. 2019, 156, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Jonas, A.; Scholz, S.; Fetter, E.; Sychrova, E.; Novakova, K.; Ortmann, J.; Benisek, M.; Adamovsky, O.; Giesy, J.P.; Hilscherova, K. Endocrine, Teratogenic and Neurotoxic Effects of Cyanobacteria Detected by Cellular in Vitro and Zebrafish Embryos Assays. Chemosphere 2015, 120, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Pipal, M.; Priebojova, J.; Koci, T.; Blahova, L.; Smutna, M.; Hilscherova, K. Field Cyanobacterial Blooms Producing Retinoid Compounds Cause Teratogenicity in Zebrafish Embryos. Chemosphere 2020, 241, 125061. [Google Scholar] [CrossRef] [PubMed]
- Jonas, A.; Buranova, V.; Scholz, S.; Fetter, E.; Novakova, K.; Kohoutek, J.; Hilscherova, K. Retinoid-like Activity and Teratogenic Effects of Cyanobacterial Exudates. Aquat. Toxicol. 2014, 155, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Ruch, S.; Beyer, P.; Ernst, H.; Al-Babili, S. Retinal Biosynthesis in Eubacteria: In Vitro Characterization of a Novel Carotenoid Oxygenase from Synechocystis Sp. PCC 6803. Mol. Microbiol. 2005, 55, 1015–1024. [Google Scholar] [CrossRef]
- Scherzinger, D.; Ruch, S.; Kloer, D.P.; Wilde, A.; Al-Babili, S. Retinal Is Formed from Apo-Carotenoids in Nostoc Sp. PCC7120: In Vitro Characterization of an Apo-Carotenoid Oxygenase. Biochem. J. 2006, 398, 361–369. [Google Scholar] [CrossRef]
- Cui, H.; Wang, Y.; Qin, S. Genomewide Analysis of Carotenoid Cleavage Dioxygenases in Unicellular and Filamentous Cyanobacteria. Comp. Funct. Genom. 2012, 2012, 164690. [Google Scholar] [CrossRef]
- Miles, J.A.; Machattou, P.; Nevin-Jones, D.; Webb, M.E.; Millard, A.; Scanlan, D.J.; Taylor, P.C. Identification of a Cyanobacterial Aldehyde Dehydrogenase That Produces Retinoic Acid in Vitro. Biochem. Biophys. Res. Commun. 2019, 510, 27–34. [Google Scholar] [CrossRef]
- Mordi, R.C.; Ademosun, O.T.; Ajanaku, C.O.; Olanrewaju, I.O.; Walton, J.C. Free Radical Mediated Oxidative Degradation of Carotenes and Xanthophylls. Molecules 2020, 25, 1038. [Google Scholar] [CrossRef]
- Pípal, M.; Novák, J.; Rafajová, A.; Smutná, M.; Hilscherová, K. Teratogenicity of Retinoids Detected in Surface Waters in Zebrafish Embryos and Its Predictability by in Vitro Assays. Aquat. Toxicol. 2022, 246, 106151. [Google Scholar] [CrossRef]
- Priebojová, J.; Hilscherová, K.; Procházková, T.; Sychrová, E.; Smutná, M. Intracellular and Extracellular Retinoid-like Activity of Widespread Cyanobacterial Species. Ecotoxicol. Environ. Saf. 2018, 150, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Ngo, H.P.T.; Nam, H.K.; Kim, K.R.; Kang, L.W.; Oh, D.K. Alternative Biotransformation of Retinal to Retinoic Acid or Retinol by an Aldehyde Dehydrogenase from Bacillus Cereus. Appl. Environ. Microbiol. 2016, 82, 3940. [Google Scholar] [CrossRef] [PubMed]
- Trautmann, D.; Beyer, P.; Al-Babili, S. The ORF Slr0091 of Synechocystis Sp. PCC6803 Encodes a High-Light Induced Aldehyde Dehydrogenase Converting Apocarotenals and Alkanals. FEBS J. 2013, 280, 3685–3696. [Google Scholar] [CrossRef] [PubMed]
- Mordi, R.C.; Walton, J.C. Identification of Products from Canthaxanthin Oxidation. Food Chem. 2016, 197, 836–840. [Google Scholar] [CrossRef] [PubMed]
- Alder, A.; Bigler, P.; Werck-Reichhart, D.; Al-Babili, S. In Vitro Characterization of Synechocystis Cyp120a1 Revealed the First Nonanimal Retinoic Acid Hydroxylase. FEBS J. 2009, 276, 5416–5431. [Google Scholar] [CrossRef] [PubMed]
- Salgado, P.; Melin, V.; Contreras, D.; Moreno, Y.; Mansilla, H.D. Fenton Reaction Driven by Iron Ligands. J. Chil. Chem. Soc. 2013, 58, 2096–2101. [Google Scholar] [CrossRef]
- Tadolini, B.; Cabrini, L. On the Mechanism of OH. Scavenger Action. Biochem. J. 1988, 253, 931–932. [Google Scholar] [CrossRef]
- Yoshimura, Y.; Matsuzaki, Y.; Watanabe, T.; Uchiyama, K.; Ohsawa, K.; Imaeda, K. Effects of Buffer Sofutions and Chelators on the Generation of Hydroxyl Radical and the Lipid Peroxidation in the Fenton Reaction System. J. Clin. Biochem. Nutr. 1992, 13, 147–154. [Google Scholar] [CrossRef]
- Ahrazem, O.; Gómez-Gómez, L.; Rodrigo, M.J.; Avalos, J.; Limón, M.C. Carotenoid Cleavage Oxygenases from Microbes and Photosynthetic Organisms: Features and Functions. Int. J. Mol. Sci. 2016, 17, 1781. [Google Scholar] [CrossRef]
- Ribeiro, D.; Sousa, A.; Nicola, P.; Ferreira de Oliveira, J.M.P.; Rufino, A.T.; Silva, M.; Freitas, M.; Carvalho, F.; Fernandes, E. β-Carotene and Its Physiological Metabolites: Effects on Oxidative Status Regulation and Genotoxicity in in Vitro Models. Food Chem. Toxicol. 2020, 141, 111392. [Google Scholar] [CrossRef]
- Ramel, F.; Birtic, S.; Ginies, C.; Soubigou-Taconnat, L.; Triantaphylidès, C.; Havaux, M. Carotenoid Oxidation Products Are Stress Signals That Mediate Gene Responses to Singlet Oxygen in Plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5535–5540. [Google Scholar] [CrossRef] [PubMed]
- Schlösser, U.G. SAG - Sammlung von Algenkulturen at the University of Göttingen Catalogue of Strains 1994. Bot. Acta 1994, 107, 113–186. [Google Scholar] [CrossRef]
- Stein-Taylor, J.R. Handbook of Phycological Methods: Culture Methods and Growth Measurements. Camb. Univ. Press 1973, 460. [Google Scholar]
- Moosová, Z.; Šindlerová, L.; Ambrůzová, B.; Ambrožová, G.; Vašíček, O.; Velki, M.; Babica, P.; Kubala, L. Lipopolysaccharides from Microcystis Cyanobacteria-Dominated Water Bloom and from Laboratory Cultures Trigger Human Immune Innate Response. Toxins 2019, 11, 218. [Google Scholar] [CrossRef] [PubMed]
- Morin, N.; Vallaeys, T.; Hendrickx, L.; Natalie, L.; Wilmotte, A. An Efficient DNA Isolation Protocol for Filamentous Cyanobacteria of the Genus Arthrospira. J. Microbiol. Methods 2010, 80, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Schirrmeister, B.E.; Antonelli, A.; Bagheri, H.C. The Origin of Multicellularity in Cyanobacteria. BMC Evol. Biol. 2011, 11, 1–21. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Masuda, K.; Tanaka, Y.; Kanehisa, M.; Ninomiya, T.; Inoue, A.; Higuma, H.; Kawashima, C.; Nakanishi, M.; Okamoto, K.; Akiyoshi, J. Natural Reduced Water Suppressed Anxiety and Protected the Heightened Oxidative Stress in Rats. Neuropsychiatr. Dis. Treat. 2017, 13, 2357–2362. [Google Scholar] [CrossRef]
- Blough, N.V.; Zepp, R.G. Reactive Oxygen Species in Natural Waters. In Active Oxygen in Chemistry; Springer: Dordrecht, The Netherlands, 1995; pp. 280–333. [Google Scholar]
- Primo, O.; Rueda, A.; Rivero, M.J.; Ortiz, I. An Integrated Process, Fenton Reaction−Ultrafiltration, for the Treatment of Landfill Leachate: Pilot Plant Operation and Analysis. Ind. Eng. Chem. Res. 2008, 47, 946–952. [Google Scholar] [CrossRef]
- Villeneuve, D.L.; Blankenship, A.L.; Giesy, J.P. Derivation and Application of Relative Potency Estimates Based on in Vitro Bioassay Results. Environ. Toxicol. Chem. 2000, 19, 2835–2843. [Google Scholar] [CrossRef]
- Apprill, A.; Mcnally, S.; Parsons, R.; Weber, L. Minor Revision to V4 Region SSU RRNA 806R Gene Primer Greatly Increases Detection of SAR11 Bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pẽa, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Stoeck, T.; Bass, D.; Nebel, M.; Christen, R.; Jones, M.D.M.; Breiner, H.W.; Richards, T.A. Multiple Marker Parallel Tag Environmental DNA Sequencing Reveals a Highly Complex Eukaryotic Community in Marine Anoxic Water. Mol. Ecol. 2010, 19, 21–31. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Aronesty, E. Comparison of Sequencing Utility Programs. Open Bioinforma. J. 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sehnal, L.; Smutná, M.; Bláhová, L.; Babica, P.; Šplíchalová, P.; Hilscherová, K. The Origin of Teratogenic Retinoids in Cyanobacteria. Toxins 2022, 14, 636. https://doi.org/10.3390/toxins14090636
Sehnal L, Smutná M, Bláhová L, Babica P, Šplíchalová P, Hilscherová K. The Origin of Teratogenic Retinoids in Cyanobacteria. Toxins. 2022; 14(9):636. https://doi.org/10.3390/toxins14090636
Chicago/Turabian StyleSehnal, Luděk, Marie Smutná, Lucie Bláhová, Pavel Babica, Petra Šplíchalová, and Klára Hilscherová. 2022. "The Origin of Teratogenic Retinoids in Cyanobacteria" Toxins 14, no. 9: 636. https://doi.org/10.3390/toxins14090636
APA StyleSehnal, L., Smutná, M., Bláhová, L., Babica, P., Šplíchalová, P., & Hilscherová, K. (2022). The Origin of Teratogenic Retinoids in Cyanobacteria. Toxins, 14(9), 636. https://doi.org/10.3390/toxins14090636