On the Hunt for New Toxin Families Produced by a Mediterranean Strain of the Benthic Dinoflagellate Ostreopsis cf. ovata
Abstract
:1. Introduction
2. Results
2.1. Experimental Section
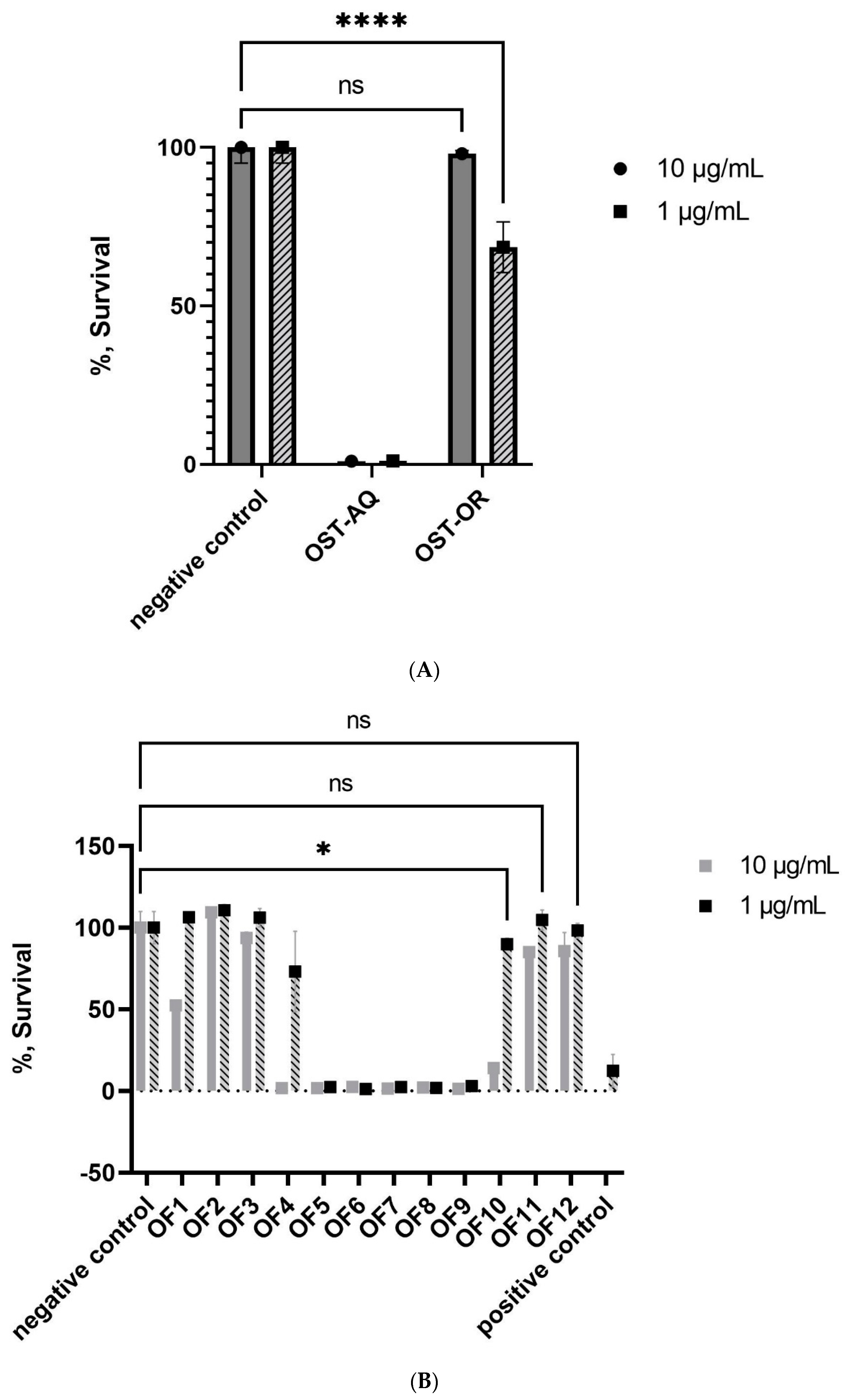
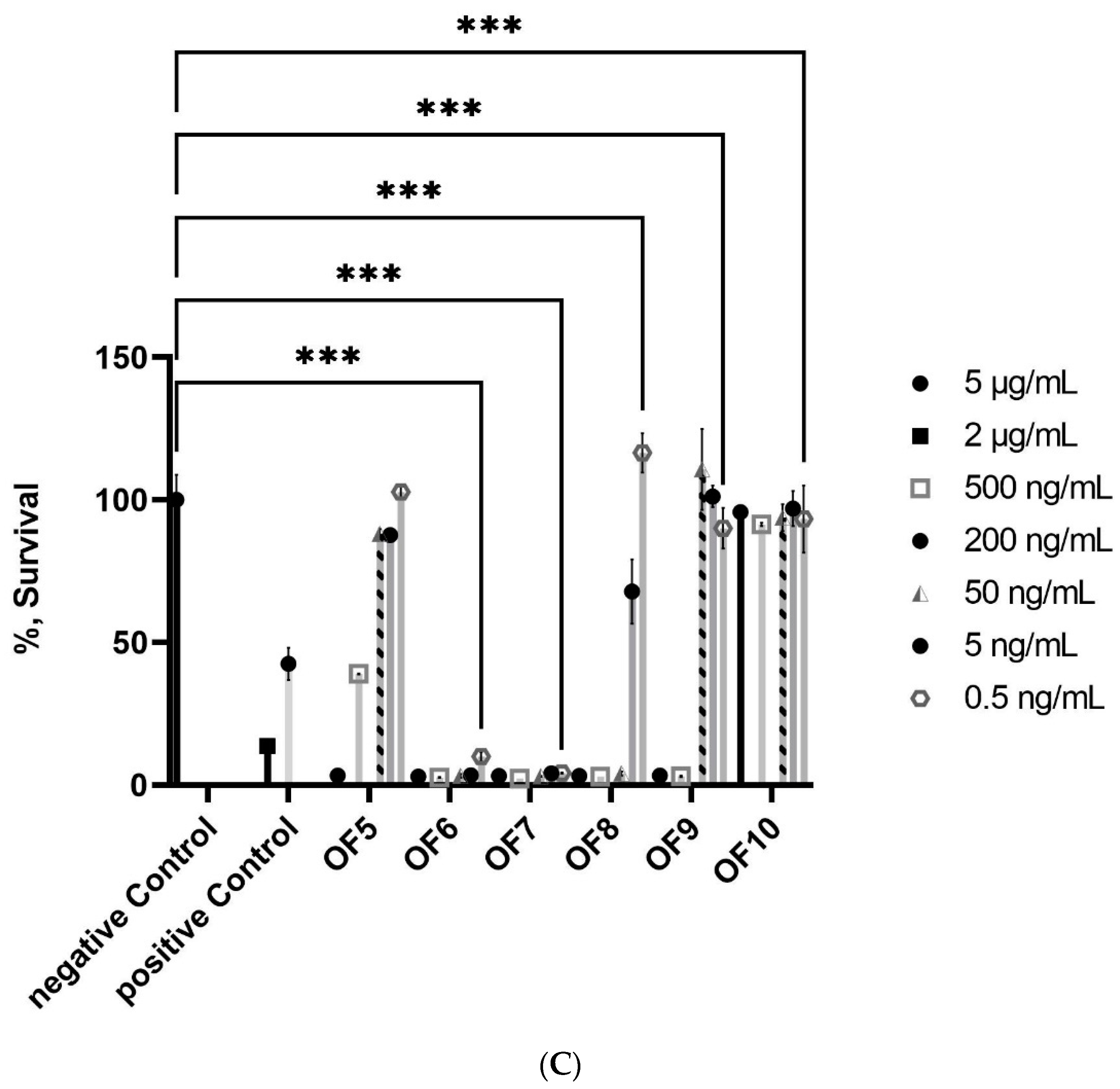
2.1.1. Bioactivity
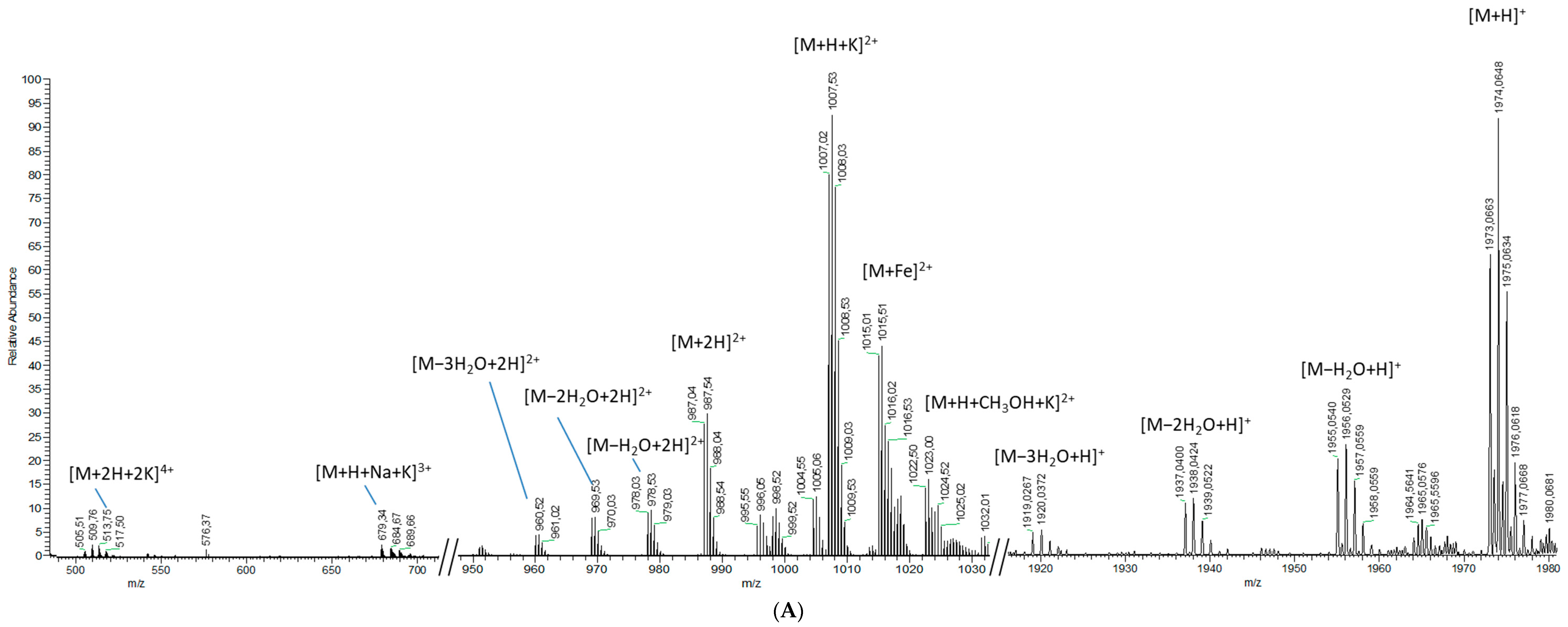
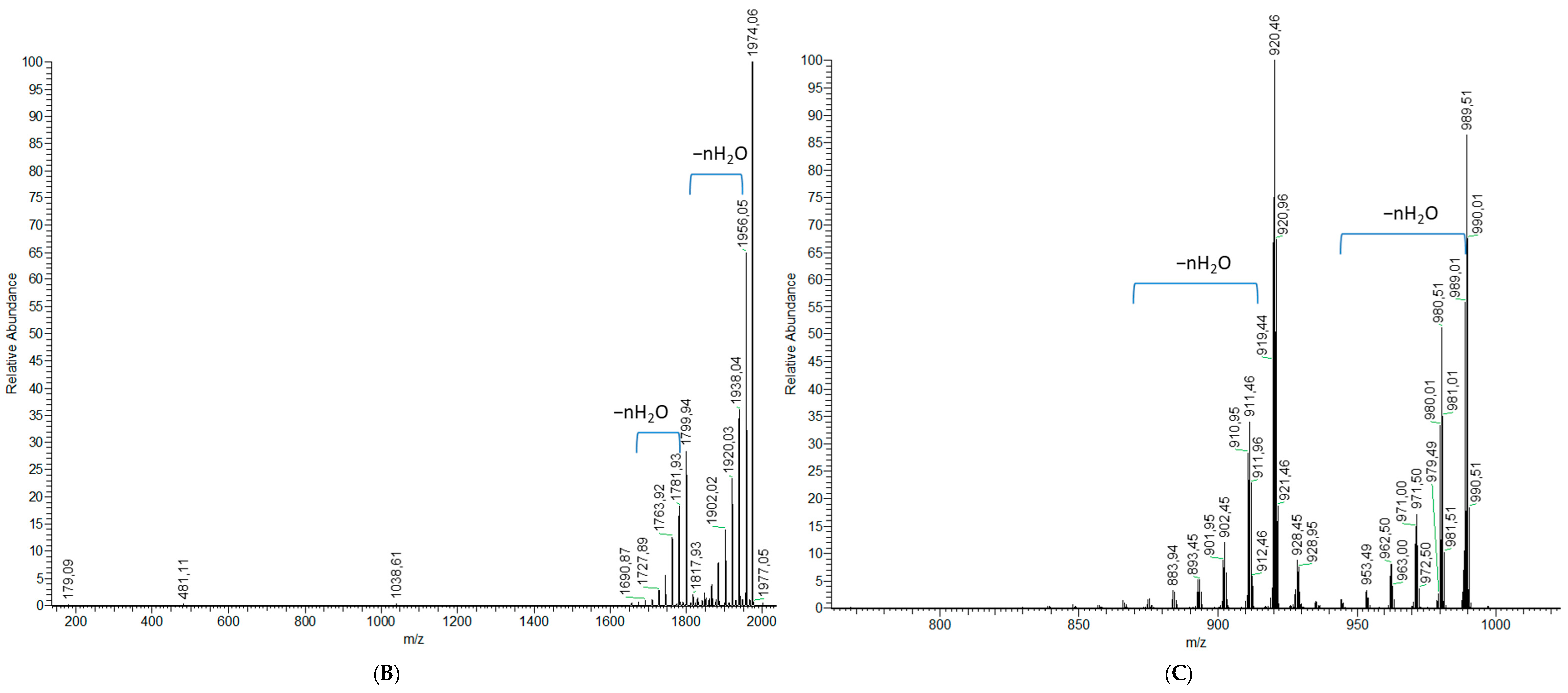
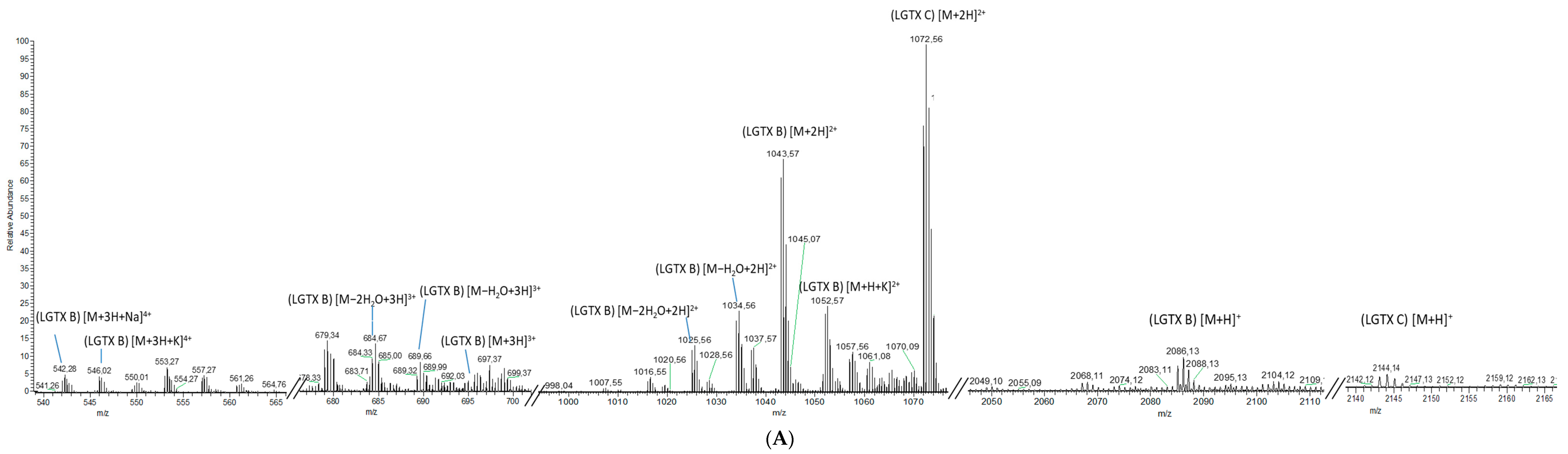
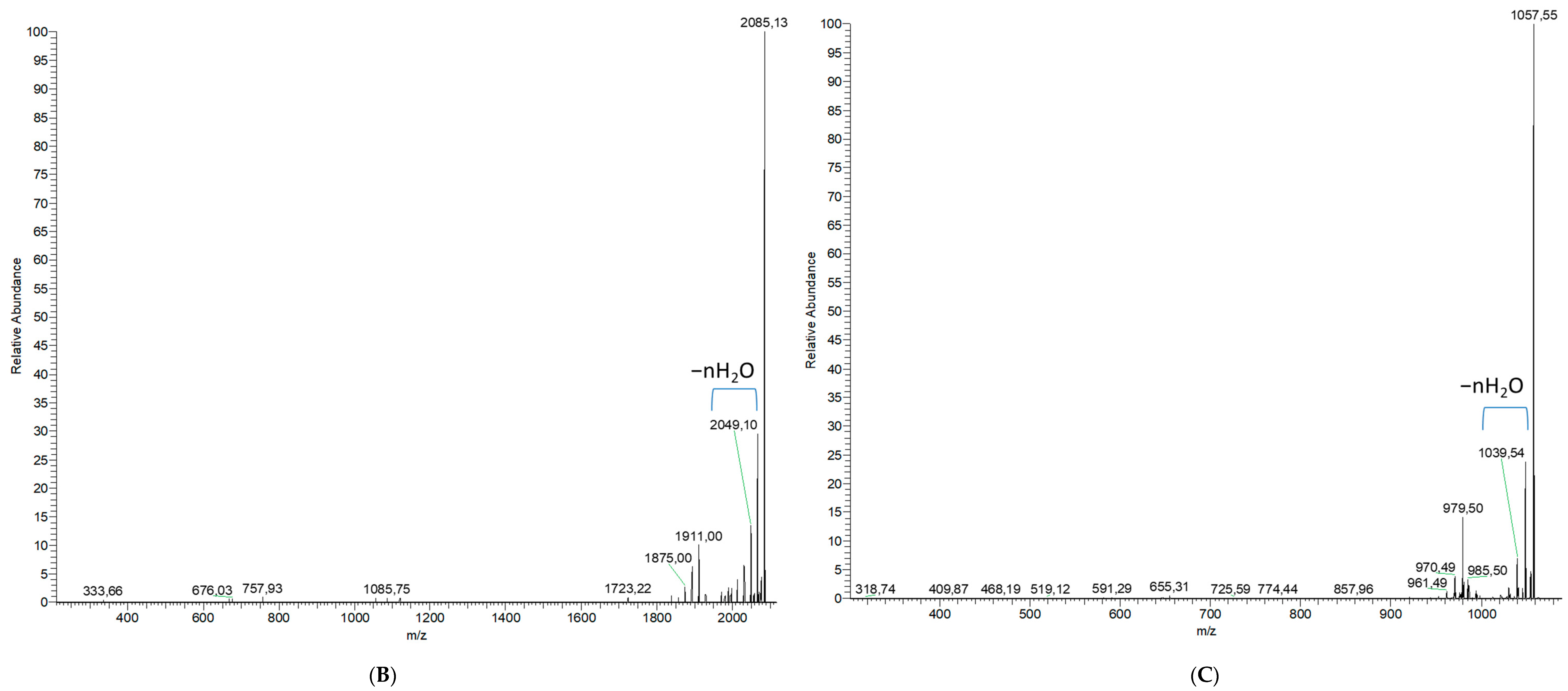
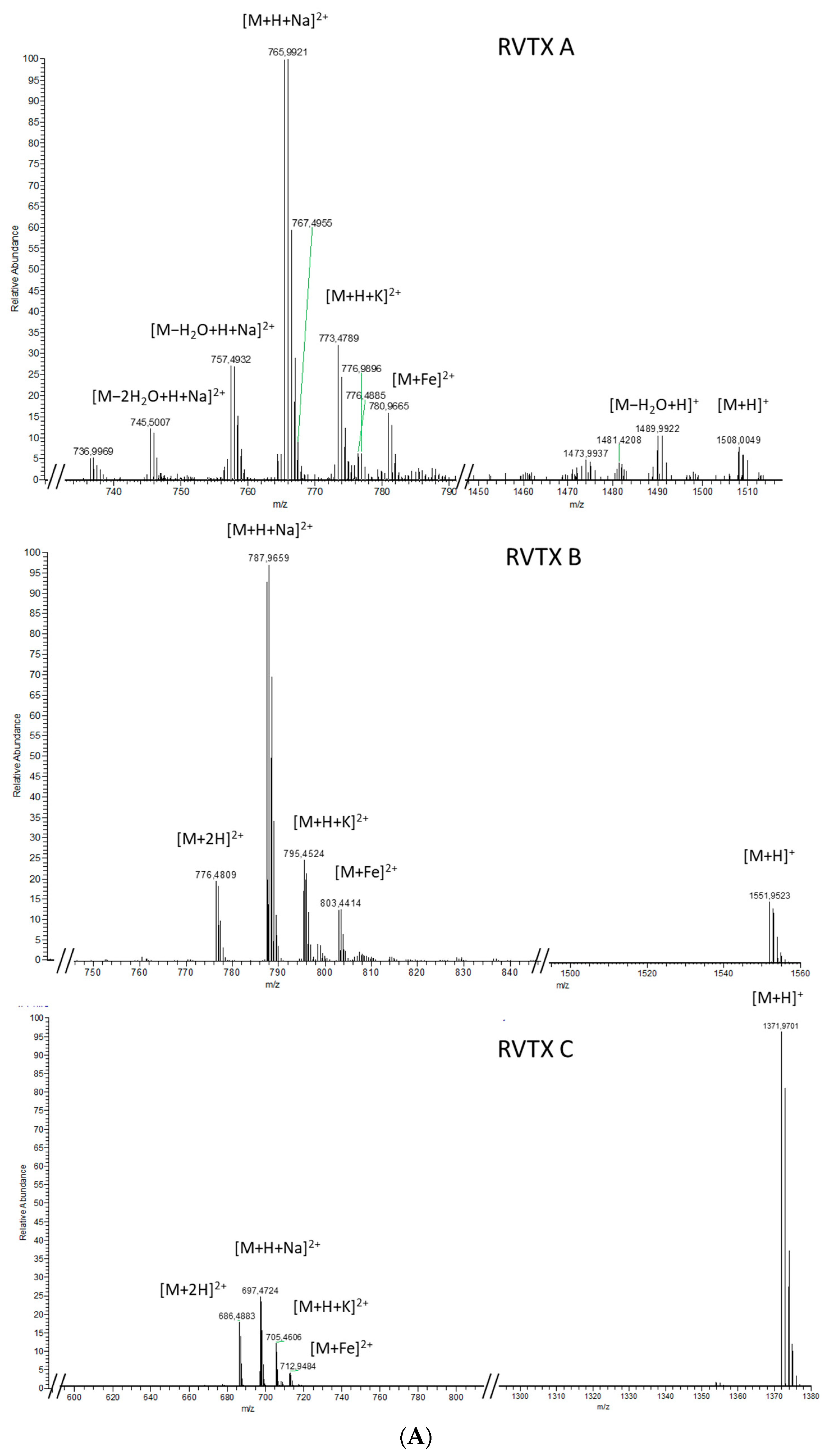
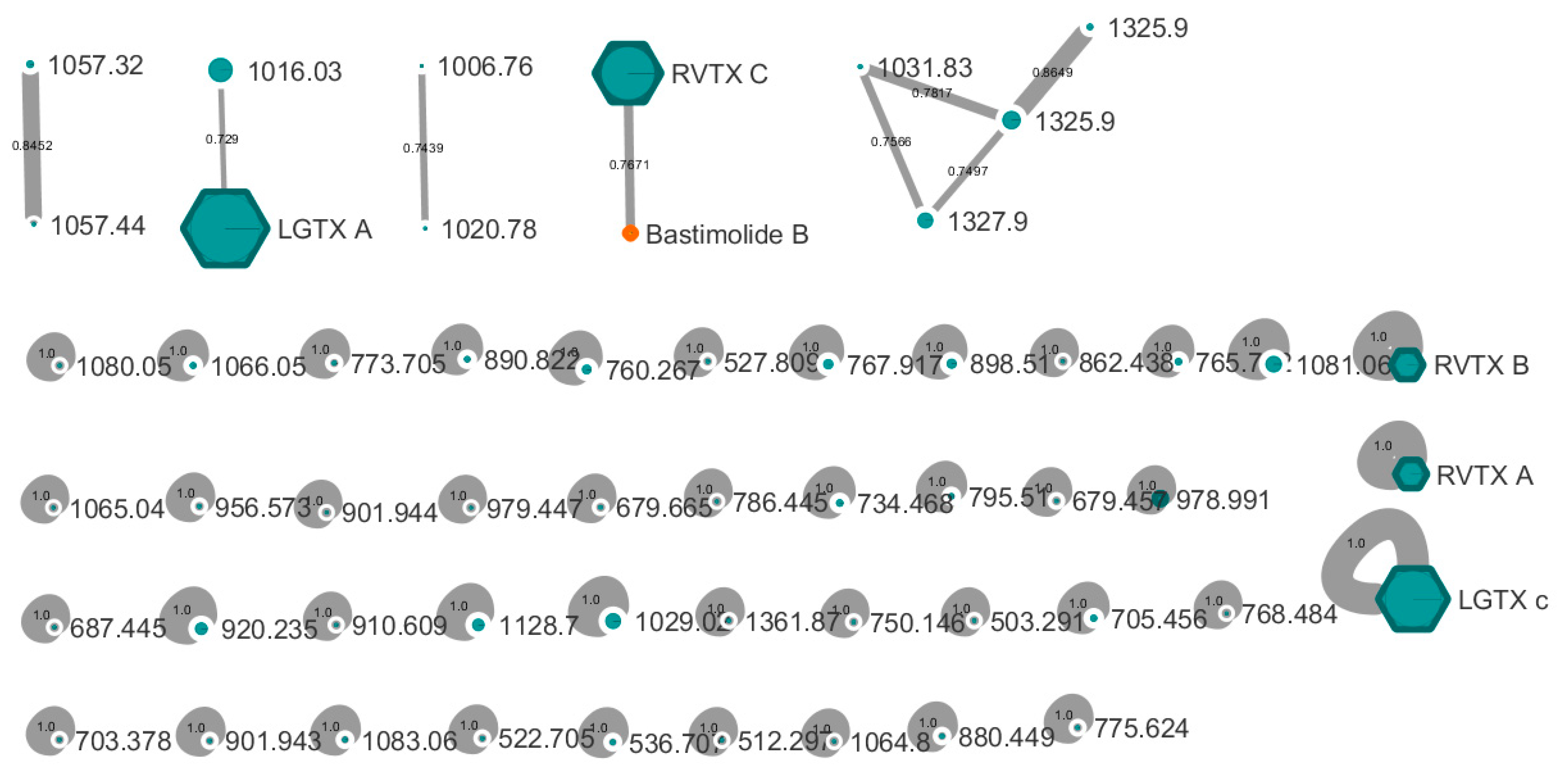
2.1.2. Analytical Chemistry Data
3. Discussion
4. Methods
4.1. Culture
4.2. Compound Purification
4.3. LC-HRMS/MS Analysis
4.4. Cytotoxicity Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ciminiello, P.; Dell’Aversano, C.; Fattorusso, E.; Forino, M.; Magno, G.S.; Tartaglione, L.; Grillo, C.; Melchiorre, N. The Genoa 2005 Outbreak. Determination of Putative Palytoxin in Mediterranean Ostreopsis Ovata by a New Liquid Chromatography Tandem Mass Spectrometry Method. Anal. Chem. 2006, 78, 6153–6159. [Google Scholar] [CrossRef] [PubMed]
- Vila, M.; Abós-Herràndiz, R.; Isern-Fontanet, J.; Àlvarez, J.; Berdalet, E. Establishing the Link between Ostreopsis Cf. Ovata Blooms and Human Health Impacts Using Ecology and Epidemiology. Sci. Mar. 2016, 80, 107–115. [Google Scholar] [CrossRef] [Green Version]
- Rossi, R.; Castellano, V.; Scalco, E.; Serpe, L.; Zingone, A.; Soprano, V. New Palytoxin-like Molecules in Mediterranean Ostreopsis cf. Ovata (Dinoflagellates) and in Palythoa tuberculosa Detected by Liquid Chromatography-Electrospray Ionization Time-of-Flight Mass Spectrometry. Toxicon 2010, 56, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Mangialajo, L.; Ganzin, N.; Accoroni, S.; Asnaghi, V.; Blanfune, A.; Cabrini, M.; Cattaneo-Vietti, R.; Chavanon, F.; Chiantore, M.; Cohu, S.; et al. Trends in Ostreopsis Proliferation along the Northern Mediterranean Coasts. Toxicon 2011, 57, 408–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, H.; Laza-Martínez, A.; Miguel, I.; Orive, E. Ostreopsis cf. siamensis and Ostreopsis cf. ovata from the Atlantic Iberian Peninsula: Morphological and Phylogenetic Characterization. Harmful Algae 2013, 30, 44–55. [Google Scholar] [CrossRef]
- Santos, M.; Oliveira, P.B.; Moita, M.T.; David, H.; Caeiro, M.F.; Zingone, A.; Amorim, A.; Silva, A. Ocurrence of Ostreopsis in Two Temperate Coastal Bays (SW Iberia): Insights from the Plankton. Harmful Algae 2019, 86, 20–36. [Google Scholar] [CrossRef]
- Moore, R.E.; Scheuer, P.J. Palytoxin: A New Marine Toxin from a Coelenterate. Science 1971, 172, 495–498. [Google Scholar] [CrossRef]
- Ciminiello, P.; Dell’Aversano, C.; Fattorusso, E.; Forino, M.; Tartaglione, L.; Grillo, C.; Melchiorre, N. Putative Palytoxin and Its New Analogue, Ovatoxin-a, in Ostreopsis ovata collected along the Ligurian Coasts during the 2006 Toxic Outbreak. J. Am. Soc. Mass Spectrom. 2008, 19, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Ternon, E.; Pavaux, A.-S.; Marro, S.; Thomas, O.P.; Lemée, R. Allelopathic Interactions between the Benthic Toxic Dinoflagellate Ostreopsis cf. ovata and a Co-Occurring Diatom. Harmful Algae 2018, 75, 35–44. [Google Scholar] [CrossRef]
- Ciminiello, P.; Dell’Aversano, C.; Dello Iacovo, E.; Fattorusso, E.; Forino, M.; Grauso, L.; Tartaglione, L.; Guerrini, F.; Pezzolesi, L.; Pistocchi, R.; et al. Isolation and Structure Elucidation of Ovatoxin-a, the Major Toxin Produced by Ostreopsis ovata. J. Am. Chem Soc. 2012, 134, 1869–1875. [Google Scholar] [CrossRef]
- Tosteson, T.R.; Ballantine, D.L.; Tosteson, C.G.; Hensley, V.; Bardales, A.T. Associated Bacterial Flora, Growth, and Toxicity of Cultured Benthic Dinoflagellates Ostreopsis lenticularis and Gambierdiscus toxicus. Appl. Environ. Microbiol. 1989, 55, 137–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenoir, S.; Ten-Hage, L.; Turquet, J.; Quod, J.-P.; Bernard, C.; Hennion, M.-C. First Evidence of Palytoxin Analgues from an Ostreopsis mascarenesis (Dinophyceae) Benthic Bloom in Southwestern Indian Ocean. J. Phycol. 2004, 40, 1042–1051. [Google Scholar] [CrossRef]
- Ukena, T.; SATAKE, M.; Usami, M.; Oshima, Y.; Naoki, H.; Fujita, T.; Kan, Y.; Yasumoto, T. Structure Elucidation of Ostreocin D, a Palytoxin Analog Isolated from the Dinoflagellate Ostreopsis siamensis. Biosci. Biotechnol. Biochem. 2001, 65, 2585–2588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, B.S.; Yoon, E.Y.; Kim, H.S.; Yih, W.; Park, J.Y.; Jeong, H.J.; Rho, J.R. Ostreol A: A New Cytotoxic Compound Isolated from the Epiphytic Dinoflagellate Ostreopsis cf. ovata from the Coastal Waters of Jeju Island, Korea. Bioorganic Med. Chem. Lett. 2013, 23, 3023–3027. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.S.; Yoon, E.Y.; Jeong, E.J.; Park, J.; Kim, E.-H.; Rho, J.-R. Determination of the Absolute Configuration of Polyhydroxy Compound Ostreol B Isolated from the Dinoflagellate Ostreopsis cf. ovata. J. Org. Chem. 2018, 83, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Ciminiello, P.; Dell’Aversano, C.; Iacovo, E.D.; Fattorusso, E.; Forino, M.; Grauso, L.; Tartaglione, L.; Guerrini, F.; Pistocchi, R. Complex Palytoxin-like Profile of Ostreopsis ovata. Identification of Four New Ovatoxins by High-Resolution Liquid Chromatography/Mass Spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 2735–2744. [Google Scholar] [CrossRef]
- Brissard, C.; Herve, F.; Sibat, M.; Sechet, V.; Hess, P.; Amzil, Z.; Herrenknecht, C. Characterization of Ovatoxin-h, a New Ovatoxin Analog, and Evaluation of Chromatographic Columns for Ovatoxin Analysis and Purification. J. Chromatogr. A 2015, 1388, 87–101. [Google Scholar] [CrossRef] [Green Version]
- García-Altares, M.; Tartaglione, L.; Dell’Aversano, C.; Carnicer, O.; de la Iglesia, P.; Forino, M.; Diogène, J.; Ciminiello, P. The Novel Ovatoxin-g and Isobaric Palytoxin (so Far Referred to as Putative Palytoxin) from Ostreopsis cf. ovata (NW Mediterranean Sea): Structural Insights by LC-High Resolution MSn. Anal. Bioanal. Chem. 2015, 407, 1191–1204. [Google Scholar] [CrossRef]
- Pavaux, A.-S.; Ternon, E.; Dufour, L.; Marro, S.; Gémin, M.-P.; Thomas, O.P.; Lemée, R. Efficient, Fast and Inexpensive Bioassay to Monitor Benthic Microalgae Toxicity: Application to Ostreopsis Species. Aquat. Toxicol. 2020, 223, 105485. [Google Scholar] [CrossRef]
- Uchida, H.; Taira, Y.; Yasumoto, T. Structural Elucidation of Palytoxin Analogs Produced by the Dinoflagellate Ostreopsis ovata IK2 Strain by Complementary Use of Positive and Negative Ion Liquid Chromatography/Quadrupole Time-of-Flight Mass Spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 1999–2008. [Google Scholar] [CrossRef]
- Ciminiello, P.; Dell’Aversano, C.; Iacovo, E.D.; Fattorusso, E.; Forino, M.; Tartaglione, L.; Battocchi, C.; Crinelli, R.; Carloni, E.; Magnani, M.; et al. Unique Toxin Profile of a Mediterranean Ostreopsis cf. ovata Strain: HR LC-MSn Characterization of Ovatoxin-f, a New Palytoxin Congener. Chem. Res. Toxicol. 2012, 25, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.-L.; Mou, X.-F.; Cao, F.; Spadafora, C.; Glukhov, E.; Gerwick, L.; Wang, C.-Y.; Gerwick, W.H. Bastimolide B, an Antimalarial 24-Membered Marine Macrolide Possessing a Tert-Butyl Group. J. Nat. Prod. 2018, 81, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Kerbrat, A.-S.; Amzil, Z.; Pawlowiez, R.; Golubic, S.; Sibat, M.; Taiana-Darius, H.; Chinain, M.; Laurent, D. First Evidence of Palytoxin and 42-Hydroxy-Palytoxin in the Marine Cyanobacterium Trichodesmium. Mar. Drugs 2011, 9, 543–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledreux, A.; Krys, S.; Bernard, C. Suitability of the Neuro-2a Cell Line for the Detection of Palytoxin and Analogues (Neurotoxic Phycotoxins). Toxicon 2009, 53, 300–308. [Google Scholar] [CrossRef]
- Bellocci, M.; Ronzitti, G.; Milandri, A.; Melchiorre, N.; Grillo, C.; Poletti, R.; Yasumoto, T.; Rossini, G.P. A Cytolytic Assay for the Measurement of Palytoxin Based on a Cultured Monolayer Cell Line. Anal. Biochem. 2008, 374, 48–55. [Google Scholar] [CrossRef]
- Pelin, M.; Forino, M.; Brovedani, V.; Tartaglione, L.; Dell’Aversano, C.; Pistocchi, R.; Poli, M.; Sosa, S.; Florio, C.; Ciminiello, P.; et al. Ovatoxin-a, A Palytoxin Analogue Isolated from Ostreopsis cf. ovata Fukuyo: Cytotoxic Activity and ELISA Detection. Environ. Sci. Technol. 2016, 50, 1544–1551. [Google Scholar] [CrossRef]
- Heinilä, L.M.P.; Fewer, D.P.; Jokela, J.K.; Wahlsten, M.; Jortikka, A.; Sivonen, K. Shared PKS Module in Biosynthesis of Synergistic Laxaphycins. Front. Microbiol. 2020, 11, 2173. [Google Scholar] [CrossRef]
- Ternon, E.; Pavaux, A.-S.; Peltekis, A.; Gemin, M.-P.; Jauzein, C.; Bailleul, B.; Lemée, R.; Thomas, O.P. Assessment of the Allelochemical Activity of Ostreopsis cf. ovata and the Ovatoxins towards Competitive Benthic Microalgae. Aquat. Ecol. 2022. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Carmichael, J.; DeGraff, W.G.; Gazdar, A.F.; Minna, J.D.; Mitchell, J.B. Evaluation of a Tetrazolium-Based Semiautomated Colorimetric Assay: Assessment of Chemosensitivity Testing1. Cancer Res. 1987, 47, 936–942. [Google Scholar]

| Compound | CC50 (µg/mL) |
|---|---|
| OF6-2 | 0.68 ± 0.1 |
| OF6-3 | 1.40 ± 0.08 |
| OF6-6 | 0.69 ± 0.16 |
| OF6-12 | 3.12 ± 0.13 |
| OF6-15 | 1.28 ± 0.24 |
| OF6-18 | 0.76 ± 0.17 |
| OF6-mix | 0.14 ± 0.02 |
| PLTX | 4.46 × 10−5 ± 0.76 × 10−5 |
| Doxorubicin | 0.18 ± 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ternon, E.; Glukhov, E.; Trytten, E.; Lemée, R.; Gerwick, W.H. On the Hunt for New Toxin Families Produced by a Mediterranean Strain of the Benthic Dinoflagellate Ostreopsis cf. ovata. Toxins 2022, 14, 234. https://doi.org/10.3390/toxins14040234
Ternon E, Glukhov E, Trytten E, Lemée R, Gerwick WH. On the Hunt for New Toxin Families Produced by a Mediterranean Strain of the Benthic Dinoflagellate Ostreopsis cf. ovata. Toxins. 2022; 14(4):234. https://doi.org/10.3390/toxins14040234
Chicago/Turabian StyleTernon, Eva, Evgenia Glukhov, Emily Trytten, Rodolphe Lemée, and William H. Gerwick. 2022. "On the Hunt for New Toxin Families Produced by a Mediterranean Strain of the Benthic Dinoflagellate Ostreopsis cf. ovata" Toxins 14, no. 4: 234. https://doi.org/10.3390/toxins14040234
APA StyleTernon, E., Glukhov, E., Trytten, E., Lemée, R., & Gerwick, W. H. (2022). On the Hunt for New Toxin Families Produced by a Mediterranean Strain of the Benthic Dinoflagellate Ostreopsis cf. ovata. Toxins, 14(4), 234. https://doi.org/10.3390/toxins14040234






