Prevalence, Antimicrobial Resistance, and Whole Genome Sequencing Analysis of Shiga Toxin-Producing Escherichia coli (STEC) and Enteropathogenic Escherichia coli (EPEC) from Imported Foods in China during 2015–2021
Abstract
:1. Introduction
2. Results
2.1. Prevalence of STEC and EPEC
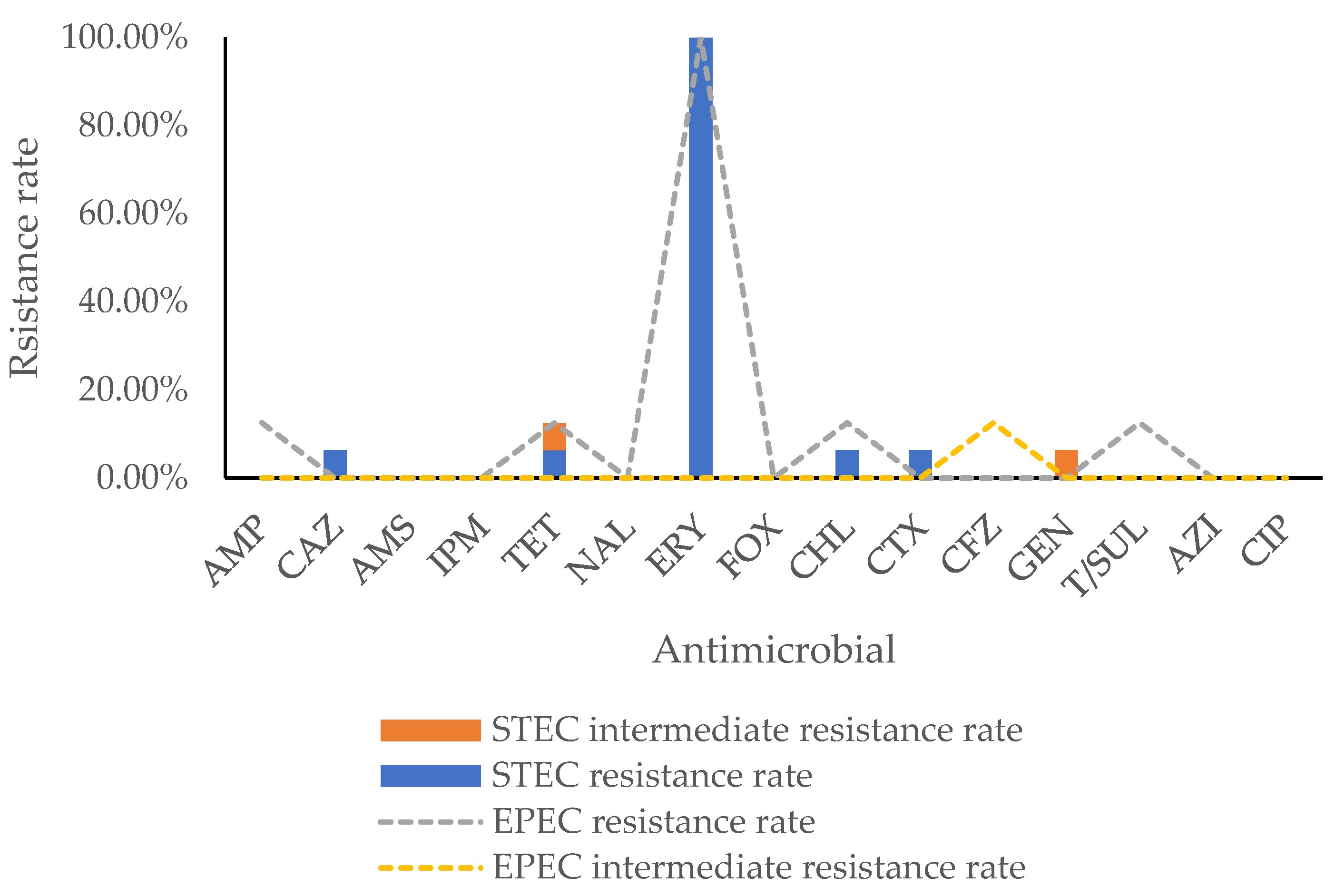
2.2. Antimicrobial Resistance Phenotype
2.3. Antimicrobial Resistance Genotype
2.4. Virulence Gene Analysis
2.5. Serotype, ST, and Phylogenetic Analysis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sample Collection
5.2. Strain Isolation and Identification
5.3. Antimicrobial Susceptibility Test
5.4. Whole Genome Sequencing Analysis
5.4.1. DNA Extraction
5.4.2. Whole Genome Sequencing and Contig Assembly
5.4.3. Molecular Characterization of the Strains Based on WGS
5.4.4. Phylogenetic Analysis
5.4.5. Nucleotide Sequence Accession Numbers
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Majowicz, S.E.; Scallan, E.; Jones-Bitton, A.; Sargeant, J.M.; Stapleton, J.; Angulo, F.J.; Yeung, D.H.; Kirk, M.D. Global incidence of human Shiga toxin-producing Escherichia coli infections and deaths: A systematic review and knowledge synthesis. Foodborne Pathog. Dis. 2014, 11, 447–455. [Google Scholar] [CrossRef] [Green Version]
- CDC. Reports of Selected E. Coli Outbreak Investigations. Available online: https://www.cdc.gov/ecoli/outbreaks.html (accessed on 22 November 2021).
- Guo, Y. Foodborne diseases surveillance in China. In Proceedings of the Propagation and Implement Training of Food Safety National Standard, Beijing, China, 12 November 2020. [Google Scholar]
- Bai, X.; Wang, H.; Xin, Y.; Wei, R.; Tang, X.; Zhao, A.; Sun, H.; Zhang, W.; Wang, Y.; Xu, Y.; et al. Prevalence and characteristics of Shiga toxin-producing Escherichia coli isolated from retail raw meats in China. Int. J. Food Microbiol. 2015, 200, 31–38. [Google Scholar] [CrossRef]
- Fn, A.; Ping, Z.A.; Jing, M.B.; Qxac, D.; Yl, A.; Ypac, D.; Yong, Z.; Hlacd, E. Characterization of Shiga-toxin producing Escherichia coli (STEC) isolated from retail raw meats in Southeast China. Food Control 2021, 126, 108061. [Google Scholar] [CrossRef]
- Fuller, C.A.; Pellino, C.A.; Flagler, M.J.; Strasser, J.E.; Weiss, A.A. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 2011, 79, 1329–1337. [Google Scholar] [CrossRef] [Green Version]
- Karmali, M.A. Host and pathogen determinants of verocytotoxin-producing Escherichia coli-associated hemolytic uremic syndrome. Kidney Int. Suppl. 2009, 75, S4–S7. [Google Scholar] [CrossRef] [Green Version]
- Franzin, F.M.; Sircili, M.P. Locus of enterocyte effacement: A pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. Biomed Res. Int. 2015, 2015, 534738. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Shiga Toxin-Producing Escherichia coli (STEC) and Food: Attribution, Characterization, and Monitoring; Food and Agriculture Organization of the United Nations and World Health Organization: Rome, Italy, 2018. [Google Scholar]
- Garmendia, J.; Ren, Z.; Tennant, S.; Midolli Viera, M.A.; Chong, Y.; Whale, A.; Azzopardi, K.; Dahan, S.; Sircili, M.P.; Franzolin, M.R.; et al. Distribution of tccP in clinical enterohemorrhagic and enteropathogenic Escherichia coli isolates. J. Clin. Microbiol. 2005, 43, 5715–5720. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Zhang, J.; Li, Y.; Xie, S.; Jiang, Y.; Wu, Y.; Ye, Y.; Yang, H.; Mo, H.; Situ, C.; et al. The 12 Gastrointestinal Pathogens Spectrum of Acute Infectious Diarrhea in a Sentinel Hospital, Shenzhen, China. Front. Microbiol. 2016, 7, 1926. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhu, X.; Hou, H.; Lu, Y.; Yu, J.; Mao, L.; Mao, L.; Sun, Z. Characteristics of diarrheagenic Escherichia coli among children under 5 years of age with acute diarrhea: A hospital based study. BMC Infect. Dis. 2018, 18, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Wu, Q.; Zhang, J.; Zhu, X. Occurrence and Characterization of Enteropathogenic Escherichia coli (EPEC) in Retail Ready-to-Eat Foods in China. Foodborne Pathog. Dis. 2016, 13, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, G.; Huang, Y.; Zhang, J.; Cui, L.; Wu, Q. Prevalence and Characterization of Atypical Enteropathogenic Escherichia coli Isolated from Retail Foods in China. J. Food Prot. 2018, 81, 1761–1767. [Google Scholar] [CrossRef] [PubMed]
- Toma, C.; Martinez Espinosa, E.; Song, T.; Miliwebsky, E.; Chinen, I.; Iyoda, S.; Iwanaga, M.; Rivas, M. Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 2004, 42, 4937–4946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wani, S.A.; Hussain, I.; Nabi, A.; Fayaz, I.; Nishikawa, Y. Variants of eae and stx genes of atypical enteropathogenic Escherichia coli and non-O157 Shiga toxin-producing Escherichia coli from calves. Lett. Appl. Microbiol. 2007, 45, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, A.; Byrne, B.; Fanning, S.; Sweeney, T.; McDowell, D.; Bolton, D.J. Serotypes and virulence profiles of atypical enteropathogenic Escherichia coli (EPEC) isolated from bovine farms and abattoirs. J. Appl. Microbiol. 2013, 114, 595–603. [Google Scholar] [CrossRef]
- Rios, E.A.; Santos, J.; García-Meniño, I.; Flament-Simon, S.C.; Blanco, J.; García-López, M.; Otero, A.; Rodríguez-Calleja, J. Characterisation, antimicrobial resistance and diversity of atypical EPEC and STEC isolated from cow’s milk, cheese and dairy cattle farm environments. Lebensm. Wiss. Technol. 2019, 108, 319–325. [Google Scholar] [CrossRef]
- Toro, M.; Rivera, D.; Jimenez, M.F.; Diaz, L.; Navarrete, P.; Reyes-Jara, A. Isolation and characterization of non-O157 Shiga toxin-producing Escherichia coli (STEC) isolated from retail ground beef in Santiago, Chile. Food Microbiol. 2018, 75, 55–60. [Google Scholar] [CrossRef]
- Ju, W.; Shen, J.; Li, Y.; Toro, M.A.; Zhao, S.; Ayers, S.; Najjar, M.B.; Meng, J. Non-O157 Shiga toxin-producing Escherichia coli in retail ground beef and pork in the Washington D.C. area. Food Microbiol. 2012, 32, 371–377. [Google Scholar] [CrossRef]
- Varcasia, B.M.; Tomassetti, F.; De Santis, L.; Di Giamberardino, F.; Lovari, S.; Bilei, S.; De Santis, P. Presence of Shiga Toxin-Producing Escherichia coli (STEC) in Fresh Beef Marketed in 13 Regions of ITALY (2017). Microorganisms 2018, 6, 126. [Google Scholar] [CrossRef] [Green Version]
- Quiros, P.; Martinez-Castillo, A.; Muniesa, M. Improving detection of Shiga toxin-producing Escherichia coli by molecular methods by reducing the interference of free Shiga toxin-encoding bacteriophages. Appl. Environ. Microbiol. 2015, 81, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelyas, N.; Poon, A.; Patterson-Fortin, L.; Johnson, R.P.; Lee, W.; Chui, L. Assessment of commercial chromogenic solid media for the detection of non-O157 Shiga toxin-producing Escherichia coli (STEC). Diagn. Microbiol. Infect. Dis. 2016, 85, 302–308. [Google Scholar] [CrossRef]
- Verhaegen, B.; De Reu, K.; Heyndrickx, M.; De Zutter, L. Comparison of Six Chromogenic Agar Media for the Isolation of a Broad Variety of Non-O157 Shigatoxin-Producing Escherichia coli (STEC) Serogroups. Int. J. Environ. Res. Public Health 2015, 12, 6965–6978. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Lou, Y.; Zhang, Z.; Pan, Y.; Zhao, Y. Mismatch between antimicrobial resistance phenotype and genotype of pathogenic Vibrio parahaemolyticus isolated from seafood. Food Control 2016, 59, 207–211. [Google Scholar] [CrossRef]
- Laura, D.M.; Scott, N.L.; Vanner, E.A.; Miller, D.; Flynn, H.W., Jr. Genotypic and Phenotypic Antibiotic Resistance in Staphylococcus Epidermidis Endophthalmitis. Ophthalmic Surg. Lasers Imaging Retin. 2020, 51, S13–S16. [Google Scholar] [CrossRef]
- Valilis, E.; Ramsey, A.; Sidiq, S.; DuPont, H.L. Non-O157 Shiga toxin-producing Escherichia coli-A poorly appreciated enteric pathogen: Systematic review. Int. J. Infect. Dis. 2018, 76, 82–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abram, K.; Udaondo, Z.; Bleker, C.; Wanchai, V.; Wassenaar, T.M.; Robeson, M.S.; Ussery, D.W. What can we learn from over 100,000 Escherichia coli genomes? bioRxiv 2020, 708131. [Google Scholar] [CrossRef] [Green Version]
- Colello, R.; Kruger, A.; Velez, M.V.; Del Canto, F.; Etcheverria, A.I.; Vidal, R.; Padola, N.L. Identification and detection of iha subtypes in LEE-negative Shiga toxin-producing Escherichia coli (STEC) strains isolated from humans, cattle and food. Heliyon 2019, 5, e03015. [Google Scholar] [CrossRef] [Green Version]
- Tozzoli, R.; Caprioli, A.; Cappannella, S.; Michelacci, V.; Marziano, M.L.; Morabito, S. Production of the subtilase AB5 cytotoxin by Shiga toxin-negative Escherichia coli. J. Clin. Microbiol. 2010, 48, 178–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Suarez, M.E.; Otero, A.; Garcia-Lopez, M.L.; Dahbi, G.; Blanco, M.; Mora, A.; Blanco, J.; Santos, J.A. Genetic characterization of Shiga toxin-producing Escherichia coli (STEC) and atypical enteropathogenic Escherichia coli (EPEC) isolates from goat’s milk and goat farm environment. Int. J. Food Microbiol. 2016, 236, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Amézquita-Montes, Z.; Tamborski, M.; Kopsombut, U.G.; Zhang, C.; Arzuza, O.S.; Gómez-Duarte, O.G. Genetic Relatedness Among Escherichia coli Pathotypes Isolated from Food Products for Human Consumption in Cartagena, Colombia. Foodborne Pathog. Dis. 2015, 12, 454. [Google Scholar] [CrossRef] [Green Version]
- Werber, D.; Scheutz, F. The importance of integrating genetic strain information for managing cases of Shiga toxin-producing E. coli infection. Epidemiol. Infect. 2019, 147, e264. [Google Scholar] [CrossRef] [Green Version]
- Rasko, D.A.; Webster, D.R.; Sahl, J.W.; Bashir, A.; Boisen, N.; Scheutz, F.; Paxinos, E.E.; Sebra, R.; Chin, C.S.; Iliopoulos, D.; et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 2011, 365, 709–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyholm, O.; Heinikainen, S.; Pelkonen, S.; Hallanvuo, S.; Haukka, K.; Siitonen, A. Hybrids of Shigatoxigenic and Enterotoxigenic Escherichia coli (STEC/ETEC) Among Human and Animal Isolates in Finland. Zoonoses Public Health 2015, 62, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.H.; Shin, E.; Jung, S.M.; Im, J.; Cho, S.H.; Hong, S.; Yoo, C.K.; Chung, G.T. First Isolation of a Hybrid Shigatoxigenic and Enterotoxigenic Escherichia coli Strain Harboring the stx2 and elt Genes in Korea. Jpn. J. Infect. Dis. 2017, 70, 347–348. [Google Scholar] [CrossRef]
- Hazen, T.H.; Michalski, J.; Luo, Q.; Shetty, A.C.; Daugherty, S.C.; Fleckenstein, J.M.; Rasko, D.A. Comparative genomics and transcriptomics of Escherichia coli isolates carrying virulence factors of both enteropathogenic and enterotoxigenic E. coli. Sci. Rep. 2017, 7, 3513. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhang, J.; Ambikan, A.; Jernberg, C.; Ehricht, R.; Scheutz, F.; Xiong, Y.; Matussek, A. Molecular Characterization and Comparative Genomics of Clinical Hybrid Shiga Toxin-Producing and Enterotoxigenic Escherichia coli (STEC/ETEC) Strains in Sweden. Sci. Rep. 2019, 9, 5619. [Google Scholar] [CrossRef] [Green Version]
- SO/TS 13136: 2012 (E); Microbiology of food and animal feed—Real-time polymerase chain reaction (PCR)-based method for the detection of food-borne pathogens—Horizontal method for the detection of Shiga toxin-producing Escherichia coli (STEC) and the determination of O157, O111, O26, O103 and O145 serogroups. ISO: Geneva, Switzerland, 2012.
- MLG 5C.00; Detection, Isolation and Identification of Top Seven Shiga Toxin-Producing Escherichia coli (STECs) from Meat Products and Carcass and Environmental Sponges. United States Department of Agriculture, Food Safety and Inspection Service (USDA FSIS): Washington, DC, USA, 2019.
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019; ISBN 978-1-68440-032-4/978-1-68440-033-1. [Google Scholar]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [Green Version]
- Ivica, L.; Peer, B. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011, 39, W475–W478. [Google Scholar] [CrossRef]

| Food Type (n) | STEC Prevalence (%) | EPEC Prevalence (%) |
|---|---|---|
| Frozen beef (n = 1066) | 12 (1.13%) | 5 (0.47%) |
| Frozen pork (n = 172) | 3 (1.74%) | 2 (1.16%) |
| Frozen and fresh aquatic products (n = 198) | 0 | 1 (0.51%) |
| Frozen mutton (n = 102) | 1 (0.98%) | 0 |
| Milk products (n = 27) | 0 | 0 |
| Frozen chicken (n = 12) | 0 | 0 |
| Total (n = 1577) | 16 (1.01%) | 8 (0.51%) |
| Strain ID | Food Type | Antimicrobial Resistance Phenotype | Antimicrobial Resistance Genotype | |
|---|---|---|---|---|
| Resistance | Intermediate | |||
| 1053l-2 | Pork | Tetracycline + chloramphenicol | Gentamicin | tet(A), tet(B), catA1, mph(B), aadA1, aph (4)-Ia 1, aac(3)-IV 1, aph(6)-Id, aph(3″)-Ib, sul1, qacE, mdf(A) 5 |
| 1095a | Pork | Chloramphenicol + trimethoprim-sulfamethoxazole | aadA1, aadA8b, aadA2, cmlA1, sul3, dfrA12, mdf(A) 5 | |
| 1509-1 | Beef | Ampicillin + tetracycline | Cefazolin | blaTEM-1B2, tet(A), qnrS3,4, mdf(A) 5 |
| 816b | Beef | Ceftazidime + cefotaxime | mdf(A) 5 | |
| 1789-1 | Beef | Tetracycline | mdf(A) 5 | |
| VG | STEC | EPEC | VG | STEC | EPEC | VG | STEC | EPEC | VG | STEC | EPEC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| stx1a | 1 | 0 | astA | 1 | 6 | cib | 4 | 0 | ibeA | 0 | 1 |
| stx1c | 1 | 0 | irp2 | 3 | 3 | iha | 5 | 1 | neuC | 0 | 2 |
| stx2d | 6 | 0 | fyuA | 3 | 3 | subA | 6 | 1 | efa1 | 0 | 1 |
| stx2a | 2 | 0 | gad | 5 | 6 | espP | 4 | 2 | vat | 0 | 1 |
| stx2e | 4 | 0 | tir | 0 | 8 | epeA | 4 | 0 | cdtB | 1 | 0 |
| stx1c + stx2b | 1 | 0 | espB | 0 | 6 | nleA | 0 | 5 | iucC | 0 | 1 |
| stx1a + stx2a + stx2c | 1 | 0 | espF | 0 | 8 | tccP | 0 | 5 | sab | 1 | 0 |
| eae | 0 | 8 | espA | 0 | 8 | toxB | 0 | 2 | celb | 2 | 0 |
| ehxA | 6 | 5 | espJ | 0 | 4 | perA | 0 | 2 | ireA | 2 | 0 |
| terC | 16 | 8 | afaD | 0 | 2 | etpD | 0 | 4 | kpsE | 2 | 0 |
| ompT | 14 | 5 | nleB | 0 | 8 | katP | 0 | 3 | mchB | 1 | 0 |
| chuA | 5 | 3 | cia | 7 | 2 | cea | 4 | 2 | mchC | 1 | 0 |
| eilA | 5 | 0 | cif | 0 | 5 | sepA | 1 | 0 | mchF | 1 | 0 |
| iss | 14 | 6 | sta1 | 2 | 1 | stb | 1 | 0 | esp1 | 1 | 1 |
| traT | 16 | 7 | cba | 3 | 0 | yfcV | 0 | 1 | cvaC | 1 | 0 |
| hra | 7 | 0 | cma | 4 | 0 | nleC | 0 | 4 | senB | 1 | 0 |
| lpfA | 8 | 3 | air | 4 | 0 | usp | 0 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, J.; Zhi, S.; Guo, D.; Jiang, Y.; Xu, X.; Zhao, L.; Lv, J. Prevalence, Antimicrobial Resistance, and Whole Genome Sequencing Analysis of Shiga Toxin-Producing Escherichia coli (STEC) and Enteropathogenic Escherichia coli (EPEC) from Imported Foods in China during 2015–2021. Toxins 2022, 14, 68. https://doi.org/10.3390/toxins14020068
Shen J, Zhi S, Guo D, Jiang Y, Xu X, Zhao L, Lv J. Prevalence, Antimicrobial Resistance, and Whole Genome Sequencing Analysis of Shiga Toxin-Producing Escherichia coli (STEC) and Enteropathogenic Escherichia coli (EPEC) from Imported Foods in China during 2015–2021. Toxins. 2022; 14(2):68. https://doi.org/10.3390/toxins14020068
Chicago/Turabian StyleShen, Jinling, Shuai Zhi, Dehua Guo, Yuan Jiang, Xuebin Xu, Lina Zhao, and Jingzhang Lv. 2022. "Prevalence, Antimicrobial Resistance, and Whole Genome Sequencing Analysis of Shiga Toxin-Producing Escherichia coli (STEC) and Enteropathogenic Escherichia coli (EPEC) from Imported Foods in China during 2015–2021" Toxins 14, no. 2: 68. https://doi.org/10.3390/toxins14020068
APA StyleShen, J., Zhi, S., Guo, D., Jiang, Y., Xu, X., Zhao, L., & Lv, J. (2022). Prevalence, Antimicrobial Resistance, and Whole Genome Sequencing Analysis of Shiga Toxin-Producing Escherichia coli (STEC) and Enteropathogenic Escherichia coli (EPEC) from Imported Foods in China during 2015–2021. Toxins, 14(2), 68. https://doi.org/10.3390/toxins14020068





