Stx2 Induces Differential Gene Expression and Disturbs Circadian Rhythm Genes in the Proximal Tubule
Abstract
:1. Introduction
2. Results
2.1. Stx2-Injected Mouse Kidney Pathology
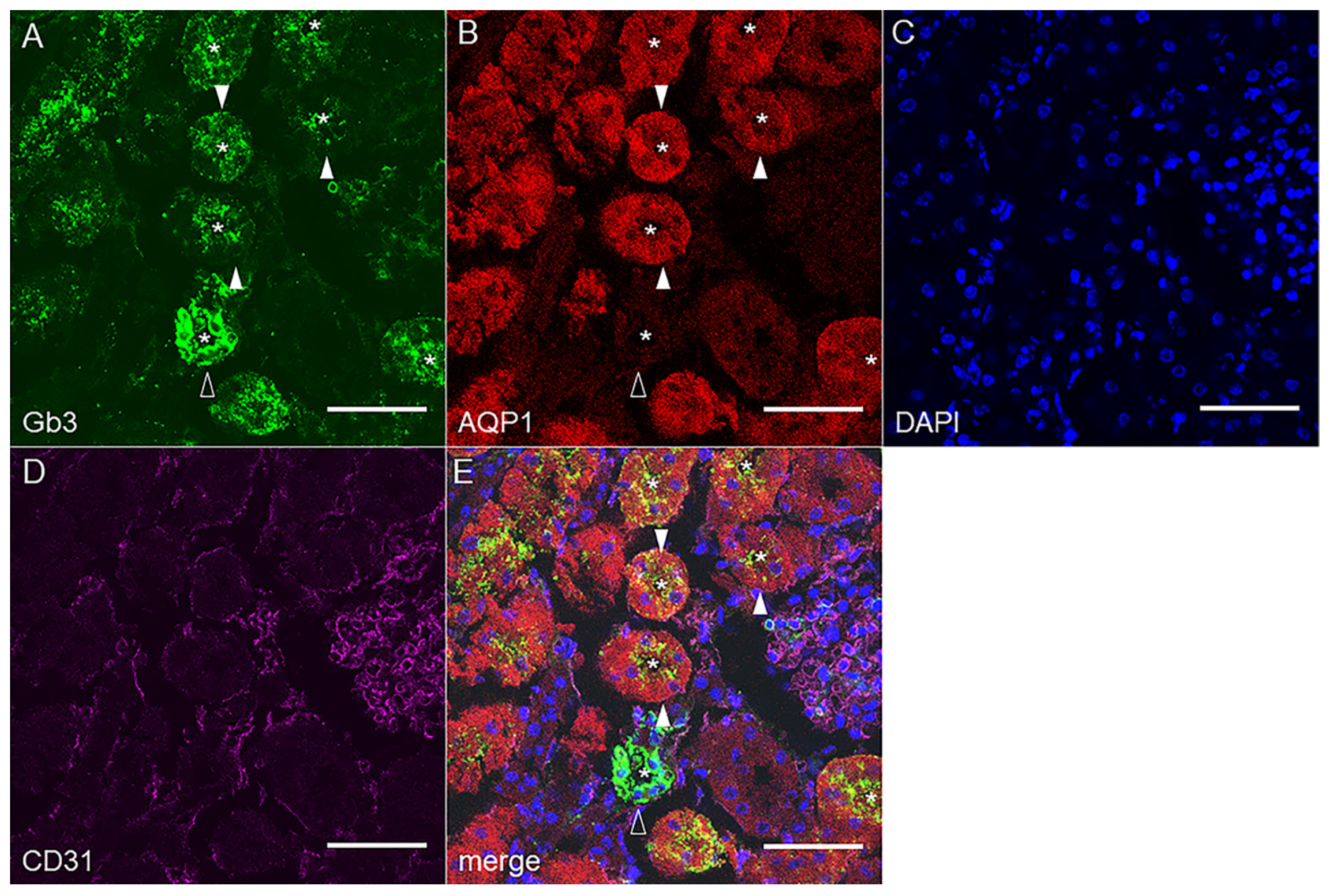
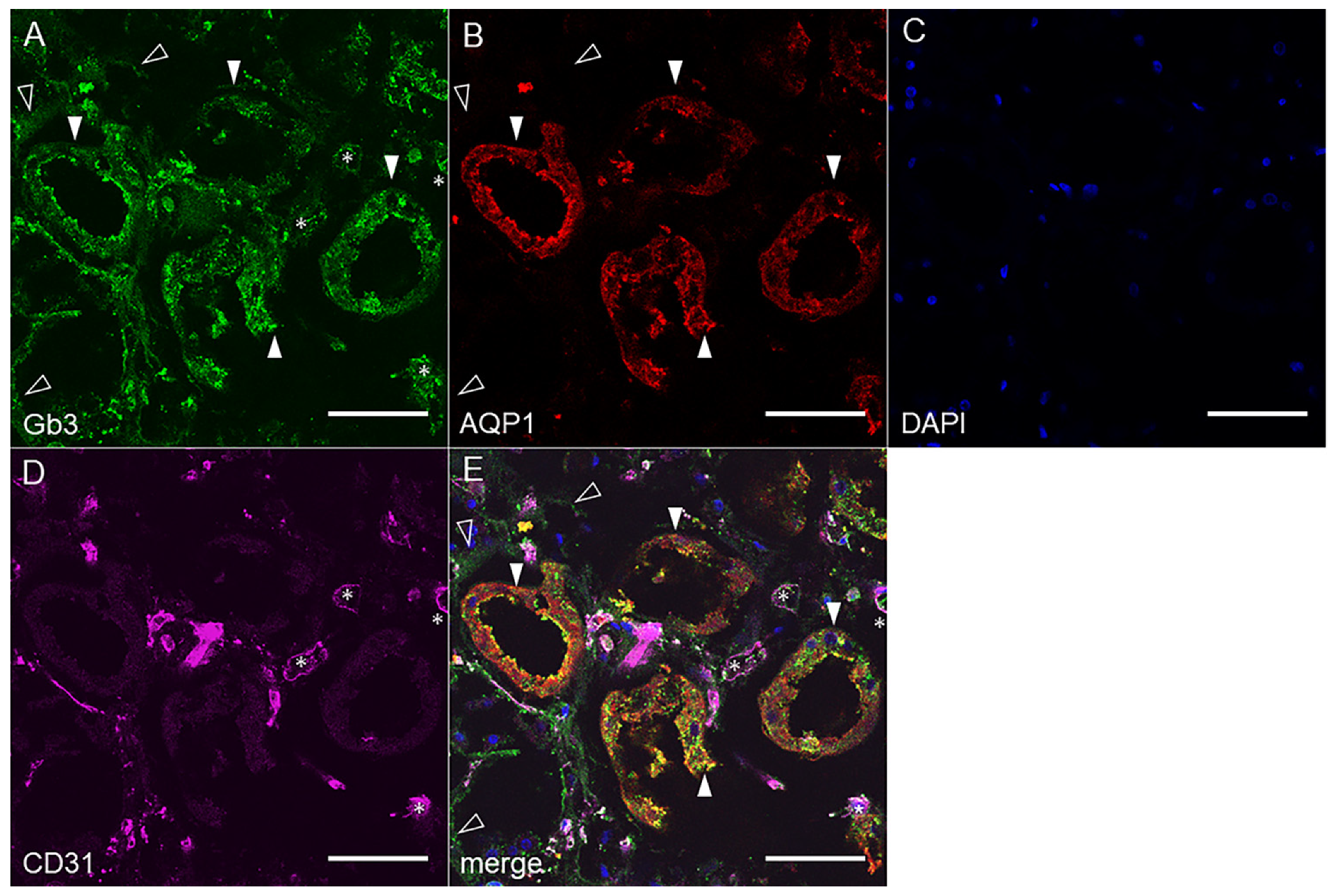
2.2. Gb3 Expression in Mouse and Human Kidneys
2.2.1. Mouse Renal Cortex Gb3 Expression
2.2.2. Human Renal Cortex Gb3 Expression
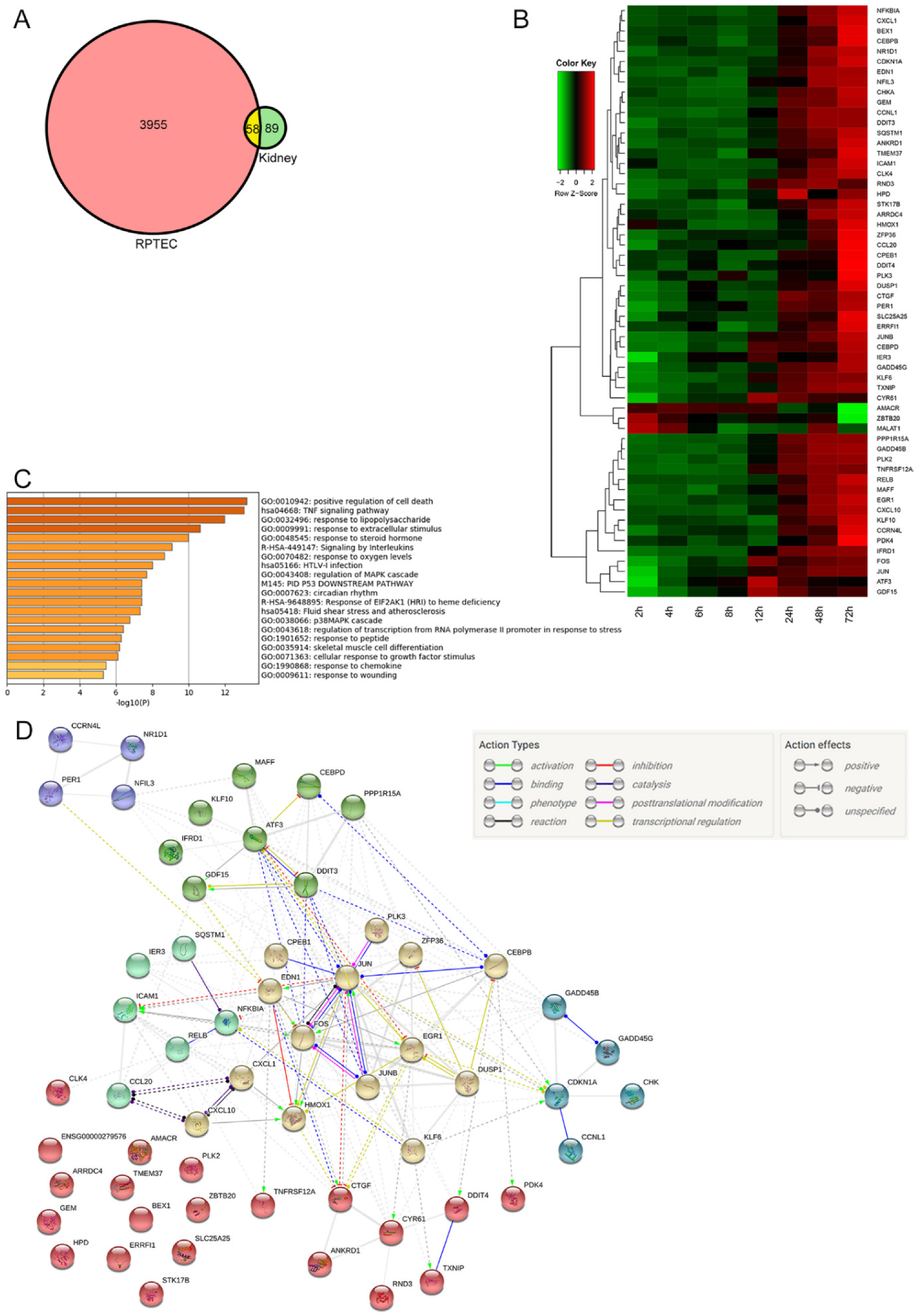
2.3. Microarray Analysis
2.3.1. Differentially Expressed Genes in Both Mouse Kidney and RPTEC
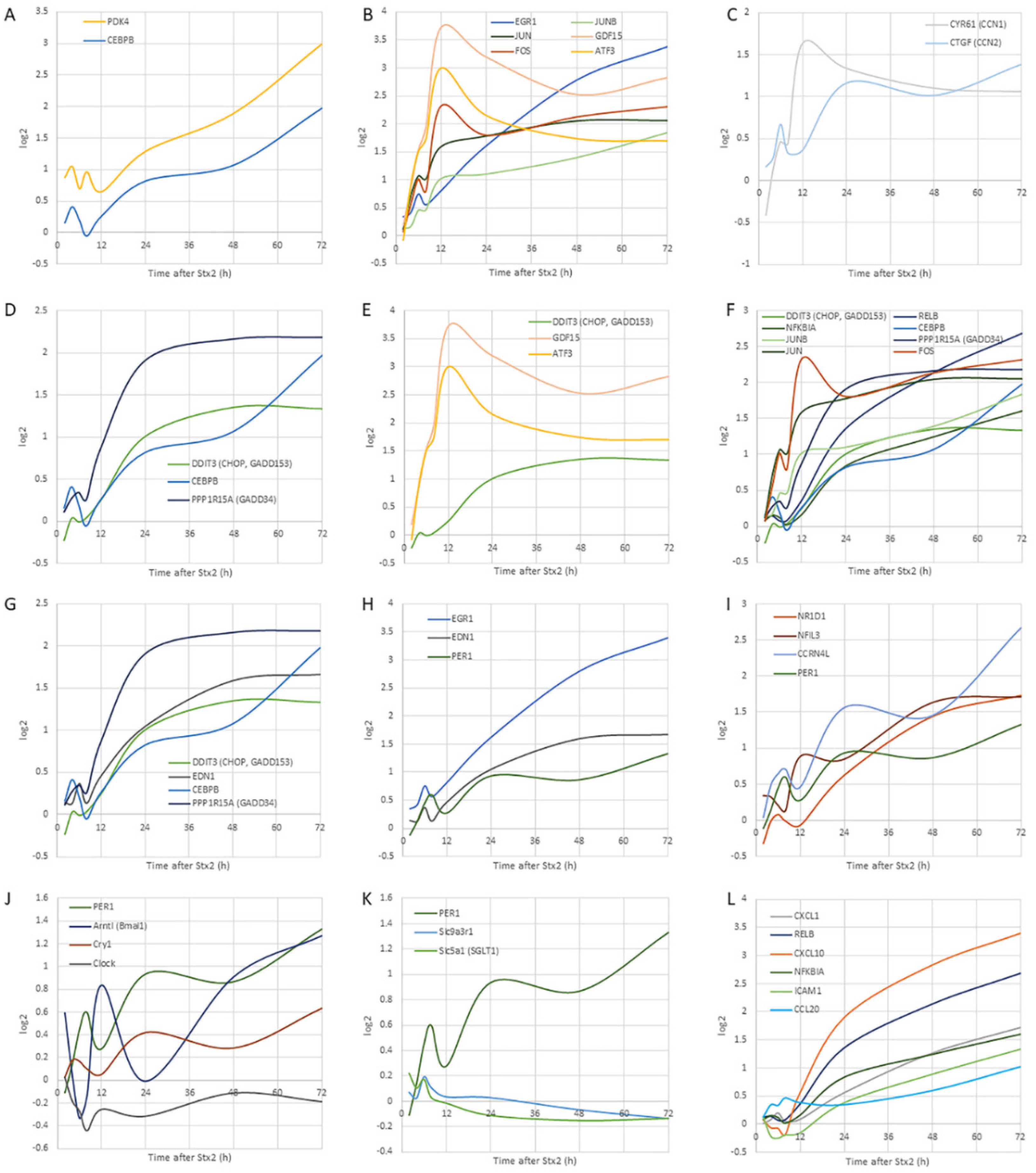
2.3.2. Trend of Gene Expression
The Earliest Responsive Genes
Immediate-Early Genes
EDN-1, EGR-1, and PER1
Circadian Genes
Circadian-Regulated Genes
Matricellular Protein Genes
UPR Genes
NF-κB Pathway-Associated Genes
3. Discussion
3.1. The Proximal Tubular Pathology in STEC Patients and Animal Models
3.2. Proximal Tubular Factors Involved in Stx2-Induced Pathology in Relation to Known Stx2 Activities in Gb3 Expressing Cells
3.2.1. Activity (I), Signal Transduction Induced by Stxs Binding to Gb3
3.2.2. Activity (II), Inducing ER-Stress/UPR
3.2.3. Activity (III), Ribosome Inactivating Protein (RIP) Activity
3.2.4. Activity (IV), Ribotoxic Stress Response (RSR) Activity
3.2.5. Activity (V), Particular Gene Transcription Leading to Cytokine Secretion
3.2.6. Activity (VI), Stxs-Induced Apoptosis
3.3. Circadian Dysregulation by Stx2
4. Conclusions
5. Materials and Methods
5.1. Animals
5.2. Human Kidney Tissue
5.3. Cell Culture
5.4. Purification of LPS-Free Stx2
5.5. Stx2 Administration to Mice
5.6. Urine Collection and Urine Glucose Measurement
5.7. Tissue Processing
5.8. Periodic Acid-Schiff (PAS) Stain
5.9. Stx2 Treatment in Human RPTEC
5.10. Free-Floating Immunofluorescence Staining
5.11. Microarray Analysis
5.12. Bioinformatic Analysis of Microarray Data and Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gianantonio, C.; Vitacco, M.; Mendilaharzu, F.; Rutty, A.; Mendilaharzu, J. The Hemolytic-Uremic Syndrome. J. Pediatr. 1964, 64, 478–491. [Google Scholar] [CrossRef]
- Vitsky, B.H.; Suzuki, Y.; Strauss, L.; Churg, J. The hemolytic-uremic syndrome: A study of renal pathologic alterations. Am. J. Pathol. 1969, 57, 627–647. [Google Scholar]
- Takeda, T.; Dohi, S.; Igarashi, T.; Yamanaka, T.; Yoshiya, K.; Kobayashi, N. Impairment by verotoxin of tubular function contributes to the renal damage seen in haemolytic uraemic syndrome. J. Infect. 1993, 27, 339–341. [Google Scholar] [CrossRef]
- Katagiri, Y.U.; Mori, T.; Nakajima, H.; Katagiri, C.; Taguchi, T.; Takeda, T.; Kiyokawa, N.; Fujimoto, J. Activation of Src family kinase yes induced by Shiga toxin binding to globotriaosyl ceramide (Gb3/CD77) in low density, detergent-insoluble microdomains. J. Biol. Chem. 1999, 274, 35278–35282. [Google Scholar] [CrossRef] [Green Version]
- Takenouchi, H.; Kiyokawa, N.; Taguchi, T.; Matsui, J.; Katagiri, Y.U.; Okita, H.; Okuda, K.; Fujimoto, J. Shiga toxin binding to globotriaosyl ceramide induces intracellular signals that mediate cytoskeleton remodeling in human renal carcinoma-derived cells. J. Cell Sci. 2004, 117, 3911–3922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garred, O.; van Deurs, B.; Sandvig, K. Furin-induced cleavage and activation of Shiga toxin. J. Biol. Chem. 1995, 270, 10817–10821. [Google Scholar] [CrossRef] [Green Version]
- Majoul, I.; Ferrari, D.; Söling, H.D. Reduction of protein disulfide bonds in an oxidizing environment. The disulfide bridge of cholera toxin A-subunit is reduced in the endoplasmic reticulum. FEBS Lett. 1997, 401, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Spooner, R.A.; Watson, P.D.; Marsden, C.J.; Smith, D.C.; Moore, K.A.H.; Cook, J.P.; Lord, J.M.; Roberts, L.M. Protein disulphide-isomerase reduces ricin to its A and B chains in the endoplasmic reticulum. Biochem. J. 2004, 383, 285–293. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Haslam, D.B. Shiga Toxin Is Transported from the Endoplasmic Reticulum following Interaction with the Luminal Chaperone HEDJ/ERdj3. Infect. Immun. 2005, 73, 2524–2532. [Google Scholar] [CrossRef] [Green Version]
- Falguieres, T.; Johannes, L. Shiga toxin B-subunit binds to the chaperone BiP and the nucleolar protein B23. Biol. Cell 2006, 98, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Iordanov, M.S.; Pribnow, D.; Magun, J.L.; Dinh, T.H.; Pearson, J.A.; Chen, S.L.; Magun, B.E. Ribotoxic stress response: Activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol. Cell. Biol. 1997, 17, 3373–3381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfson, J.J.; May, K.L.; Thorpe, C.M.; Jandhyala, D.M.; Paton, J.C.; Paton, A.W. Subtilase cytotoxin activates PERK, IRE1 and ATF6 endoplasmic reticulum stress-signalling pathways. Cell. Microbiol. 2008, 10, 1775–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, J.; Wood, K.; Matsuda, F.; Carneiro-Filho, B.A.; Schlegel, K.H.; Yutsudo, T.; Binnington-Boyd, B.; Lingwood, C.A.; Obata, F.; Kim, K.S.; et al. Shiga toxin 2 causes apoptosis in human brain microvascular endothelial cells via C/EBP homologous protein. Infect. Immun. 2008, 76, 3679–3689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.Y.; Lee, M.S.; Cherla, R.P.; Tesh, V.L. Shiga toxin 1 induces apoptosis through the endoplasmic reticulum stress response in human monocytic cells. Cell. Microbiol. 2008, 10, 770–780. [Google Scholar] [CrossRef]
- Smith, W.E.; Kane, A.V.; Campbell, S.T.; Acheson, D.W.; Cochran, B.H.; Thorpe, C.M. Shiga toxin 1 triggers a ribotoxic stress response leading to p38 and JNK activation and induction of apoptosis in intestinal epithelial cells. Infect. Immun. 2003, 71, 1497–1504. [Google Scholar] [CrossRef] [Green Version]
- Ehrlenbach, S.; Rosales, A.; Posch, W.; Wilflingseder, D.; Hermann, M.; Brockmeyer, J.; Karch, H.; Satchell, S.C.; Würzner, R.; Orth-Höller, D. Shiga toxin 2 reduces complement inhibitor CD59 expression on human renal tubular epithelial and glomerular endothelial cells. Infect. Immun. 2013, 81, 2678–2685. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.K.; Stricklett, P.K.; Kohan, D.E. Cytotoxic effect of Shiga toxin-1 on human proximal tubule cells. Kidney Int. 1998, 54, 426–437. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.K.; Stricklett, P.K.; Kohan, D.E. Shiga toxin-1 regulation of cytokine production by human proximal tubule cells. Kidney Int. 1998, 54, 1093–1106. [Google Scholar] [CrossRef] [Green Version]
- Karpman, D.; Håkansson, A.; Perez, M.T.; Isaksson, C.; Carlemalm, E.; Caprioli, A.; Svanborg, C. Apoptosis of renal cortical cells in the hemolytic-uremic syndrome: In vivo and in vitro studies. Infect. Immun. 1998, 66, 636–644. [Google Scholar] [CrossRef] [Green Version]
- Kodama, T.; Nagayama, K.; Yamada, K.; Ohba, Y.; Akeda, Y.; Honda, T. Induction of apoptosis in human renal proximal tubular epithelial cells by Escherichia coli verocytotoxin 1 in vitro. Med. Microbiol. Immunol. 1999, 188, 73–78. [Google Scholar] [CrossRef]
- Rutjes, N.W.; Binnington, B.A.; Smith, C.R.; Maloney, M.D.; Lingwood, C.A. Differential tissue targeting and pathogenesis of verotoxins 1 and 2 in the mouse animal model. Kidney Int. 2002, 62, 832–845. [Google Scholar] [CrossRef] [Green Version]
- Isogai, E.; Isogai, H.; Kimura, K.; Hayashi, S.; Kubota, T.; Fujii, N.; Takeshi, K. Role of tumor necrosis factor alpha in gnotobiotic mice infected with an Escherichia coli O157:H7 strain. Infect. Immun. 1998, 66, 197–202. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, H.; Takahashi, M.; Yamaguchi, H.; Osaki, T.; Komatsu, A.; Fujioka, Y.; Kamiya, S. Experimental infection of germ-free mice with hyper-toxigenic enterohaemorrhagic Escherichia coli O157:H7, strain 6. J. Med. Microbiol. 2002, 51, 336–343. [Google Scholar] [CrossRef] [Green Version]
- Wadolkowski, E.A.; Sung, L.M.; Burris, J.A.; Samuel, J.E.; O’Brien, A.D. Acute renal tubular necrosis and death of mice orally infected with Escherichia coli strains that produce Shiga-like toxin type II. Infect. Immun. 1990, 58, 3959–3965. [Google Scholar] [CrossRef] [Green Version]
- Tesh, V.L.; Burris, J.A.; Owens, J.W.; Gordon, V.M.; Wadolkowski, E.A.; O’Brien, A.D.; Samuel, J.E. Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect. Immun. 1993, 61, 3392–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindgren, S.W.; Samuel, J.E.; Schmitt, C.K.; O’Brien, A.D. The specific activities of Shiga-like toxin type II (SLT-II) and SLT-II-related toxins of enterohemorrhagic Escherichia coli differ when measured by Vero cell cytotoxicity but not by mouse lethality. Infect. Immun. 1994, 62, 623–631. [Google Scholar] [CrossRef] [Green Version]
- Wolski, V.M.; Soltyk, A.M.; Brunton, J.L. Tumour necrosis factor alpha is not an essential component of verotoxin 1-induced toxicity in mice. Microb. Pathog. 2002, 32, 263–271. [Google Scholar] [CrossRef]
- Psotka, M.A.; Obata, F.; Kolling, G.L.; Gross, L.K.; Saleem, M.A.; Satchell, S.C.; Mathieson, P.W.; Obrig, T.G. Shiga toxin 2 targets the murine renal collecting duct epithelium. Infect. Immun. 2009, 77, 959–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attia, R.R.; Sharma, P.; Janssen, R.C.; Friedman, J.E.; Deng, X.; Lee, J.S.; Elam, M.B.; Cook, G.A.; Park, E.A. Regulation of pyruvate dehydrogenase kinase 4 (PDK4) by CCAAT/enhancer-binding protein beta (C/EBPbeta). J. Biol. Chem. 2011, 286, 23799–23807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef]
- Johnston, J.G.; Pollock, D.M. Circadian regulation of renal function. Free Radic. Biol. Med. 2018, 119, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Solocinski, K.; Richards, J.; All, S.; Cheng, K.-Y.; Khundmiri, S.J.; Gumz, M.L. Transcriptional regulation of NHE3 and SGLT1 by the circadian clock protein Per1 in proximal tubule cells. Am. J. Physiol. Renal Physiol. 2015, 309, F933–F942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, H.; Fujimoto, J.; Takeda, T. Primary tubular impairment by verocytotoxin in hemolytic uremic syndrome. Nippon Rinsho Kekkaku 1997, 55, 726–730. [Google Scholar]
- Eaton, K.A.; Friedman, D.I.; Francis, G.J.; Tyler, J.S.; Young, V.B.; Haeger, J.; Abu-Ali, G.; Whittam, T.S. Pathogenesis of renal disease due to enterohemorrhagic Escherichia coli in germ-free mice. Infect. Immun. 2008, 76, 3054–3063. [Google Scholar] [CrossRef] [Green Version]
- Kamarajugadda, S.; Stemboroski, L.; Cai, Q.; Simpson, N.E.; Nayak, S.; Tan, M.; Lu, J. Glucose oxidation modulates anoikis and tumor metastasis. Mol. Cell. Biol. 2012, 32, 1893–1907. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Zheng, J.; Cheng, S.; Feng, D.; He, J. EBP50 phosphorylation by Cdc2/Cyclin B kinase affects actin cytoskeleton reorganization and regulates functions of human breast cancer cell line MDA-MB-231. Mol. Cells 2013, 36, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Keepers, T.R.; Psotka, M.A.; Gross, L.K.; Obrig, T.G. A murine model of HUS: Shiga toxin with lipopolysaccharide mimics the renal damage and physiologic response of human disease. J. Am. Soc. Nephrol. 2006, 17, 3404–3414. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, K.; Wu, L.-W.; Grivennikov, S.I.; de Jong, P.R.; Lian, I.; Yu, F.-X.; Wang, K.; Ho, S.B.; Boland, B.S.; Chang, J.T.; et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 2015, 519, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Chen, C.C.; Lau, L.F. Adhesion of human skin fibroblasts to Cyr61 is mediated through integrin alpha 6beta 1 and cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2000, 275, 24953–24961. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Du, X.-Y. Functional properties and intracellular signaling of CCN1/Cyr61. J. Cell. Biochem. 2007, 100, 1337–1345. [Google Scholar] [CrossRef]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Yu, J.; Lin, J.D.; Wang, C.-Y.; Chinnaiyan, A.M.; et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Q.; Cheng, Y.; Chang, H.-M.; Deguchi, M.; Hsueh, A.J.; Leung, P.C.K. Sphingosine-1-phosphate promotes ovarian cancer cell proliferation by disrupting Hippo signaling. Oncotarget 2017, 8, 27166–27176. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.-X.; Guan, K.-L. The Hippo pathway: Regulators and regulations. Genes Dev. 2013, 27, 355–371. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.C.; Mo, F.E.; Lau, L.F. The angiogenic factor Cyr61 activates a genetic program for wound healing in human skin fibroblasts. J. Biol. Chem. 2001, 276, 47329–47337. [Google Scholar] [CrossRef] [Green Version]
- Hazes, B.; Read, R.J. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry 1997, 36, 11051–11054. [Google Scholar] [CrossRef]
- LaPointe, P.; Wei, X.; Gariepy, J. A role for the protease-sensitive loop region of Shiga-like toxin 1 in the retrotranslocation of its A1 domain from the endoplasmic reticulum lumen. J. Biol. Chem. 2005, 280, 23310–23318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Bailly-Maitre, B.; Reed, J.C. Endoplasmic reticulum stress: Cell life and death decisions. J. Clin. Investig. 2005, 115, 2656–2664. [Google Scholar] [CrossRef] [Green Version]
- Oyadomari, S.; Mori, M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004, 11, 381–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coll, A.P.; Chen, M.; Taskar, P.; Rimmington, D.; Patel, S.; Tadross, J.A.; Cimino, I.; Yang, M.; Welsh, P.; Virtue, S.; et al. GDF15 mediates the effects of metformin on body weight and energy balance. Nature 2020, 578, 444–448. [Google Scholar] [CrossRef]
- Hsu, J.-Y.; Crawley, S.; Chen, M.; Ayupova, D.A.; Lindhout, D.A.; Higbee, J.; Kutach, A.; Joo, W.; Gao, Z.; Fu, D.; et al. Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 2017, 550, 255–259. [Google Scholar] [CrossRef]
- Tsai, S.-F.; Tao, M.; Ho, L.-I.; Chiou, T.-W.; Lin, S.-Z.; Su, H.-L.; Harn, H.-J. Isochaihulactone-induced DDIT3 causes ER stress-PERK independent apoptosis in glioblastoma multiforme cells. Oncotarget 2017, 8, 4051–4061. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Vanderlaag, K.; Ireland, C.; Ortiz, J.; Grage, H.; Safe, S.; Frankel, A.E. 1,1-Bis(3’-indolyl)-1-(p-biphenyl)methane inhibits basal-like breast cancer growth in athymic nude mice. Breast Cancer Res. 2007, 9, R56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novoa, I.; Zeng, H.; Harding, H.P.; Ron, D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 2001, 153, 1011–1022. [Google Scholar] [CrossRef] [Green Version]
- Whitmarsh, A.J. A central role for p38 MAPK in the early transcriptional response to stress. BMC Biol. 2010, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Jandhyala, D.M.; Ahluwalia, A.; Schimmel, J.J.; Rogers, A.B.; Leong, J.M.; Thorpe, C.M. Activation of the classical mitogen-activated protein kinases is part of the Shiga toxin-induced ribotoxic stress response and may contribute to Shiga toxin-induced inflammation. Infect. Immun. 2016, 84, 138–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondoh, K.; Nishida, E. Regulation of MAP kinases by MAP kinase phosphatases. Biochim. Biophys. Acta 2007, 1773, 1227–1237. [Google Scholar] [CrossRef] [Green Version]
- Kojima, S.; Yanagihara, I.; Kono, G.; Sugahara, T.; Nasu, H.; Kijima, M.; Hattori, A.; Kodama, T.; Nagayama, K.I.; Honda, T. mkp-1 encoding mitogen-activated protein kinase phosphatase 1, a verotoxin 1 responsive gene, detected by differential display reverse transcription-PCR in Caco-2 cells. Infect. Immun. 2000, 68, 2791–2796. [Google Scholar] [CrossRef] [Green Version]
- Thorpe, C.M.; Hurley, B.P.; Lincicome, L.L.; Jacewicz, M.S.; Keusch, G.T.; Acheson, D.W. Shiga toxins stimulate secretion of interleukin-8 from intestinal epithelial cells. Infect. Immun. 1999, 67, 5985–5993. [Google Scholar] [CrossRef] [Green Version]
- Roche, J.K.; Keepers, T.R.; Gross, L.K.; Seaner, R.M.; Obrig, T.G. CXCL1/KC and CXCL2/MIP-2 are critical effectors and potential targets for therapy of Escherichia coli O157:H7-associated renal inflammation. Am. J. Pathol. 2007, 170, 526–537. [Google Scholar] [CrossRef] [Green Version]
- Tam, A.B.; Mercado, E.L.; Hoffmann, A.; Niwa, M. ER stress activates NF-κB by integrating functions of basal IKK activity, IRE1 and PERK. PLoS ONE 2012, 7, e45078. [Google Scholar] [CrossRef] [Green Version]
- Sobbe, I.V.; Krieg, N.; Dennhardt, S.; Coldewey, S.M. Involvement of NF-κB1 and the Non-canonical NF-κB Signaling Pathway in the Pathogenesis of Acute Kidney Injury in Shiga-toxin-2-induced Hemolytic-uremic Syndrome in Mice. Shock 2021, 56, 573–581. [Google Scholar] [CrossRef]
- Fujii, J.; Matsui, T.; Heatherly, D.P.; Schlegel, K.H.; Lobo, P.I.; Yutsudo, T.; Ciraolo, G.M.; Morris, R.E.; Obrig, T. Rapid apoptosis induced by Shiga toxin in HeLa cells. Infect. Immun. 2003, 71, 2724–2735. [Google Scholar] [CrossRef] [Green Version]
- Zinszner, H.; Kuroda, M.; Wang, X.; Batchvarova, N.; Lightfoot, R.T.; Remotti, H.; Stevens, J.L.; Ron, D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998, 12, 982–995. [Google Scholar] [CrossRef]
- Nishitoh, H.; Matsuzawa, A.; Tobiume, K.; Saegusa, K.; Takeda, K.; Inoue, K.; Hori, S.; Kakizuka, A.; Ichijo, H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002, 16, 1345–1355. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Nagayama, K.; Satomura, K.; Honda, T.; Okada, S. Increased serum IL-10 and endothelin levels in hemolytic uremic syndrome caused by Escherichia coli O157. Nephron 2000, 84, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Morigi, M.; Buelli, S.; Zanchi, C.; Longaretti, L.; Macconi, D.; Benigni, A.; Moioli, D.; Remuzzi, G.; Zoja, C. Shigatoxin-induced endothelin-1 expression in cultured podocytes autocrinally mediates actin remodeling. Am. J. Pathol. 2006, 169, 1965–1975. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Qi, Q.; Men, L.; Wang, S.; Li, M.; Xiao, M.; Chen, X.; Wang, S.; Wang, G.; Jia, H.; et al. Berberine protects Kawasaki disease-induced human coronary artery endothelial cells dysfunction by inhibiting of oxidative and endoplasmic reticulum stress. Vascul. Pharmacol. 2020, 127, 106660. [Google Scholar] [CrossRef] [PubMed]
- Simo-Cheyou, E.R.; Vardatsikos, G.; Srivastava, A.K. Src tyrosine kinase mediates endothelin-1-induced early growth response protein-1 expression via MAP kinase-dependent pathways in vascular smooth muscle cells. Int. J. Mol. Med. 2016, 38, 1879–1886. [Google Scholar] [CrossRef]
- Tao, W.; Wu, J.; Zhang, Q.; Lai, S.-S.; Jiang, S.; Jiang, C.; Xu, Y.; Xue, B.; Du, J.; Li, C.-J. EGR1 regulates hepatic clock gene amplitude by activating Per1 transcription. Sci. Rep. 2015, 5, 15212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Fu, Z. Time-dependent glucocorticoid administration differently affects peripheral circadian rhythm in rats. Acta Biochim. Biophys. Sin. 2017, 49, 1122–1128. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Ni, Y.; Dong, Y.; Xu, J.; Song, X.; Kato, H.; Fu, Z. Regulation of circadian gene expression in the kidney by light and food cues in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 298, R635–R641. [Google Scholar] [CrossRef]
- Obata, F.; Tohyama, K.; Bonev, A.D.; Kolling, G.L.; Keepers, T.R.; Gross, L.K.; Nelson, M.T.; Sato, S.; Obrig, T.G. Shiga toxin 2 affects the central nervous system through receptor globotriaosylceramide localized to neurons. J. Infect. Dis. 2008, 198, 1398–1406. [Google Scholar] [CrossRef] [Green Version]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Del Carratore, F.; Jankevics, A.; Eisinga, R.; Heskes, T.; Hong, F.; Breitling, R. RankProd 2.0: A refactored bioconductor package for detecting differentially expressed features in molecular profiling datasets. Bioinformatics 2017, 33, 2774–2775. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Nueda, M.J.; Ferrer, A.; Talon, M. MaSigPro: A method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics 2006, 22, 1096–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. gplots: Various R Programming Tools for Plotting Data. R Package Version 3.0.3. Available online: https://CRAN.R-project.org/package=gplots (accessed on 4 January 2022).
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Time after Stx2 (h) | Number of Common Genes log2 > 1 | Name of Genes |
|---|---|---|
| 2 | 0 | n.a. |
| 4 | 1 | PDK4 |
| 6 | 4 | ATF3, FOS, GDF15, JUN |
| 8 | 3 | ATF3, GDF15, JUN |
| 12 | 11 | ATF3, CEBPD, CYR61, FOS, GDF15, IER3, IFRD1, JUN, JUNB, KLF6, TNFRSF12A |
| 24 | 28 | ATF3, CCRN4L, CEBPD, CTGF, CXCL10, CYR61, DDIT3, EDN1, EGR1, FOS, GADD45B, GADD45G, GDF15, HPD, IFRD1, JUN, JUNB, KLF10, KLF6, MAFF, PDK4, PLK2, PPP1R15A, RELB, RND3, SLC25A25, TNFRSF12A, TXNIP |
| 48 | 39 | ATF3, CCNL1, CCRN4L, CDKN1A, CEBPB, CEBPD, CHKA, CLK4, CTGF, CXCL1, CXCL10, CYR61, DDIT3, DUSP1, EDN1, EGR1, ERRFI1, FOS, GADD45B, GADD45G, GDF15, GEM, IFRD1, JUN, JUNB, KLF10, KLF6, MAFF, NFIL3, NFKBIA, NR1D1, PDK4, PLK2, PPP1R15A, RELB, RND3, SLC25A25, TNFRSF12A, TXNIP |
| 72 | 55 | ANKRD1, ARRDC4, ATF3, BEX1, CCL20, CCNL1, CCRN4L, CDKN1A, CEBPB, CEBPD, CHKA, CLK4, CPEB1, CTGF, CXCL1, CXCL10, CYR61, DDIT3, DDIT4, DUSP1, EDN1, EGR1, ERRFI1, FOS, GADD45B, GADD45G, GDF15, GEM, HMOX1, HPD, ICAM1, IER3, IFRD1, JUN, JUNB, KLF10, KLF6, MAFF, NFIL3, NFKBIA, NR1D1, PDK4, PLK2, PLK3, PPP1R15A, RELB, RND3, SLC25A25, SQSTM1, STK17B, TMEM37, TNFRSF12A, TXNIP, ZFP36 |
| Gene Name | Description | Database |
|---|---|---|
| Nr1d1 | nuclear receptor subfamily 1, group D, member 1 | a |
| Nfil3 | nuclear factor, interleukin 3, regulated | a, b |
| Cdkn1a | cyclin-dependent kinase inhibitor 1A (P21) | a |
| Slc25a25 | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 | a |
| Dusp1 | dual specificity phosphatase 1 | a |
| Chka | choline kinase alpha | a |
| Ctgf | connective tissue growth factor | b |
| Ccrn4l | CCR4 carbon catabolite repression 4-like (S. cerevisiae) | a |
| Per1 | period homolog 1 (Drosophila) | a |
| Amacr | alpha-methylacyl-CoA racemase | a |
| Edn1 | endothelin 1 | a |
| Gadd45g | growth arrest and DNA-damage-inducible 45 gamma | b |
| Zfp36 | zinc finger protein 36 | a |
| Pdk4 | pyruvate dehydrogenase kinase, isoenzyme 4 | a |
| Time after Stx2 (h) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mouse ID | 0 | 2 | 6 | 8 | 12 | 24 | 36 | 48 | 60 | 84 |
| 1 | 20 | 19 | 30 | 24 | 15 | 20 | 18 | 15 | 94 | |
| 2 | 19 | 20 | 18 | 17 | 20 | 10 | 15 | 43 | 118 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obata, F.; Ozuru, R.; Tsuji, T.; Matsuba, T.; Fujii, J. Stx2 Induces Differential Gene Expression and Disturbs Circadian Rhythm Genes in the Proximal Tubule. Toxins 2022, 14, 69. https://doi.org/10.3390/toxins14020069
Obata F, Ozuru R, Tsuji T, Matsuba T, Fujii J. Stx2 Induces Differential Gene Expression and Disturbs Circadian Rhythm Genes in the Proximal Tubule. Toxins. 2022; 14(2):69. https://doi.org/10.3390/toxins14020069
Chicago/Turabian StyleObata, Fumiko, Ryo Ozuru, Takahiro Tsuji, Takashi Matsuba, and Jun Fujii. 2022. "Stx2 Induces Differential Gene Expression and Disturbs Circadian Rhythm Genes in the Proximal Tubule" Toxins 14, no. 2: 69. https://doi.org/10.3390/toxins14020069
APA StyleObata, F., Ozuru, R., Tsuji, T., Matsuba, T., & Fujii, J. (2022). Stx2 Induces Differential Gene Expression and Disturbs Circadian Rhythm Genes in the Proximal Tubule. Toxins, 14(2), 69. https://doi.org/10.3390/toxins14020069





