Mycotoxin Production and the Relationship between Microbial Diversity and Mycotoxins in Pyrus bretschneideri Rehd cv. Huangguan Pear
Abstract
1. Introduction
2. Results
2.1. Microbial Community Composition in Pear Fruits
2.2. Comparison of Microbial Diversity in Pear Fruit
2.3. Correlation Analysis of Fungi and Bacteria
2.4. Metabolic Pathways of Microorganisms
2.5. Mycotoxin Content and Its Relationship with Microbial Composition
3. Discussion
4. Materials and Methods
4.1. Storage Conditions and Sample Preparation of Pear Fruit
4.2. DNA Extraction and Amplicon Sequencing
4.3. Bioinformatic Pipeline for Analysis of Microbial Diversity
4.4. Determination of Mycotoxin Production
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, F.; Saito, S.; Michailides, T.J.; Xiao, C.L. Phylogenetic, morphological, and pathogenic characterization of Alternaria species associated with fruit rot of Mandarin in California. Plant Dis. 2021, 105, 2606–2617. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, T.C.; Ferreira, M.C.; Rosa, L.H.; de Oliveira, A.M.; Oliveira Júnior, E.N. Penicillium citrinum and Penicillium mallochii: New phytopathogens of orange fruit and their control using chitosan. Carbohydr. Polym. 2020, 234, 115918. [Google Scholar] [CrossRef] [PubMed]
- Saif, F.A.; Yaseen, S.A.; Alameen, A.S.; Mane, S.B.; Undre, P.B. Identification and characterization of Aspergillus species of fruit rot fungi using microscopy, FT-IR, Raman and UV-Vis spectroscopy. Spectrochim. Acta A Mol. Biomol. 2021, 246, 119010. [Google Scholar] [CrossRef]
- Petrasch, S.; Silva, C.J.; Mesquida-Pesci, S.D.; Gallegos, K.; van den Abeele, C.; Papin, V.; Fernandez-Acero, F.J.; Knapp, S.J.; Blanco-Ulate, B. Infection strategies deployed by Botrytis cinerea, Fusarium acuminatum, and Rhizopus stolonifer as a function of tomato fruit ripening stage. Front. Plant Sci. 2019, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Dowling, M.; Peres, N.; Villani, S.; Schnabel, G. Managing Colletotrichum on fruit crops: A “complex” challenge. Plant Dis. 2020, 104, 2301–2316. [Google Scholar] [CrossRef] [PubMed]
- Balsells-Llauradó, M.; Silva, C.J.; Usall, J.; Vall-Llaura, N.; Serrano-Prieto, S.; Teixidó, N.; Mesquida-Pesci, S.D.; de Cal, A.; Blanco-Ulate, B.; Torres, R. Depicting the battle between nectarine and Monilinia laxa: The fruit developmental stage dictates the effectiveness of the host defenses and the pathogen’s infection strategies. Hortic. Res. 2020, 7, 167. [Google Scholar] [CrossRef]
- Chen, L.; Ingham, B.H.; Ingham, S.C. Survival of Penicillium expansum and patulin production on stored apples after wash treatments. J. Food Sci. 2004, 69, C669–C675. [Google Scholar]
- Katerere, D.R.; Stockenström, S.; Thembo, K.M.; Balducci, G.; Shephard, G.S. Investigation of patulin contamination in apple juice sold in retail outlets in Italy and South Africa. Food Addit. Contam. 2007, 24, 630–634. [Google Scholar] [CrossRef]
- Ostry, V. Alternaria mycotoxins: An overview of chemical characterization, producers, toxicity, analysis and occurrence in foodstuffs. World Mycotoxin J. 2008, 1, 175–188. [Google Scholar] [CrossRef]
- Daou, R.; Joubrane, K.; Maroun, R.G.; Khabbaz, L.R.; Ismail, A.; Khoury, A.E. Mycotoxins: Factors influencing production and control strategies. AIMS Agric. Food 2021, 6, 416–447. [Google Scholar] [CrossRef]
- Ji, C.; Fan, Y.; Zhao, L. Review on biological degradation of mycotoxins. Anim. Nutr. 2016, 2, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Paterson, R.; Venancio, A.; Lima, N. Solutions to Penicillium taxonomy crucial to mycotoxin research and health. Res. Microbiol. 2004, 155, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.S.; Brasel, J.M. Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology 2001, 167, 101–134. [Google Scholar] [CrossRef]
- Guan, J.; He, J.; Shen, C.; Li, L.; Wang, Y.; Cheng, Y. How cultivars influence fruit composition: Total phenols, flavonoids contents, and antioxidant activity in the pulp of selected asian pears. In Processing and Impact on Active Components in Food; Preedy, V., Ed.; Academic Press: Cambridge, MA, USA, 2015; pp. 139–145. [Google Scholar]
- Wiseman, M.S.; Kim, Y.K.; Dugan, F.M.; Rogers, J.D.; Xiao, C.L. A new postharvest fruit rot in apple and pear caused by Phacidium lacerum. Plant Dis. 2016, 100, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Wenneker, M.; Pham, K.T.K.; Lemmers, M.E.C.; De Boer, F.A.; Van Leeuwen, P.J.; Hollinger, T.C.; van de Geijn, F.G.; Thomma, B.P.H.J. Fibulorhizoctonia psychrophila is the causal agent of lenticel spot on apple and pear fruit in the Netherlands. Eur. J. Plant Pathol. 2017, 148, 213–217. [Google Scholar] [CrossRef]
- Oyom, W.; Li, Y.-C.; Prusky, D.; Zhang, Z.; Bi, Y.; Tahergorabi, R. Recent advances in postharvest technology of Asia pears fungi disease control: A review. Physiol. Mol. Plant Pathol. 2022, 117, 101771. [Google Scholar] [CrossRef]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Freilich, S.; Bartuv, R.; Zhimo, V.Y.; Kumar, A.; Biasi, A.; Salim, S.; Feygenberg, O.; Burchard, E.; Dardick, C.; et al. Global analysis of the apple fruit microbiome: Are all apples the same? Environ. Microbiol. 2021, 23, 6038–6055. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M. The fruit microbiome: A new frontier for postharvest biocontrol and postharvest biology. Postharvest Biol. Technol. 2018, 140, 107–112. [Google Scholar] [CrossRef]
- Zhang, H.; Serwah Boateng, N.A.; Ngolong Ngea, G.L.; Shi, Y.; Lin, H.; Yang, Q.; Wang, K.; Zhang, X.; Zhao, L.; Droby, S. Unravelling the fruit microbiome: The key for developing effective biological control strategies for postharvest diseases. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4906–4930. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Guan, J.F.; Ma, S.J.; Liu, L.L.; Feng, Y.X.; Cheng, Y.D. Calcium content and its correlated distribution with skin browning spot in bagged Huangguan pear. Protoplasma 2015, 252, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Guan, J. The key technologies of commercial handling and storage of postharvest Huangguan pear. Storage Process 2013, 13, 59–61. [Google Scholar]
- Zhang, Z.-Q.; Chen, T.; Li, B.; Qin, G.-Z.; Tian, S.-P. Molecular basis of pathogenesis of postharvest pathogenic fungi and control strategy in fruits: Progress and prospect. Mol. Hort. 2021, 1, 2. [Google Scholar] [CrossRef]
- Leng, J.; Yu, L.; Dai, Y.; Leng, Y.; Wang, C.; Chen, Z.; Wisniewski, M.; Wu, X.; Liu, J.; Sui, Y. Recent advances in research on biocontrol of postharvest fungal decay in apples. Crit. Rev. Food Sci. Nutr. 2022, 1–14. [Google Scholar] [CrossRef]
- Carneiro, G.A.; Baric, S. Colletotrichum fioriniae and Colletotrichum godetiae causing postharvest bitter rot of apple in south tyrol (Northern Italy). Plant Dis. 2021, 105, 3118–3126. [Google Scholar] [CrossRef]
- Peng, H.X.; Wei, X.Y.; Xiao, Y.X.; Sun, Y.; Biggs, A.R.; Gleason, M.L.; Shang, S.P.; Zhu, M.Q.; Guo, Y.Z.; Sun, G.Y. Management of Valsa canker on apple with adjustments to potassium nutrition. Plant Dis. 2016, 100, 884–889. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, B.; Gai, Y.; Sun, X.; Chung, K.R.; Li, H. Cell-wall-degrading enzymes required for virulence in the host selective toxin-producing necrotroph Alternaria alternata of citrus. Front. Microbiol. 2019, 10, 2514. [Google Scholar] [CrossRef]
- Mansilla, J.P.; Aguín, O.; Salinero, M.C. First report of a root rot caused by Rosellinia necatrix on Camellia in Spain. Plant Dis. 2002, 86, 813. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, M.; Pan, L.; Fu, Y.; Xiang, M. First report of fruit rot on ‘Cuiguan’ pear caused by Fusarium proliferatum in China. Plant Dis. 2019, 104, 1554–1555. [Google Scholar] [CrossRef]
- Chen, T.; Liu, Y.-X.; Huang, L. ImageGP: An easy-to-use data visualization web server for scientific researchers. iMeta 2022, 1, e5. [Google Scholar] [CrossRef]
- Khan, J.; Ooka, J.J.; Miller, S.A.; Madden, L.V.; Hoitink, H.A.J. Systemic resistance induced by Trichoderma hamatum 382 in cucumber against Phytophthora crown rot and leaf blight. Plant Dis. 2004, 88, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Begum, M.F.; Rahman, M.A.; Alam, M.F. Biological control of Alternaria fruit rot of chili by Trichoderma species under field conditions. Mycobiology 2010, 38, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Intana, W.; Kheawleng, S.; Sunpapao, A. Trichoderma asperellum T76-14 released volatile organic compounds against postharvest fruit rot in muskmelons (cucumis melo) caused by Fusarium incarnatum. J. Fungi 2021, 7, 46. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, C.; Masum, M.M.I.; Cheng, Y.; Wei, C.; Guan, Y.; Guan, J. Dynamic microbiome changes reveal the effect of 1-methylcyclopropene treatment on reducing post-harvest fruit decay in “doyenne du comice” pear. Front. Microbiol. 2021, 12, 729014. [Google Scholar] [CrossRef]
- Bevardi, M.; Frece, J.; Mesarek, D.; Bošnir, J.; Mrvčić, J.; Delaš, F.; Markov, K. Antifungal and antipatulin activity of Gluconobacter oxydans isolated from apple surface. Arh. Hig. Rada. Toksikol. 2013, 64, 93–98. [Google Scholar] [CrossRef]
- Van Keer, C.; Vanden Abeele, P.; Swings, J.; Gosselé, F.; De Ley, J. Acetic acid bacteria as causal agents of browning and rot of apples and pears. Syst. Appl. Microbiol. 1981, 2, 197–204. [Google Scholar] [CrossRef]
- Malimas, T.; Yukphan, P.; Lundaa, T.; Muramatsu, Y.; Takahashi, M.; Kaneyasu, M.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Suzuki, K.; et al. Gluconobacter kanchanaburiensis sp. nov., a brown pigment-producing acetic acid bacterium for Thai isolates in the Alphaproteobacteria. J. Gen. Appl. Microbiol. 2009, 55, 247–254. [Google Scholar] [CrossRef]
- Zhu, B.; Zhang, Z.; Wang, H.; Ma, X. Isolation and culture conditions optimization of a new bacterial cellulose producing strain Komagataeibacter intermedius 6-5. IOP Conf. Ser. Earth Environ. Sci. 2021, 632, 32040. [Google Scholar] [CrossRef]
- Verce, M.; Schoonejans, J.; Hernandez Aguirre, C.; Molina-Bravo, R.; De Vuyst, L.; Weckx, S. A combined metagenomics and metatranscriptomics approach to unravel costa rican cocoa box fermentation processes reveals yet unreported microbial species and functionalities. Front. Microbiol. 2021, 12, 641185. [Google Scholar] [CrossRef]
- He, X.; Meng, H.; Song, H.; Deng, S.; He, T.; Wang, S.; Wei, D.; Zhang, Z. Novel bacterial cellulose membrane biosynthesized by a new and highly efficient producer Komagataeibacter rhaeticus TJPU03. Carbohydr. Res. 2020, 493, 108030. [Google Scholar] [CrossRef] [PubMed]
- Gosselé, F.; Swings, J. Identification of Acetobacter liquefaciens as causal agent of pink-disease of pineapple fruit. J. Phytopathol. 1986, 116, 167–175. [Google Scholar] [CrossRef]
- Nunes, C.; Usall, J.; Teixidó, N.; Viñas, I. Biological control of postharvest pear diseases using a bacterium, Pantoea agglomerans CPA-2. Int. J. Food Microbiol. 2001, 70, 53–61. [Google Scholar] [CrossRef]
- Morales, H.; Sanchis, V.; Usall, J.; Ramos, A.J.; Marín, S. Effect of biocontrol agents Candida sake and Pantoea agglomerans on Penicillium expansum growth and patulin accumulation in apples. Int. J. Food Microbiol. 2008, 122, 61–67. [Google Scholar] [CrossRef]
- Xu, S.; Liu, Y.-X.; Cernava, T.; Wang, H.; Zhou, Y.; Xia, T.; Cao, S.; Berg, G.; Shen, X.-X.; Wen, Z.; et al. Fusarium fruiting body microbiome member Pantoea agglomerans inhibits fungal pathogenesis by targeting lipid rafts. Nat. Microbiol. 2022, 7, 831–843. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, I.D.; Pizzutti, I.R.; Dias, J.V.; Fontana, M.E.Z.; Brackmann, A.; Anese, R.O.; Thewes, F.R.; Marques, L.N.; Cardoso, C.D. Patulin accumulation in apples under dynamic controlled atmosphere storage. Food Chem. 2018, 255, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Sadhasivam, S.; Barda, O.; Zakin, V.; Reifen, R.; Sionov, E. Rapid detection and quantification of Patulin and Citrinin contamination in fruits. Molecules 2021, 26, 4545. [Google Scholar] [CrossRef]
- Zhong, L.; Carere, J.; Lu, Z.; Lu, F.; Zhou, T. Patulin in apples and apple-based food products: The burdens and the mitigation strategies. Toxins 2018, 10, 475. [Google Scholar] [CrossRef]
- Menniti, A.M.; Neri, F.; Gregori, R.; Maccaferri, M. Some factors influencing patulin production by Penicillium expansum in pome fruits. J. Sci. Food Agric. 2010, 90, 2183–2187. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The genus alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Illueca, F.; Vila-Donat, P.; Calpe, J.; Luz, C.; Meca, G.; Quiles, J.M. Antifungal activity of biocontrol agents in vitro and potential application to reduce mycotoxins (aflatoxin b1 and ochratoxin a). Toxins 2021, 13, 752. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Jeong, J.C.; Lee, J.S.; Park, J.M.; Yang, J.W.; Lee, M.H.; Choi, S.H.; Kim, C.Y.; Kim, D.H.; Kim, S.W.; et al. Potential of Pantoea dispersa as an effective biocontrol agent for black rot in sweet potato. Sci. Rep. 2019, 9, 16354. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Heck, K.L., Jr.; van Belle, G.; Simberloff, D. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 1975, 56, 1459–1461. [Google Scholar] [CrossRef]
- Kemp, P.F.; Aller, J.Y. Bacterial diversity in aquatic and other environments: What 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 2004, 47, 161–177. [Google Scholar] [CrossRef]
- Chao, A. Non-parametric estimation of the classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, N.; Xian, H.; Wei, D.; Shi, L.; Feng, X. A single-step solid phase extraction for the simultaneous determination of 8 mycotoxins in fruits by ultra-high performance liquid chromatography tandem mass spectrometry. J. Chromatogr. A 2016, 1429, 22–29. [Google Scholar] [CrossRef] [PubMed]
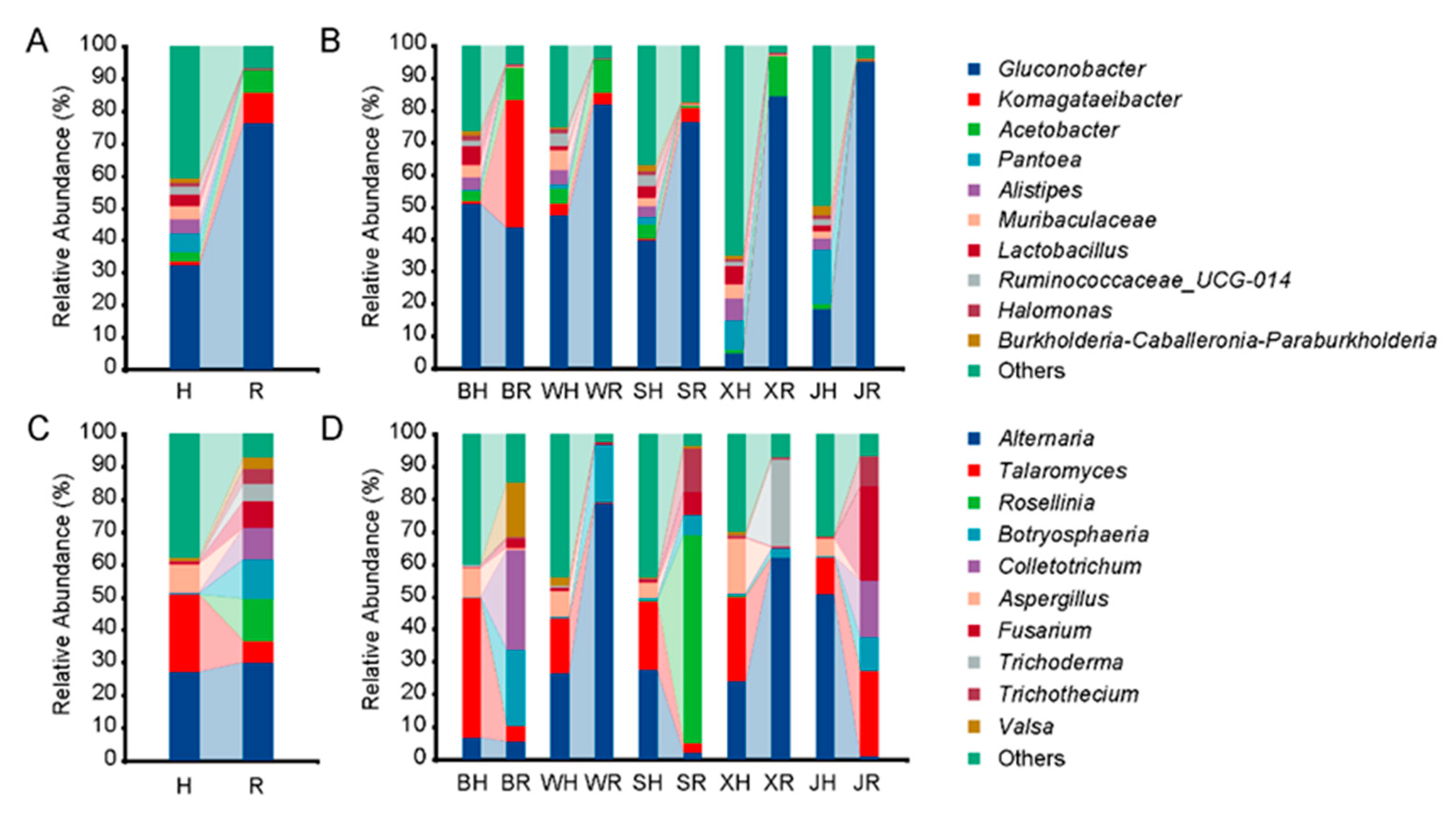




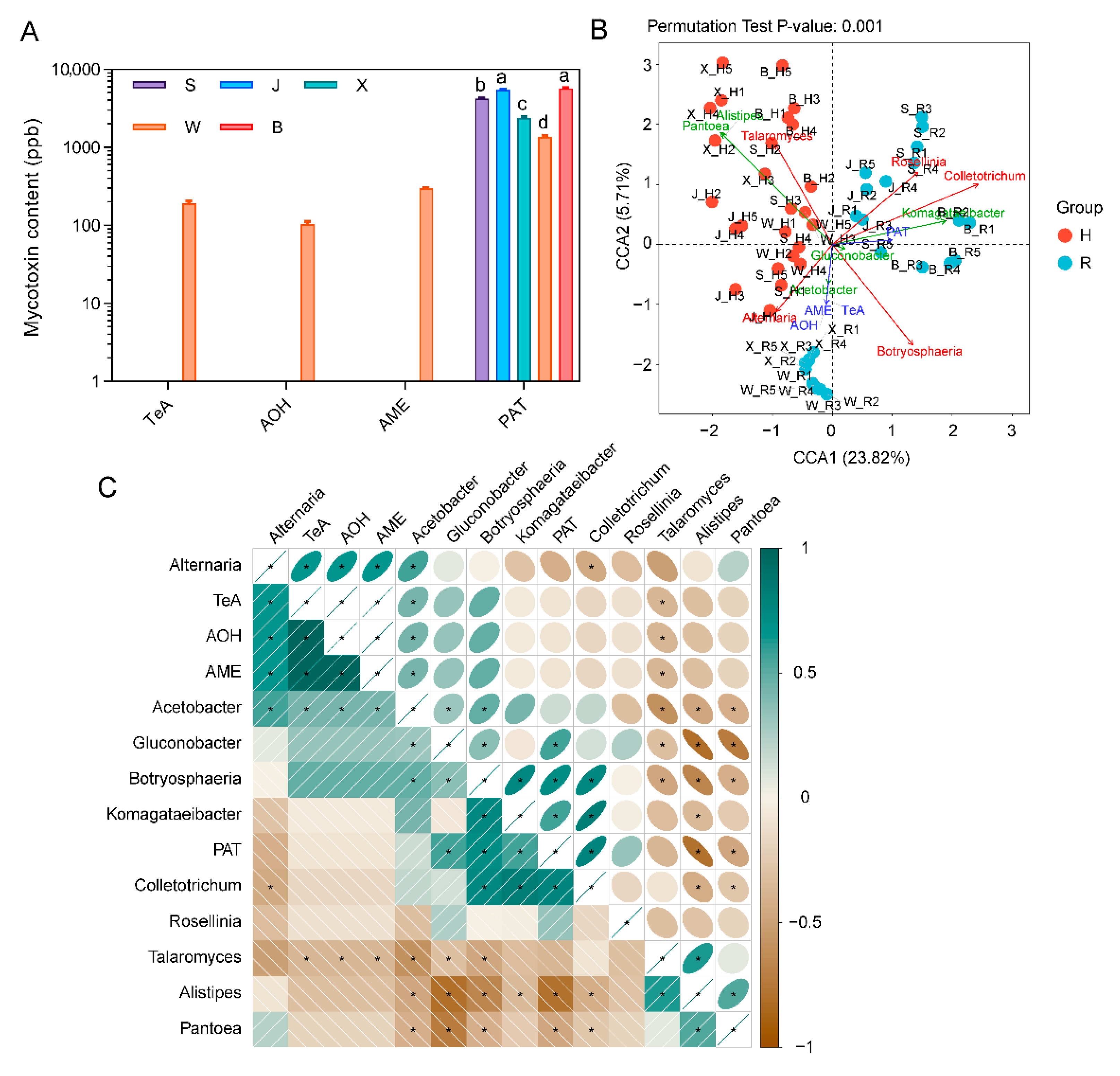
| Pairs | Bacteria | Fungi | ||||
|---|---|---|---|---|---|---|
| R2 | P-Adjusted | Sig | R2 | P-Adjusted | Sig | |
| BH vs. WH | 0.591978 | 0.011053 | * | 0.321574 | 0.011053 | * |
| BH vs. SH | 0.212455 | 0.0465 | * | 0.312754 | 0.011053 | * |
| BH vs. XH | 0.721622 | 0.011053 | * | 0.215719 | 0.015714 | * |
| BH vs. JH | 0.409482 | 0.011053 | * | 0.532116 | 0.011053 | * |
| BH vs. BR | 0.813081 | 0.011053 | * | 0.702342 | 0.011053 | * |
| BH vs. WR | 0.81491 | 0.011053 | * | 0.749095 | 0.011053 | * |
| BH vs. XR | 0.679522 | 0.011053 | * | 0.747734 | 0.011053 | * |
| BH vs. JR | 0.721766 | 0.011053 | * | 0.680768 | 0.011053 | * |
| WH vs. SH | 0.396408 | 0.011053 | * | 0.171005 | 0.055739 | |
| WH vs. XH | 0.71682 | 0.011053 | * | 0.162669 | 0.070833 | |
| WH vs. JH | 0.359256 | 0.011786 | * | 0.348047 | 0.011063 | * |
| WH vs. BR | 0.827208 | 0.011053 | * | 0.716855 | 0.011053 | * |
| WH vs. WR | 0.658041 | 0.011053 | * | 0.681124 | 0.011053 | * |
| WH vs. SR | 0.72838 | 0.011053 | * | 0.688894 | 0.011053 | * |
| WH vs. XR | 0.843609 | 0.011053 | * | 0.677885 | 0.011053 | * |
| WH vs. JR | 0.859374 | 0.01186 | * | 0.72708 | 0.011053 | * |
| SH vs. XH | 0.517982 | 0.011786 | * | 0.164089 | 0.027209 | * |
| SH vs. JH | 0.278789 | 0.011053 | * | 0.316119 | 0.011053 | * |
| SH vs. BR | 0.651772 | 0.011053 | * | 0.738717 | 0.011053 | * |
| SH vs. WR | 0.616787 | 0.011053 | * | 0.695862 | 0.011053 | * |
| SH vs. SR | 0.586405 | 0.011053 | * | 0.697443 | 0.011053 | * |
| SH vs. XR | 0.586903 | 0.011053 | * | 0.693377 | 0.011053 | * |
| SH vs. JR | 0.631411 | 0.011053 | * | 0.735392 | 0.011053 | * |
| XH vs. JH | 0.40517 | 0.016193 | * | 0.382327 | 0.011053 | * |
| XH vs. BR | 0.887939 | 0.011053 | * | 0.712806 | 0.011053 | * |
| XH vs. WR | 0.89198 | 0.011053 | * | 0.688394 | 0.011053 | * |
| XH vs. SR | 0.878855 | 0.011053 | * | 0.682479 | 0.011053 | * |
| XH vs. XR | 0.89183 | 0.011053 | * | 0.685562 | 0.011053 | * |
| XH vs. JR | 0.892796 | 0.011053 | * | 0.715241 | 0.011053 | * |
| JH vs. BR | 0.621556 | 0.011053 | * | 0.809491 | 0.011053 | * |
| JH vs. WR | 0.551114 | 0.011053 | * | 0.606817 | 0.011053 | * |
| JH vs. SR | 0.558436 | 0.011053 | * | 0.761766 | 0.011053 | * |
| JH vs. XR | 0.631405 | 0.011053 | * | 0.607294 | 0.011053 | * |
| JH vs. JR | 0.638654 | 0.011053 | * | 0.830115 | 0.011053 | * |
| BR vs. WR | 0.96884 | 0.011053 | * | 0.894688 | 0.011053 | * |
| BR vs. SR | 0.945308 | 0.011538 | * | 0.775024 | 0.011053 | * |
| BR vs. XR | 0.975101 | 0.011053 | * | 0.920598 | 0.011053 | * |
| BR vs. JR | 0.977461 | 0.011053 | * | 0.83395 | 0.011053 | * |
| WR vs. SR | 0.908071 | 0.011786 | * | 0.851352 | 0.011341 | * |
| WR vs. XR | 0.983926 | 0.011053 | * | 0.90034 | 0.011053 | * |
| WR vs. JR | 0.987103 | 0.011053 | * | 0.958482 | 0.011053 | * |
| SR vs. XR | 0.961185 | 0.011053 | * | 0.858435 | 0.011053 | * |
| SR vs. JR | 0.955762 | 0.011053 | * | 0.760137 | 0.011063 | * |
| XR vs. JR | 0.890881 | 0.011053 | * | 0.961663 | 0.011053 | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Zhang, Y.; Gao, C.; Gao, Q.; Cheng, Y.; Zhao, M.; Guan, J. Mycotoxin Production and the Relationship between Microbial Diversity and Mycotoxins in Pyrus bretschneideri Rehd cv. Huangguan Pear. Toxins 2022, 14, 699. https://doi.org/10.3390/toxins14100699
Li H, Zhang Y, Gao C, Gao Q, Cheng Y, Zhao M, Guan J. Mycotoxin Production and the Relationship between Microbial Diversity and Mycotoxins in Pyrus bretschneideri Rehd cv. Huangguan Pear. Toxins. 2022; 14(10):699. https://doi.org/10.3390/toxins14100699
Chicago/Turabian StyleLi, Huimin, Yang Zhang, Congcong Gao, Qi Gao, Yudou Cheng, Min Zhao, and Junfeng Guan. 2022. "Mycotoxin Production and the Relationship between Microbial Diversity and Mycotoxins in Pyrus bretschneideri Rehd cv. Huangguan Pear" Toxins 14, no. 10: 699. https://doi.org/10.3390/toxins14100699
APA StyleLi, H., Zhang, Y., Gao, C., Gao, Q., Cheng, Y., Zhao, M., & Guan, J. (2022). Mycotoxin Production and the Relationship between Microbial Diversity and Mycotoxins in Pyrus bretschneideri Rehd cv. Huangguan Pear. Toxins, 14(10), 699. https://doi.org/10.3390/toxins14100699





