Biological Control and Mitigation of Aflatoxin Contamination in Commodities
Abstract
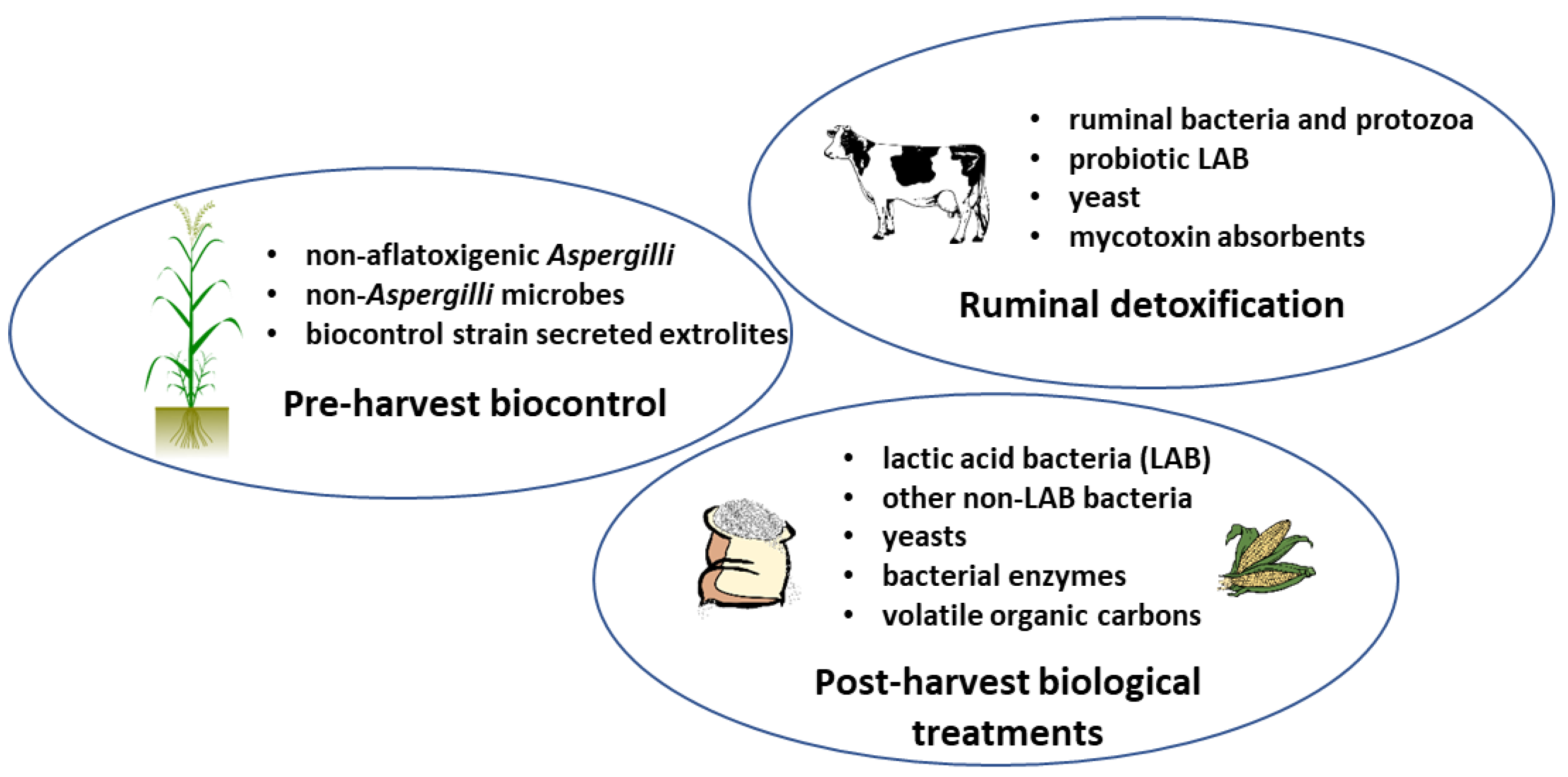
:1. Introduction
2. Document Analysis
3. Ruminal Detoxification of Aflatoxins
4. Pre-Harvest Biocontrol
4.1. Pre-Harvest Biocontrol by Competitive Exclusion
4.2. Pre-Harvest Biocontrol by Microbial Biofungicides
5. Post-Harvest Management of Aflatoxin Contamination
5.1. Bacteria
5.2. Yeasts
5.3. Fungal Biomass, Enzymes, and Antifungal Proteins
6. Conclusions and Future Trends
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Aflatoxin | Bacteria | Strain | Toxin Elimination (%) | References |
|---|---|---|---|---|
| B1 | Bifidobacterium animalis subsp. animalis (formerly Bifidobacterium animalis) | CSCC 1941 | 45.7 | [118] |
| B1 | Bifidobacterium animalis subsp. lactis (formerly Bifidobacterium lactis) | E-94508 | 18 | |
| CSCC 5094 | 34.7 | |||
| CSCC 1906 | 48.7 | |||
| B1 | Bifidobacterium longum | CSCC 5304 | 37.5 | |
| B1 | Enterococcus faecium | M74 | 19.3–30.5 | [73] |
| EF031 | 23.4–37.5 | |||
| B1 | Lactobacillus acidophilus | ATCC 4356 | 48.3 | [119] |
| E-94507 | 18.2 | [118] | ||
| CSCC 5361 | 20.7 | |||
| Chr. Hansen, Denmark | 56.6 | [120] | ||
| CH-2 | 36 | [121] | ||
| B1 | Lactobacillus amylovorus | CSCC 5160 | 59.7 | [118] |
| CSCC 5197 | 57.8 | |||
| B1 | Lactobacillus bulgaricus | Chr. Hansen, Denmark | 16.3 | [120] |
| B1 | Lacticaseibacillus casei (formerly Lactobacillus casei) Shirota | YIT 901, commercial | 21.8 | [119] |
| Yakult | 98 | [122] | ||
| B1 | Lacticaseibacillus casei (formerly Lactobacillus casei) | Chr. Hansen, Denmark | 22.4 | [120] |
| Iranian dairy products | 43 | [123] | ||
| B1 | Lactobacillus delbrueckii | MK9 | 17.3 | [118] |
| B1 | Lactobacillus delbrueckii subsp. bulgaricus | Australian Starter Culture Research Centre Collection, Werribee, Australia | 15.6 | [119] |
| B1 | Limosilactobacillus fermentum (formerly Lactobacillus fermentum) | CSCC 5362 | 22.6 | [118] |
| N.A. | ≥60 | [93] | ||
| Iranian sourdough | 61 | [123] | ||
| PTCC 1744 | 99.9 | [124] | ||
| B1 | Lactobacillus helveticus | Australian Starter Culture Research Centre Collection, Werribee, Australia | 17.5 | [119] |
| Aki4 | 30.1 | [118] | ||
| Chr. Hansen, Denmark | 17.8 | [120] | ||
| B1 | Lactobacillus johnsonii | CSCC 5142 | 30.1 | [118] |
| B1 | Lactiplantibacillus plantarum (formerly Lactobacillus plantarum) | ATCC 8014 | 29.9 | [119] |
| E-79098 | 28.4 | [118] | ||
| N.A. | ≥40 | [93] | ||
| Iranian dairy products | 56 | [123] | ||
| EMCC-1039 | 39.2 | [121] | ||
| B1 | Lacticaseibacillus rhamnosus (formerly Lactobacillus rhamnosus) | DSM 7061 | 76.5 | [119] |
| ATCC 53103 | 54 | [125] | ||
| 78.9 | [119] | |||
| E-97800 | 22.7 | [118] | ||
| Lc 1/3 | 54.6 | |||
| NRRL B-445 | 17.2 | [121] | ||
| B1 | Lactococcus lactis subsp. lactis | Australian Starter Culture Research Centre Collection, Werribee, Australia | 59 | [119] |
| E-90414 | 31.6 | [118] | ||
| Dairy products | 54.3 | [126] | ||
| B1 | Lactococcus lactis subsp. cremoris | Australian Starter Culture Research Centre Collection, Werribee, Australia | 26.9 | [119] |
| MK4 | 5.6 | [118] | ||
| ARH74 (Valio Ltd, Finland) | 41.1 | |||
| B1 | Lactobacillus selangorensis (formerly Paralactobacillus selangorensis) | N.A. | <39 | [93] |
| B1 | Pediococcus acidilactici | N.A. | 45–59 | [93] |
| B1 | Propionibacterium freudenreichii subsp. shermanii JS | Valio Ltd. Finland | 22–82 | [125] |
| [119] | ||||
| B1 | Streptococcus thermophilus | Australian Starter Culture Research Centre Collection, Werribee, Australia | 32.7 | [119] |
| Dairy products | 81 | [126] | ||
| CH-1 | 26.9 | [121] | ||
| B1 | Weissella confuse | N.A. | 15–39 | [93] |
| M1 | Bifidobacterium bifidum | Bb13 | 23.48 | [127] |
| NCC 381 | 18.11 | |||
| PTCC 1644 | 99.9 | [124] | ||
| M1 | Bifidobacterium lactis | FLORA-FIT BI07 (Danisco Ltd.) | 16.89 | [128] |
| M1 | Enterococcus avium | CTC 469 (Tecnolat, Brazil) | 7.36 | [128] |
| M1 | Lactobacillus acidophilus | NCC 12 | 16.05 | [127] |
| NCC 36 | 22.23 | [127] | ||
| NCC 68 | 14.22 | [127] | ||
| LA-5 (Chr. Hansen, Denmark) | 95 | [62] | ||
| M1 | Lactobacillus delbrueckii subsp. bulgaricus | LB340 (Danisco Ltd.) | 30.22 | [128] |
| CH-2 (Chr. Hansen, Denmark) | 18.7 | [129] | ||
| M1 | Lactobacillus gasseri | ATCC 33323 | 21.37 | [128] |
| M1 | Lactiplantibacillus plantarum (formerly Lactobacillus plantarum) | CTC368 | 5.6 | [128] |
| M1 | Lacticaseibacillus rhamnosus (formerly Lactobacillus rhamnosus) | Ezal, France | 20.21 | [127] |
| HOWARU (Danisco Ltd.) | 17.13 | [128] | ||
| M1 | Pediococcus pentosaceus | TR570 (Tecnolat, Brazil) | 8.68 | [128] |
| M1 | Streptococcus thermophilus | ST-36 (Chr. Hansen, Denmark) | 29.42 | [129] |
| Toxin | Bacteria | Strain | Toxin Elimination (%) | References |
|---|---|---|---|---|
| B1 | Bacillus licheniformis | Thai fermented soybean | 74 | [130] |
| B1 | Bacillus stearothermophilus | N.A. | 87 | [131] |
| B1 | Bacillus subtilis | UTBSP1 | 85.66 | [74] |
| B1, G1, M1 | ANSB060 | 81.5, 81, 60 | [132] | |
| B1 | Brachybacterium spp. | Rabbit feces | 74.83 | [133] |
| B1 | Brevundimonas spp. | Yellow cheek feces | 76.86 | [133] |
| B1 | Cellulosimicrobium funkei | Soil around coal factories | 97 | [78] |
| B1 | Enterobacter spp. | Hog deer feces | 76 | [133] |
| B1 | Escherichia coli | Strain CG1061 | 18–62 | [80] |
| B1 | Klebsiella spp. | Rabbit feces | 78 | [133] |
| B1 | Mycolicibacterium fluoranthenivorans (formerly Mycobacterium fluoranthenivorans) | DSM 44556 | >90 | [134] |
| B1 | Mycolicibacterium smegmatis (formerly Mycobacterium smegmatis) | N.A. | >90 | [82] |
| B1, G1, M1 | Myxococcus fulvus | ANSM068, Deer feces | 72, 68, 64 | [83] |
| B1 | Nocardia corynebacterioides (formerly Flavobacterium aurantiacum) | DSM 12,676 | 74.5 | [135] |
| NRRL B-184 | 85 | [136] | ||
| B1, B2M1 | Pseudomonas aeruginosa | Grain kernels and soils | 83, 47, 32 | [137] |
| B1 | Pseudomonas stutzeri | F4 strain, Budorcas taxicolor feces | 90 | [138] |
| B1 | Rhodococcus erythropolis | Strain 4.1491 | 96 | [139] |
| ATCC 4277 | [140] | |||
| 18 strains | 70–100 | [141] | ||
| DSM 14303 | 83 | [134] | ||
| B1 | Streptomyces aureofaciens | ATCC 10762 | 88 | [140] |
| B1 | Streptomyces lividans | TK 24 | 86 | [140] |
| B1 | Stenotrophomonas maltophilia | South American tapir feces | 85 | [133] |
| Toxin | Fungi | Strain | Toxin Elimination (%) | References |
|---|---|---|---|---|
| B1 | Aureobasidium pullulans | H1 | 68.61 | [142] |
| B1 | Candida albicans | AA17 | 50.34 | [142] |
| AA19 | 64.61 | |||
| B1 | Diutina catenulata (formerly Candida catenulata) | N.A. | <15 | [93] |
| B1 | Candida krusei | N.A. | 15–60 | [93] |
| AUMC 8161 | 100 | [143] | ||
| B1, B2, G1, G2 | Candida parapsilosis | N.A. | 15–72 | [93] |
| IP1698 | 99.94 | [144] | ||
| B1 | Wickerhamiella sorbophila (formerly Candida sorbophila) | ECF16 | 52.59 | [142] |
| ECF85 | 51.49 | |||
| B1 | Debaryomyces hansenii | N.A. | 15–39 | [93] |
| M1 | Kluyveromyces lactis | N.A. | 60–69 | [145] |
| B1 | Komagataella pastoris | EW1 | 71.5 | [142] |
| EW3 | 51.5 | |||
| EW6 | 50.5 | |||
| B1 | Wickerhamomyces anomalus (formerly Pichia anomala) | N.A. | 15 | [93] |
| AUMC 2674 | 100 | [143] | ||
| AFs | Meyerozyma guilliermondii (formerly Pichia guilliermondii) | AUMC 2663 | 80 | [143] |
| B1 | Pichia membranifaciens | N.A. | 40–59 | [93] |
| B1 | Rhodotorula mucilaginosa | various strains | 52.77–70.2 | [142] |
| B1 | Saccharomyces cerevisiae | N.A. | 10–60 | [93] |
| CECT 1891 | - * | [146] | ||
| A18 | 53 | [105] | ||
| 26.1.11 | 48.8 | [105] | ||
| RC 016 | - | [101] | ||
| EB34 | 52.25 | [142] | ||
| EB57 | 51.12 | |||
| M1 | Saccharomyces cerevisiae | SAFLAGER W37/70 | 90.3 | [147] |
| N.A. | 81.3 | [107] | ||
| ATCC 9763 | 75 | |||
| B1 | Saccharomycopsis fibuligera | N.A. | <15 | [93] |
| B1 | Saccharomycodes ludwigii | N.A. | 40–59 | |
| B1 | Schizosaccharomyces pombe | N.A. | 40–59 | |
| B1 | Cutaneotrichosporon mucoides (formerly Trichosporon mucoides) | N.A. | <15 | |
| B1 | Zygosaccharomyces bailii | N.A. | 15–39 |
References
- Frisvad, J.C.; Hubka, V.; Ezekiel, C.N.; Hong, S.B.; Nováková, A.; Chen, A.J.; Arzanlou, M.; Larsen, T.O.; Sklenář, F.; Mahakarnchanakul, W.; et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2019, 93, 1–63. [Google Scholar] [CrossRef]
- Lizárraga-Paulín, E.G.; Miranda-Castro, S.P.; Moreno-Martínez, E.; Torres-Pacheco, I.; Lara-Sagahón, A.V. Novel methods for preventing and controlling aflatoxins in food: A worldwide daily challenge. In Aflatoxins-Recent Advances and Future Prospects; Razzaghi-Abyaneh, M., Ed.; InTech: Rijeka, Croatia, 2013; pp. 93–128. [Google Scholar]
- Caceres, I.; Al Khoury, A.; El Khoury, R.; Lorber, S.; Oswald, I.P.; El Khoury, A.; Atoui, A.; Puel, O.; Bailly, J.-D. Aflatoxin biosynthesis and genetic regulation: A Review. Toxins 2020, 12, 150. [Google Scholar] [CrossRef] [Green Version]
- Pfliegler, V.; Pócsi, I.; Győri, Z.; Pusztahelyi, T. The Aspergilli and their mycotoxins: Metabolic interactions with plants and the soil biota. Front. Microbiol. 2020, 10, 1–45. [Google Scholar] [CrossRef] [Green Version]
- Peles, F.; Sipos, P.; Győri, Z.; Pfliegler, W.P.; Giacometti, F.; Serraino, A.; Pagliuca, G.; Gazzotti, T.; Pócsi, I. Adverse effects, transformation, and channeling of aflatoxins into food raw materials in livestock. Front. Microbiol. 2019, 10, 2861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ráduly, Z.; Szabó, L.; Madar, A.; Pócsi, I.; Csernoch, L. Toxicological and medical aspects of Aspergillus-derived mycotoxins entering the feed and food chain. Front. Microbiol. 2020, 10, 2908. [Google Scholar] [CrossRef] [Green Version]
- Jalili, M. A review on aflatoxins reduction in food. Iran. J. Health Saf. Environ. 2015, 3, 445–459. [Google Scholar]
- Torres, A.M.; Barros, G.G.; Palacios, S.A.; Chulze, S.N.; Battilani, P. Review on pre- and post-harvest management of peanuts to minimize aflatoxin contamination. Food Res. Int. 2014, 62, 11–19. [Google Scholar] [CrossRef]
- Norlia, M.; Jinap, S.; Nor-Khaizura, M.; Radu, S.; Samsudin, N.; Azri, F.A. Aspergillus section Flavi and aflatoxins: Occurrence, detection, and identification in raw peanuts and peanut-based products along the supply chain. Front. Microbiol. 2019, 10, 2602. [Google Scholar] [CrossRef] [Green Version]
- Ketney, O.; Santini, A.; Oancea, S. Recent aflatoxin survey data in milk and milk products: A review. Int. J. Dairy Technol. 2017, 70, 320–331. [Google Scholar] [CrossRef]
- Serraino, A.; Bonilauri, P.; Kerekes, K.; Farkas, Z.; Giacometti, F.; Canever, A.; Zambrini, A.V.; Ambrus, Á. Occurrence of aflatoxin M1 in raw milk marketed in Italy: Exposure assessment and risk characterization. Front. Microbiol. 2019, 10, 2516. [Google Scholar] [CrossRef] [PubMed]
- Maleki, F.; Abdi, S.; Davodian, E.; Haghani, K.; Bakhtiyari, S. Exposure of infants to aflatoxin M1 from mother’s breast milk in Ilam, Western Iran. Osong Public Health Res. Perspec. 2015, 6, 283–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warth, B.; Braun, D.; Ezekiel, C.N.; Turner, P.C.; Degen, G.H.; Marko, D. Biomonitoring of mycotoxins in human breast milk: Current state and future perspectives. Chem. Res. Toxicol. 2016, 29, 1087–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uka, V.; Moore, G.G.; Arroyo-Manzanares, N.; Nebija, D.; De Saeger, S.; Di Mavungu, D.J. Secondary metabolite dereplication and phylogenetic analysis identify various emerging mycotoxins and reveal the high intra-species diversity in Aspergillus flavus. Front. Microbiol. 2019, 10, 667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, W.J.; Schilter, B.; Tritsher, A.M.; Stadler, R.H. Environmental contaminants. Contaminants of milk and dairy products. In Encyclopedia of Dairy Science, 2nd ed.; Fuquay, J.W., Fox, P.F., McSweeney, P.L.H., Eds.; Academic Press: London, UK, 2011; pp. 898–905. [Google Scholar]
- Costamagna, D.; Gaggiotti, M.; Chiericatti, C.A.; Costabel, L.; Audero, G.M.L.; Taverna, M.; Signorini, M.L. Quantification of aflatoxin M1 carry-over rate from feed to soft cheese. Toxicol. Rep. 2019, 6, 782–787. [Google Scholar] [CrossRef]
- Jasutiene, I.; Garmiene, G.; Kulikauskiene, M. Pasteurisation and fermentation effects on Aflatoxin M1 stability. Milchwissenschaft 2006, 61, 75–79. [Google Scholar]
- Raters, M.; Matissek, R. Thermal stability of aflatoxin B1 and ochratoxin A. Mycotoxin Res. 2008, 24, 130–134. [Google Scholar] [CrossRef]
- Assaf, J.C.; Nahle, S.; Chokr, A.; Louka, N.; Atoui, A.; El Khoury, A. Assorted methods for decontamination of aflatoxin M1 in milk using microbial adsorbents. Toxins 2019, 11, 304. [Google Scholar] [CrossRef] [Green Version]
- Ahlberg, S.H.; Joutsjoki, V.; Korhonen, H.J. Potential of lactic acid bacteria in aflatoxin risk mitigation. Int. J. Food Microbiol. 2015, 207, 87–102. [Google Scholar] [CrossRef]
- Tian, F.; Chun, H.S. Natural products for preventing and controlling aflatoxin contamination of food. In Aflatoxin-Control, Analysis, Detection and Health Risks; Abdulra’uf, L., Ed.; IntechOpen: London, UK, 2017; pp. 13–44. [Google Scholar]
- Medeiros, F.H.V.; de Martins, S.J.; Zucchi, T.D.; Melo, I.S.; de Batista, L.R.; Machado, J.D.C. Biological control of mycotoxin-producing molds. Ciênc. Agrotec. 2012, 36, 483–497. [Google Scholar] [CrossRef]
- Tsitsigiannis, D.I.; Dimakopoulou, M.; Antoniou, P.P.; Tjamos, E.C. Biological control strategies of mycotoxigenic fungi and associated mycotoxins in Mediterranean basin crops. Phytopathol. Mediterr. 2012, 51, 158–174. [Google Scholar]
- Lagogianni, C.S.; Tsitsigiannis, D.I. Effective biopesticides and biostimulants to reduce aflatoxins in maize fields. Front. Microbiol. 2019, 10, 2645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, V.G.; Hagemeister, H. 1978 Untersuchungenueber den verblieb von aflatoxin B1 im Verdaaundtarkt von Kuehen. In Biological Detoxification of Fungal Toxin and its use in Plant Breeding, Feed and food production. Nat. Toxins 1999, 7, 1–23. [Google Scholar]
- Upadhaya, S.D.; Sung, H.G.; Lee, C.H.; Lee, S.Y.; Kim, S.W.; Jo, K.J.; Ha, J.K. Comparative study on the aflatoxin B1 degradation ability of rumen fluid from Holstein steers and Korean native goats. J. Vet. Sci. 2009, 10, 29–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jouany, J.-P.; Yiannikouris, A.; Bertin, G. Risk assessment of mycotoxins in ruminants and ruminant products. Opt. Mediterr. A 2009, 85, 205–224. [Google Scholar]
- Khodabandehloo, M.; Malecky, M.; Aliarabi, H.; Saki, A.A.; Alipour, D. In Vitro evaluation of aflatoxin B1 effect on gas production and ruminal fermentation parameters. Iran. J. Vet. Res. 2019, 20, 263–269. [Google Scholar]
- Singh, R.; Park, S.; Koo, J.; Balasubramanian, B. Influence of various concentrations of aflatoxin B1 on in vitro rumen fermentation of a buffalo diet. Korean J. Agric. Sci. 2020, 47, 131–138. [Google Scholar]
- Pantaya, D.; Morgavi, D.P.; Silberberg, M.; Chaucheyras-Durand, F.; Martin, C.; Wiryawan, K.G.; Boudra, H. Bioavailability of aflatoxin B1 and ochratoxin A, but not fumonisin B1 or deoxynivalenol, is increased in starch-induced low ruminal pH in nonlactating dairy cows. J. Dairy Sci. 2016, 99, 9759–9767. [Google Scholar] [CrossRef] [Green Version]
- Kiessling, K.H.; Pettersson, H.; Sandholm, K.; Olsen, M. Metabolism of aflatoxin, ochratoxin, zearalenone, and three trichothecenes by intact rumen fluid, rumen protozoa, and rumen bacteria. Appl. Environ. Microbiol. 1984, 47, 1070–1073. [Google Scholar] [CrossRef] [Green Version]
- Westlake, K.; Mackie, R.I.; Dutton, M.F. In Vitro metabolism of mycotoxins by bacterial, protozoal, and ovine ruminal fluid preparations. Anim. Feed Sci. Technol. 1989, 25, 169–178. [Google Scholar] [CrossRef]
- Hruska, Z.; Rajasekaran, K.; Yao, H.; Kincaid, R.; Darlington, D.; Brown, R.L.; Bhatnagar, D.; Cleveland, T.E. Co-inoculation of aflatoxigenic and non-aflatoxigenic strains of Aspergillus flavus to study fungal invasion, colonization, and competition in maize kernels. Front. Microbiol. 2014, 5, 122. [Google Scholar] [CrossRef]
- Atehnkeng, J.; Ojiambo, P.S.; Cotty, P.J.; Bandyopadhyay, R. Field efficacy of a mixture of atoxigenic Aspergillus flavus Link:Fr vegetative compatibility groups in preventing aflatoxin contamination in maize (Zea mays L.). Biol. Control. 2014, 72, 62–70. [Google Scholar] [CrossRef]
- Bandyopadhyay, R.; Ortega-Beltran, A.; Akande, A.; Mutegi, C.; Atehnkeng, J.; Kaptoge, L.; Senghor, A.L.; Adhikari, B.N.; Cotty, P.J. Biological control of aflatoxins in Africa: Current status and potential challenges in the face of climate change. World Mycotoxin J. 2016, 9, 771–789. [Google Scholar] [CrossRef] [Green Version]
- Lewis, M.H.; Carbone, I.; Luis, J.M.; Payne, G.A.; Bowen, K.L.; Hagan, A.K.; Kemerait, R.; Heiniger, R.; Ojiambo, P.S. Biocontrol strains differentially shift the genetic structure of indigenous soil populations of Aspergillus flavus. Front. Microbiol. 2019, 10, 1738. [Google Scholar] [CrossRef] [Green Version]
- Chulze, S.N.; Palazzini, J.M.; Torres, A.M.; Barros, G.; Ponsone, M.L.; Geisen, R.; Schmidt-Heydt, M.; Köhl, J. Biological control as a strategy to reduce the impact of mycotoxins in peanuts, grapes and cereals in Argentina. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2015, 32, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Agbetiameh, D.; Ortega-Beltran, A.; Awuah, R.T.; Atehnkeng, J.; Islam, M.-S.; Callicott, K.A.; Cotty, P.J.; Bandyopadhyay, R. Potential of atoxigenic Aspergillus flavus vegetative compatibility groups associated with maize and groundnut in Ghana as biocontrol agents for aflatoxin management. Front. Microbiol. 2019, 10, 2069. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Mycotoxins in Australia: Biocontrol of aflatoxin in peanuts. Mycopathologia 2006, 162, 233–243. [Google Scholar] [CrossRef]
- Dorner, J.W.; Cole, R.J.; Wicklow, D.T. Aflatoxin reduction in corn through field application of competitive fungi. J. Food Prot. 1999, 62, 650–656. [Google Scholar] [CrossRef]
- Probst, C.; Bandyopadhyay, R.; Price, L.E.; Cotty, P.J. Identification of atoxigenic Aspergillus flavus isolates to reduce aflatoxin contamination of maize in Kenya. Plant. Dis. 2011, 95, 212–218. [Google Scholar] [CrossRef] [Green Version]
- Unnevehr, L.; Grace, D. (Eds.) Tackling Aflatoxins: An Overview of Challenges and Solutions in Aflatoxins. In Finding Solutions for Improved Food Safety; International Food Policy Research Institute: Washington, DC, USA, 2013. [Google Scholar]
- Mohale, S.; Medina, A.; Magan, N. Effect of environmental factors on in vitro and in situ interactions between atoxigenic and toxigenic A. flavus strains and control of aflatoxin contamination of maize. Biocontrol Sci. Technol. 2013, 23, 776–793. [Google Scholar] [CrossRef]
- Moore, G.G.; Lebar, M.D.; Carter-Wientjes, C.H. The role of extrolites secreted by non-aflatoxigenic Aspergillus flavus in biocontrol efficacy. J. Appl. Microbiol. 2018, 126, 1257–1264. [Google Scholar] [CrossRef]
- Abbas, H.K.; Zablotowicz, R.M.; Horn, B.W.; Phillips, N.A.; Johnson, B.J.; Jin, X.; Abel, C.A. Comparison of major biocontrol strains of non-aflatoxigenic Aspergillus flavus for the reduction of aflatoxins and cyclopiazonic acid in maize. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2011, 28, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Dorner, J.W.; Cole, R.J.; Connick, W.J.; Daigle, D.J.; McGuire, M.R.; Shasha, B.S. Evaluation of biological control formulations to reduce aflatoxin contamination in peanuts. Biol. Control 2003, 26, 318–324. [Google Scholar] [CrossRef]
- Accinelli, C.; Abbas, H.K.; Vicaria, A.; Shier, W.T. Evaluation of recycled bioplastic pellets and a sprayable formulation for application of an Aspergillus flavus biocontrol strain. Crop. Prot. 2015, 72, 9–15. [Google Scholar] [CrossRef]
- Weaver, M.A.; Abbas, H.K.; Jin, X.; Elliott, B. Efficacy of water-dispersible formulations of biological control strains of Aspergillus flavus for aflatoxin management in corn. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2016, 33, 346–351. [Google Scholar] [PubMed]
- Accinelli, C.; Abbas, H.K.; Vicaria, A.; Shier, W.T. Leaf application of a sprayable bioplastic-based formulation of biocontrol Aspergillus flavus strains for reduction of aflatoxins in corn. Pest Manag. Sci. 2016, 72, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Accinelli, C.; Abbas, H.K.; Little, N.; Kotowicz, J.K.; Shier, W.T. Biological control of aflatoxin production in corn using non-aflatoxigenic Aspergillus flavus administered as a bioplastic-based seed coating. Crop. Prot. 2018, 107, 87–92. [Google Scholar] [CrossRef]
- Senghor, L.A.; Ortega-Beltran, A.; Atehnkeng, J.; Callicott, K.A.; Cotty, P.J.; Bandyopadhyay, R. The atoxigenic biocontrol product Aflasafe SN01 is a valuable tool to mitigate aflatoxin contamination of both maize and groundnut cultivated in Senegal. Plant Dis. 2020, 104, 510–520. [Google Scholar] [CrossRef]
- Atehnkeng, J.; Ojiambo, P.S.; Donner, M.; Ikotun, T.; Sikora, R.A.; Cotty, P.J.; Bandyopadhyay, R. Distribution and toxigenicity of Aspergillus species isolated from maize kernels from three agro-ecological zones in Nigeria. Int. J. Food Microbiol. 2008, 122, 74–84. [Google Scholar] [CrossRef]
- Hell, K.; Mutegi, C. Aflatoxin control and prevention strategies in key crops of Sub-Saharan Africa. Afr. J. Microbiol. Res. 2011, 5, 459–466. [Google Scholar]
- Grace, D.; Mahuku, G.; Hoffmann, V.; Atherstone, C.; Upadhyaya, H.D.; Bandyopadhyay, R. International agricultural research to reduce food risks: Case studies on aflatoxins. Food Secur. 2015, 7, 569–582. [Google Scholar] [CrossRef] [Green Version]
- Udomkun, P.; Wiredu, A.N.; Nagle, M.; Müller, J.; Vanlauwe, B.; Bandyopadhyay, R. Innovative technologies to manage aflatoxins in foods and feeds and the profitability of application—A review. Food Control 2017, 76, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Shenge, K.C.; Adhikari, B.N.; Akande, A.; Callicott, K.A.; Atehnkeng, J.; Ortega-Beltran, A.; Kumar, P.L.; Bandyopadhyay, R.; Cotty, P.J. Monitoring Aspergillus flavus genotypes in a multi-genotype aflatoxin biocontrol product with quantitative pyrosequencing. Front. Microbiol. 2019, 10, 2529. [Google Scholar] [CrossRef] [PubMed]
- Mauro, A.; Garcia-Cela, E.; Pietri, A.; Cotty, P.J.; Battilani, P. Biological control products for aflatoxin prevention in Italy: Commercial field evaluation of atoxigenic Aspergillus flavus active ingredients. Toxins 2018, 10, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kifle, M.H.; Yobo, K.S.; Laing, M.D. Biocontrol of Aspergillus flavus in groundnut using Trichoderma harzianum strain kd. J. Plant Dis. Prot. 2017, 124, 51–56. [Google Scholar] [CrossRef]
- Sivparsad, B.J.; Laing, M.D. Pre-harvest silk treatment with Trichoderma harzianum reduces aflatoxin contamination in sweetcorn. J. Plant Dis. Prot. 2016, 123, 285–293. [Google Scholar] [CrossRef]
- Ren, X.; Zhang, Q.; Zhang, W.; Mao, J.; Li, P. Control of aflatoxigenic molds by antagonistic microorganisms: Inhibitory behaviors, bioactive compounds, related mechanisms, and influencing factors. Toxins 2020, 12, 24. [Google Scholar] [CrossRef] [Green Version]
- Magan, N.; Aldred, D. Post-harvest control strategies: Minimizing mycotoxins in the food chain. Int. J. Food Microbiol. 2007, 119, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adibpour, N.; Soleimanian-Zad, S.; Sarabi-Jamab, M.; Tajalli, F. Effect of storage time and concentration of aflatoxin M1 on toxin binding capacity of L. acidophilus in fermented milk product. J. Agric. Sci. Technol. 2016, 18, 1209–1220. [Google Scholar]
- Asurmendi, P.; Pascual, L.; Dalcero, A.; Barberis, L. Incidence of lactic acid bacteria and Aspergillus flavus in brewer’s grains and evaluation of potential antifungal activity of these bacteria. J. Stored Prod. Res. 2014, 56, 33–37. [Google Scholar] [CrossRef]
- Saladino, F.; Luz, C.; Manyes, L.; Fernandez-Franzon, M.; Meca, G. In Vitro antifungal activity of lactic acid bacteria against mycotoxigenic fungi and their application in loaf bread shelf-life improvement. Food Control 2016, 67, 273–277. [Google Scholar] [CrossRef]
- Assaf, J.C.; Khoury, A.; Chokr, A.; Louka, N.; Atoui, A. A novel method for elimination of aflatoxin M1 in milk using Lactobacillus rhamnosus G.G. biofilm. Int. J. Dairy Technol. 2019, 72, 248–256. [Google Scholar] [CrossRef]
- Gu, H.; Ren, D. Materials, and surface engineering to control bacterial adhesion and biofilm formation: A review of recent advances. Front. Chem. Sci. Eng. 2014, 8, 20–33. [Google Scholar] [CrossRef]
- Wacoo, A.P.; Atukunda, P.; Muhoozi, G.; Braster, M.; Wagner, M.; Broek, T.J.V.D.; Sybesma, W.; Westerberg, A.C.; Iversen, P.O.; Kort, R. Aflatoxins: Occurrence, exposure, and binding to Lactobacillus species from the gut microbiota of rural Ugandan children. Microorganisms 2020, 8, 347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadiq, F.A.; Yan, B.; Tian, F.; Zhao, J.; Zhang, H.; Wei, C. Lactic acid bacteria as antifungal and anti-mycotoxigenic agents: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1403–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalié, D.; Deschamps, A.; Richard-Forget, F. Lactic acid bacteria–Potential for control of mold growth and mycotoxins: A review. Food Control 2010, 21, 370–380. [Google Scholar] [CrossRef]
- Crowley, S.; Mahony, J.; van Sinderen, D. Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Trends Food Sci. Technol. 2013, 33, 93–109. [Google Scholar] [CrossRef]
- Le Lay, C.; Coton, E.; Le Blay, G.; Chobert, J.M.; Haertlé, T.; Choiset, Y.; Longa, N.G.V.; Meslet-Cladièrea, L.; Mounie, J. Identification and quantification of antifungal compounds produced by lactic acid bacteria and propionibacteria. Int. J. Food Microbiol. 2016, 239, 79–85. [Google Scholar] [CrossRef]
- Leyva Salas, M.; Mounier, J.; Valence, F.; Coton, M.; Thierry, A.; Coton, E. Antifungal microbial agents for food biopreservation—A review. Microorganisms 2017, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Topcu, A.; Bulat, T.; Wishah, R.; Boyaci, I.H. Detoxification of aflatoxin B1 and patulin by Enterococcus faecium strains. Int. J. Food Microbiol. 2010, 139, 202–205. [Google Scholar] [CrossRef]
- Farzaneh, M.; Shi, Z.Q.; Ghassempour, A.; Sedaghat, N.; Ahmadzadeh, M.; Mirabolfathy, M.; Javan-Nikkhah, M. Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control 2012, 23, 100–106. [Google Scholar] [CrossRef]
- Fan, Y.; Zhao, L.; Ma, Q.; Li, X.; Shi, H.; Zhou, T.; Zhang, J.; Ji, C. Effects of Bacillus subtilis ANSB060 on growth performance, meat quality and aflatoxin residues in broilers fed moldy peanut meal naturally contaminated with aflatoxins. Food Chem. Toxicol. 2013, 59, 748–753. [Google Scholar] [CrossRef]
- Razzaghi-Abyaneh, M.; Shams-Ghahfarokhi, M.; Chang, P.K. Aflatoxins: Mechanisms of inhibition by antagonistic plants and microorganisms. In Aflatoxins: Biochemistry and Molecular Biology; Guevara-Gonzalez, R.G., Ed.; IntechOpen: London, UK, 2011; pp. 285–304. [Google Scholar]
- Chandra, H.; Kumari, P.; Bisht, R.; Prasad, R.; Yadav, S. Plant growth promoting Pseudomonas aeruginosa from Valeriana wallichii displays antagonistic potential against three phytopathogenic fungi. Mol. Biol. Rep. 2020, 47, 6015–6026. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.H.; Zhang, N.; Sun, R.R.; Gao, X.; Gu, C.; Krumm, C.S.; Qi, D.S. A novel strain of Cellulosimicrobium funkei can biologically detoxify aflatoxin B1 in ducklings. Microb. Biotechnol. 2015, 8, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Shams-Ghahfarokhi, M.; Kalantari, S.; Razzaghi-Abyaneh, M. Terrestrial bacteria from agricultural soils: Versatile weapons against aflatoxigenic fungi. In Aflatoxins–Recent Advances and Future Prospects; Razzaghi-Abyaneh, M., Ed.; InTech: Rijeka, Croatia, 2013; pp. 23–39. [Google Scholar]
- Wang, L.; Wu, J.; Liu, Z.; Shi, Y.; Liu, J.; Xu, X.; Hao, S.; Mu, P.; Deng, F.; Deng, Y. Aflatoxin B1 degradation and detoxification by Escherichia coli CG1061 isolated from chicken cecum. Front. Pharmacol. 2019, 9, 1548. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.C.; Jackson, C.J.; Tattersall, D.B.; French, N.; Peat, T.S.; Newman, J. Identification, and characterization of two families of F420H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 2010, 78, 561–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.H.; Li, W.Y.; Hsu, I.N.; Liao, Y.Y.; Yang, C.Y.; Taylor, M.C.; Liu, Y.F.; Huang, W.H.; Chang, H.H.; Huang, H.L.; et al. Recombinant aflatoxin-degrading F420H2-dependent reductase from Mycobacterium smegmatis protects mammalian cells from aflatoxin toxicity. Toxins 2019, 11, 259. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.H.; Guan, S.; Gao, X.; Ma, Q.G.; Lei, Y.P.; Bai, X.M. Preparation, purification, and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 2011, 110, 147–155. [Google Scholar] [CrossRef]
- Gong, A.-D.; Wu, N.-N.; Kong, X.-W.; Zhang, Y.-M.; Hu, M.-J.; Gong, S.-J.; Dong, F.-Y.; Wang, J.-H.; Zhao, Z.-Y.; Liao, Y.-C. Inhibitory effect of volatiles emitted from Alcaligenes faecalis N1-4 on Aspergillus flavus and aflatoxins in storage. Front. Microbiol. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Gong, A.-D.; Dong, F.-Y.; Hu, M.-J.; Kong, X.-W.; Wei, F.-F.; Gong, S.-J.; Zhang, Y.-M.; Zhang, J.-B.; Wu, A.-B. Antifungal activity of volatile emitted from Enterobacter asburiae Vt-7 against Aspergillus flavus and aflatoxins in peanuts during storage. Food Control 2019, 106, 106718. [Google Scholar] [CrossRef]
- Lyu, A.; Yang, L.; Wu, M.; Zhang, J.; Li, G. High efficacy of the volatile organic compounds of Streptomyces yanglinensis 3-10 in suppression of Aspergillus contamination on peanut kernels. Front. Microbiol. 2020, 11, 142. [Google Scholar] [CrossRef]
- Zhang, H.; Kim, M.S.; Krishnamachari, V.; Payton, P.; Sun, Y.; Grimson, M.; Farag, M.A.; Ryu, C.-M.; Allen, R.; Melo, I.S.; et al. Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arab. Planta 2007, 226, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Kanchiswamy, C.N.; Malnoy, M.; Maffei, M.E. Bioprospecting bacterial and fungal volatiles for sustainable agriculture. Trends Plant Sci. 2015, 20, 206–211. [Google Scholar] [CrossRef]
- Fincheira, P.; Quiroz, A. Microbial volatiles as plant growth inducers. Microbiol. Res. 2018, 208, 63–75. [Google Scholar] [CrossRef]
- Intanoo, M.; Kongkeitkajorn, M.B.; Suriyasathaporn, W.; Phasuk, Y.; Bernard, J.K.; Pattarajinda, V. Effect of supplemental Kluyveromyces marxianus and Pichia kudriavzevii on aflatoxin M1 excretion in milk of lactating dairy cows. Animals 2020, 10, 709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jouany, J.-P.; Morgavi, D. Use of “natural” products as alternative to antibiotic feed additives in ruminant production. Animal 2007, 1, 1443–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaucheyras-Durand, F.; Walker, N.; Bach, A. Effects of active dry yeasts on the rumen microbial ecosystem: Past, present, and future. Anim. Feed Sci. Tech. 2008, 145, 5–26. [Google Scholar] [CrossRef]
- Shetty, P.H.; Jespersen, L. Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontaminating agents. Trends Food Sci. Technol. 2006, 17, 48–55. [Google Scholar] [CrossRef]
- Xiong, J.L.; Wang, Y.M.; Nennich, T.D.; Li, Y.; Liu, J.X. Transfer of dietary aflatoxin B1, to milk aflatoxin M1, and effect of inclusion of adsorbent in the diet of dairy cows. J. Dairy Sci. 2015, 98, 2545–2554. [Google Scholar] [CrossRef]
- Diaz, D.E.; Hagler, W.M.; Blackwelder, J.T.; Eve, J.A.; Hopkins, B.A.; Anderson, K.L.; Jones, F.T.; Whitlow, L.W. Aflatoxin binders II: Reduction of aflatoxin M1 in milk by sequestering agents of cows consuming aflatoxin in feed. Mycopathologia 2004, 157, 233–241. [Google Scholar] [CrossRef]
- Queiroz, O.C.M.; Han, J.H.; Staples, C.R.; Adesogan, A.T. Effect of adding a mycotoxin-sequestering agent on milk aflatoxin M1 concentration and the performance and immune response of dairy cattle fed an aflatoxin B1-contaminated diet. J. Dairy Sci. 2012, 95, 5901–5908. [Google Scholar] [CrossRef]
- Anjos, F.R.D.; Ledoux, D.R.; Rottinghaus, G.E.; Chimonyo, M. Efficacy of Mozambican bentonite and diatomaceous earth in reducing the toxic effects of aflatoxins in chicks. World Mycotoxin J. 2016, 9, 63–72. [Google Scholar] [CrossRef]
- Hua, S.S.T.; Beck, J.J.; Sarreal, S.B.L.; Gee, W. A major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthesis genes of Aspergillus flavus. Mycotoxin Res. 2014, 30, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Çelýk, K.; Denlý, M.; Savas, T. Reduction of toxic effects of aflatoxin B1 by using baker yeast (Saccharomyces cerevisiae) in growing broiler chicks’ diets. Revista brasileira de zootecnia 2003, 32, 615–619. [Google Scholar] [CrossRef] [Green Version]
- Santin, E.; Paulillo, A.C.; Maiorka, A.; Nakaghi, L.S.O.; Macari, M.; Silva, A.V.F.; Alessi, A.C. Evaluation of the efficacy of Saccharomyces cerevisiae cell wall to ameliorate the toxic effects of aflatoxin in broilers. Int. J. Poult. Sci. 2003, 2, 341–344. [Google Scholar]
- Armando, M.R.; Dogi, C.A.; Pizzolitto, R.P.; Escobar, F.; Peirano, M.S.; Salvano, M.A.; Sabini, L.I.; Combina, M.; Dalcero, A.M.; Cavaglieri, L.R. Saccharomyces cerevisiae strains from animal environment with in vitro aflatoxin B1 binding ability and anti-pathogenic bacterial influence. World Mycotoxin J. 2011, 4, 59–68. [Google Scholar] [CrossRef]
- Dogi, C.A.; Armando, R.; Luduena, R.; de Moreno de LeBlanc, A.; Rosa, C.A.; Dalcero, A.; Cavaglieri, L. Saccharomyces cerevisiae strains retain their viability and aflatoxin B1 binding ability under gastrointestinal conditions and improve ruminal fermentation. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2011, 28, 1705–1711. [Google Scholar]
- Pizzolitto, R.P.; Armando, M.R.; Combina, M.; Cavaglieri, L.R.; Dalcero, A.M.; Salvano, M.A. Evaluation of Saccharomyces cerevisiae strains as probiotic agent with aflatoxin B1 adsorption ability for use in poultry feedstuffs. J. Environ. Sci. Health B 2012, 47, 933–941. [Google Scholar] [CrossRef]
- Kim, S.; Lee, H.; Lee, S.; Lee, J.; Ha, J.; Choi, Y.; Yoon, Y.; Choi, K.-Y. Microbe-mediated aflatoxin decontamination of dairy products and feeds. J. Dairy Sci. 2017, 100, 871–880. [Google Scholar] [CrossRef]
- Shetty, P.H.; Hald, B.; Jespersen, L. Surface binding of aflatoxin B1 by Saccharomyces cerevisiae strains with potential decontaminating abilities in indigenous fermented foods. Int. J. Food Microbiol. 2007, 113, 41–46. [Google Scholar] [CrossRef]
- Inoue, T.; Nagatomi, Y.; Uyama, A.; Naoki, M. Degradation of aflatoxin B1 during the fermentation of alcoholic beverages. Toxins 2013, 5, 1219–1229. [Google Scholar] [CrossRef]
- Foroughi, M.; Sarabi Jamab, M.; Keramat, J.; Foroughi, M. Immobilization of Saccharomyces cerevisiae on perlite beads for the decontamination of aflatoxin M1 in milk. J. Food Sci. 2018, 83, 2008–2013. [Google Scholar] [CrossRef] [PubMed]
- Saki, A.; Rahmani, A.; Mahmoudi, H.; Tabatabaei, M.M.; Zamani, P.; Khosravi, A.R. The ameliorative effect of Mycosorb in aflatoxin contaminated diet of broiler chickens. J. Livest. Sci. Technol. 2018, 6, 39–47. [Google Scholar]
- Haidukowski, M.; Casamassima, E.; Cimmarusti, M.T.; Branà, M.T.; Longobardi, F.; Acquafredda, P.; Logrieco, A.; Altomare, C. Aflatoxin B1-adsorbing capability of Pleurotus eryngii mycelium: Efficiency and modeling of the process. Front. Microbiol. 2019, 10, 1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branà, M.T.; Sergio, L.; Haidukowski, M.; Logrieco, A.F.; Altomare, C. Degradation of aflatoxin B1 by a sustainable enzymatic extract from spent mushroom substrate of Pleurotus eryngii. Toxins 2020, 12, 49. [Google Scholar] [CrossRef] [Green Version]
- Motomura, M.; Toyomasu, T.; Mizuno, K.; Shinozawa, T. Purification and characterization of an aflatoxin degradation enzyme from Pleurotus ostreatus. Microbiol. Res. 2003, 158, 237–242. [Google Scholar] [CrossRef]
- Wang, J.; Ogata, M.; Hirai, H.; Kawagishi, H. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 2011, 314, 164–169. [Google Scholar] [CrossRef]
- Wu, Y.Z.; Lu, F.P.; Jiang, H.L.; Tan, C.P.; Yao, D.S.; Xie, C.F. The furofuran-ring selectivity, hydrogen peroxide-production and low Km value are the three elements for highly effective detoxification of aflatoxin oxidase. Food Chem. Toxicol. 2015, 76, 125–131. [Google Scholar] [CrossRef]
- Delgado, J.; Owens, R.A.; Doyle, S.; Asensio, M.A.; Núñez, F. Impact of the antifungal protein PgAFP from Penicillium chrysogenum on the protein profile in Aspergillus flavus. Appl. Microbiol. Biotechnol. 2015, 99, 8701–8715. [Google Scholar] [CrossRef] [Green Version]
- Delgado, J.; Owens, R.A.; Doyle, S.; Núñez, F.; Asensio, M.A. Quantitative proteomics reveals new insights into calcium-mediated resistance mechanisms in Aspergillus flavus against the antifungal protein PgAFP in cheese. Food Microbiol. 2017, 66, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Leiter, É.; Gáll, T.; Csernoch, L.; Pócsi, I. Biofungicide utilizations of antifungal proteins of filamentous ascomycetes: Current and foreseeable developments. BioControl 2017, 62, 125–138. [Google Scholar] [CrossRef]
- Ahlberg, S.; Randolph, D.; Okoth, S.; Lindahl, J. Aflatoxin binders in foods for human consumption-can this be promoted safely and ethically? Toxins 2019, 11, 410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peltonen, K.; EI-Nezami, H.; Haskard, C.; Ahokas, J.; Salminen, S. Aflatoxin B1 binding by dairy strains of lactic acid bacteria and bifidobacteria. J. Dairy Sci. 2001, 84, 2152–2156. [Google Scholar] [CrossRef]
- Haskard, C.A.; El-Nezami, H.S.; Kankaanpaa, P.E.; Salminen, S.; Ahokas, J.T. Surface binding of aflatoxin B1 by lactic acid bacteria. Appl. Environ. Microbiol. 2001, 67, 3086–3091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azab, R.M.; Tawakkol, W.M.; Hamad, A.R.M.; Abou-Elmagd, M.K.; El-Agrab, H.M.; Refai, M.K. Detection and estimation of aflatoxin B1 in feeds and its biodegradation by bacteria and fungi. Egypt. J. Nat. Toxins 2005, 2, 39–54. [Google Scholar]
- Marrez, D.A.; Shahy, E.M.; El-Sayed, H.S.; Sultan, Y.Y. Detoxification of aflatoxin B1 in milk using lactic acid bacteria. J. Biol. Sci. 2018, 18, 144–151. [Google Scholar] [CrossRef] [Green Version]
- Liew, W.P.P.; Nurul-Adilah, Z.; Than, L.T.L.; Mohd-Redzwan, S. The binding efficiency and interaction of Lactobacillus casei Shirota toward aflatoxin B1. Front. Microbiol. 2018, 9, 1503. [Google Scholar] [CrossRef] [Green Version]
- Fazeli, M.R.; Hajimohammadali, M.; Moshkani, A.; Samadi, N.; Jamalifar, H.; Khoshayand, M.R. Aflatoxin B1 binding capacity of autochthonous strains of lactic acid bacteria. J. Food Protect. 2009, 72, 189–192. [Google Scholar] [CrossRef]
- Ghazvini, R.D.; Kouhsari, E.; Zibafar, E.; Hashemi, S.J.; Amini, A.; Niknejad, F. Antifungal activity and aflatoxin degradation of Bifidobacterium bifidum and Lactobacillus fermentum against toxigenic Aspergillus parasiticus. Open Microbiol. J. 2016, 10, 1–5. [Google Scholar] [CrossRef]
- EI-Nezarni, H.; Mykkiinen, H.; Kankaanpi, H.; Salminen, S.; Ahokas, J. Ability of Lactobacillus and Propionibacterium strains to remove aflatoxin B1 from the chicken duodenum. J. Food Protect. 2000, 63, 549–552. [Google Scholar]
- Shahin, A.A.M. Removal of aflatoxin B1 from contaminated liquid media by dairy lactic acid bacteria. Int. J. Agric. Biol. 2007, 9, 71–75. [Google Scholar]
- Kabak, B.; Var, I. Factors affecting the removal of aflatoxin M1 from food model by Lactobacillus and Bifidobacterium strains. J. Environ. Sci. Health B 2008, 43, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Bovo, F.; Corassin, C.H.; Rosim, R.E.; Oliveira, C.A.F. Efficiency of lactic acid bacteria strains for decontamination of aflatoxin M1 in phosphate buffer saline solution and in skimmed milk. Food Bioproc. Technol. 2013, 6, 2230–2234. [Google Scholar] [CrossRef]
- Sarimehmetoglu, B.; Küplülü, Ö. Binding ability of aflatoxin M1 to yoghurt bacteria. Vet. Fak. Derg. 2004, 51, 195–198. [Google Scholar]
- Petchkongkaew, A.; Taillandier, P.; Gasaluck, P.; Lebrihi, A. Isolation of Bacillus spp. from Thai fermented soybean (Thuanao): Screening for aflatoxin B1 and ochratoxin A detoxification. J. Appl. Microbiol. 2008, 104, 1495–1502. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.E.; Harran, G. Microbial degradation of mycotoxins. Intern. Biodeter. Biodeg. 1993, 32, 205–211. [Google Scholar] [CrossRef]
- Gao, X.; Ma, Q.; Zhao, L.; Lei, Y.; Shan, Y.; Cheng, J. Isolation of Bacillus subtilis: Screening for aflatoxins B1, M1, and G1 detoxication. Eur. Food Res. Technol. 2011, 232, 957–962. [Google Scholar] [CrossRef]
- Guan, S.; Ji, C.; Zhou, T.; Li, J.; Ma, Q.; Niu, T. Aflatoxin B1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 2008, 9, 1489–1503. [Google Scholar] [CrossRef] [Green Version]
- Teniola, O.D.; Addo, P.A.; Brost, I.M.; Farber, P.; Jany, K.D.; Alberts, J.F.; van Zyl, W.H.; Steyn, P.S.; Holzapfel, W.H. Degradation of aflatoxin B1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp. nov. DSM44556(T). Int. J. Food Microbiol. 2005, 105, 111–117. [Google Scholar] [CrossRef]
- Smiley, R.D.; Draughon, F.A. Preliminary evidence that degradation of aflatoxin B1 by Flavobacterium aurantiacum is enzymatic. J. Food Protect. 2000, 63, 415–418. [Google Scholar] [CrossRef]
- D’Souza, D.H.; Brackett, R.E. Aflatoxin B1 degradation by Flavobacterium aurantiacum in the presence of reducing conditions and seryl and sulfhydryl group inhibitors. J. Food Protect. 2001, 64, 268–271. [Google Scholar] [CrossRef]
- Sangare, L.; Zhao, Y.; Folly, Y.M.; Chang, J.; Li, J.; Selvaraj, J.N.; Xing, F.; Zhou, L.; Wang, Y.; Liu, Y. Aflatoxin B1 degradation by a Pseudomonas strain. Toxins 2014, 6, 3028–3040. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, W.; Yang, W.; Li, H.; Liu, X.; Cao, Y. Isolation and characterisation of an aflatoxin B1-degrading bacterium. Wei Sheng Wu Xue Bao 2012, 52, 1129–1136. [Google Scholar] [PubMed]
- Kong, Q.; Zhai, C.; Guan, B.; Li, C.; Shan, S.; Yu, J. Mathematic modeling for optimum conditions on aflatoxin B1 degradation by the aerobic bacterium Rhodococcus erythropolis. Toxins 2012, 4, 1181–1195. [Google Scholar] [CrossRef] [Green Version]
- Eshelli, M.; Harvey, L.; Edrada-Ebel, R.; McNeil, B. Metabolomics of the bio-degradation process of aflatoxin B1 by actinomycetes at an initial pH of 6.0. Toxins 2015, 7, 439–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cserhati, M.; Kriszt, B.; Krifaton, C.; Szoboszlay, S.; Hahn, J.; Toth, S.; Nagy, I.; Kukolya, J. Mycotoxin-degradation profile of Rhodococcus strains. Int. J. Food Microbiol. 2013, 166, 176–185. [Google Scholar] [CrossRef]
- García-Béjar, B.; Arévalo-Villena, M.; Guisantes-Batan, E.; Rodríguez-Flores, J.; Briones, A. Study of the bioremediatory capacity of wild yeasts. Sci. Rep. 2020, 10, 11265. [Google Scholar] [CrossRef]
- Zohri, A.A.; Abdel-Kareem, M.M. Four strains of yeasts: As effective biocontrol agents against both growth and mycotoxins formation by selected 11 toxigenic fungi. Glob. Adv. Res. J. Microbiol. 2018, 7, 132–135. [Google Scholar]
- Niknejad, F.; Zaini, F.; Faramarzi, M.; Amini, M.; Kordbacheh, P.; Mahmoudi, M.; Safara, M. Candida parapsilosis as a potent biocontrol agent against growth and aflatoxin production by Aspergillus species. Iran. J. Public Health 2012, 41, 72–80. [Google Scholar]
- Ghanbari, R.; Rezaie, S.; Noorbakhsh, F.; Khaniki, G.J.; Soleimani, M.; Aghaee, E.M. Biocontrol effect of Kluyveromyces lactis on aflatoxin expression and production in Aspergillus parasiticus. FEMS Microbiol. Lett. 2019, 366, fnz114. [Google Scholar] [CrossRef]
- Bueno, D.; Casale, C.H.; Pizzolitto, R.P.; Salano, M.A.; Olivier, G. Physical adsorption of aflatoxin B1 by lactic acid bacteria and Saccharomyces cerevisiae: A theoretical model. J. Food Protect. 2007, 70, 2148–2154. [Google Scholar] [CrossRef]
- Corassin, C.H.; Bovo, F.; Rosim, R.E.; Oliveira, C.A.F. Efficiency of Saccharomyces cerevisiae and lactic acid bacteria strains to bind aflatoxin M1 in UHT skim milk. Food Control 2013, 31, 80–83. [Google Scholar] [CrossRef] [Green Version]
| Date | Country | Crop | Non-Aflatoxigenic Strains | Target | Success Rate (%) | References |
|---|---|---|---|---|---|---|
| 1994–1997 | USA | Maize, peanuts, and cotton | A. flavus strains (NRRL 21882 and NRRL 21368), A. parasiticus (NRRL 21369) | AFB1, AFB2, AFG1, AFG2 * | 66–96 | [40] |
| 1996–1997 | USA | Peanuts | atoxigenic A. flavus (NRRL21368) and A. parasiticus (NRRL 21369), | AFB1, AFB2, AFG1, AFG2 | 86–92 | [46] |
| 2004–2006 | Kenya | Maize | 12 atoxigenic A. flavus isolates | AFB1 | 64–90 | [41] |
| 2007–2008 | Nigeria | Maize (ACCR-9931-SR) | mixture of four atoxigenic strains of A. flavus | AFB1 and AFB2 by A. flavus L- and SBG-morphotypes, A. parasiticus and A. tamarii | 67–95 | [34] |
| 2007–2009 | USA | Maize (Pioneer 32R25 hybrid) | Afla-Guard, A. flavus NRRL 21882. AF36, A. flavus NRRL 18543. A. flavus K42 | AFB1, AFB2, AFG1, AFG2 by toxigenic A. flavus F3W4 (NRRL 30796), K54 (NRRL 58987), NRRL 58976, NRRL 58988, and NRRL 58974 | 83–98 | [40] |
| 2010–2014 | Senegal | Groundnut and maize | Aflasafe SN01, mixture of 4 atoxigenic isolates of A. flavus | AFB1, AFB2, AFG1, AFG2 by A. aflatoxiformans, A. flavus L-morphotype, A. parasiticus, A. tamarii | 58–100 | [51] |
| 2012–2013 | USA | Maize | Afla-Guard, A. flavus NRRL 21882. AF36, A. flavus NRRL 18543 | AFB1 by A. flavus, A. parasiticus, A. caelatus, A. nomius, and A. tamarii | 0–97 | [36] |
| 2012–2013 | Italy | Maize | AF-X1™, A. flavus A2085 and A2321 | AFB1 | 84–95 | [57] |
| 2014 | Ghana | Maize and groundnut | 13 atoxigenic A. flavus isolates | AFB1 | 87–98 | [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peles, F.; Sipos, P.; Kovács, S.; Győri, Z.; Pócsi, I.; Pusztahelyi, T. Biological Control and Mitigation of Aflatoxin Contamination in Commodities. Toxins 2021, 13, 104. https://doi.org/10.3390/toxins13020104
Peles F, Sipos P, Kovács S, Győri Z, Pócsi I, Pusztahelyi T. Biological Control and Mitigation of Aflatoxin Contamination in Commodities. Toxins. 2021; 13(2):104. https://doi.org/10.3390/toxins13020104
Chicago/Turabian StylePeles, Ferenc, Péter Sipos, Szilvia Kovács, Zoltán Győri, István Pócsi, and Tünde Pusztahelyi. 2021. "Biological Control and Mitigation of Aflatoxin Contamination in Commodities" Toxins 13, no. 2: 104. https://doi.org/10.3390/toxins13020104
APA StylePeles, F., Sipos, P., Kovács, S., Győri, Z., Pócsi, I., & Pusztahelyi, T. (2021). Biological Control and Mitigation of Aflatoxin Contamination in Commodities. Toxins, 13(2), 104. https://doi.org/10.3390/toxins13020104









