Antivenom Neutralization of Coagulopathic Snake Venom Toxins Assessed by Bioactivity Profiling Using Nanofractionation Analytics
Abstract
1. Introduction
2. Results
2.1. Correlation of Coagulopathic Toxins to Coagulant Peaks
2.2. Effect of Nanofractionated Venom Toxins on Plasma Coagulation
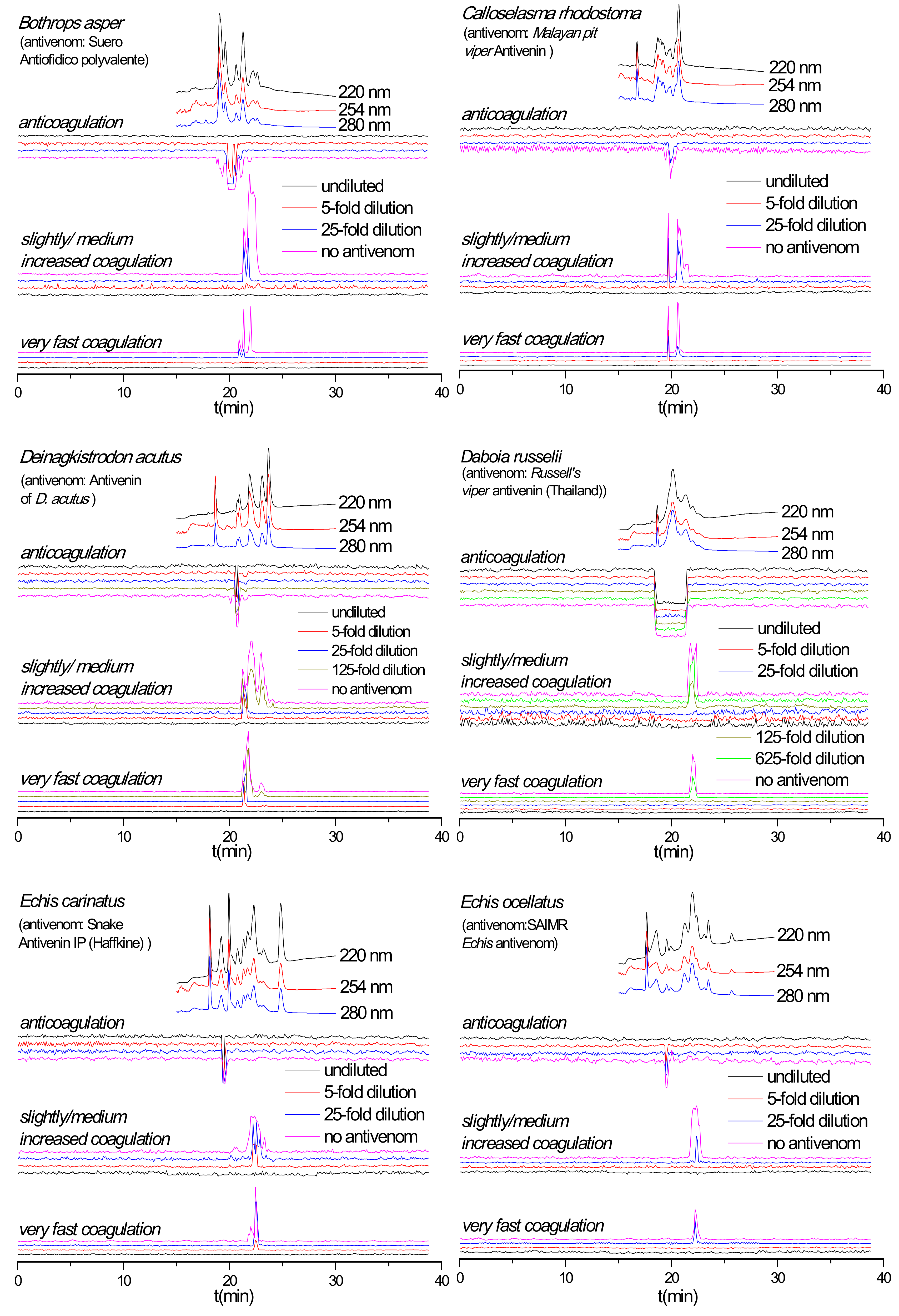
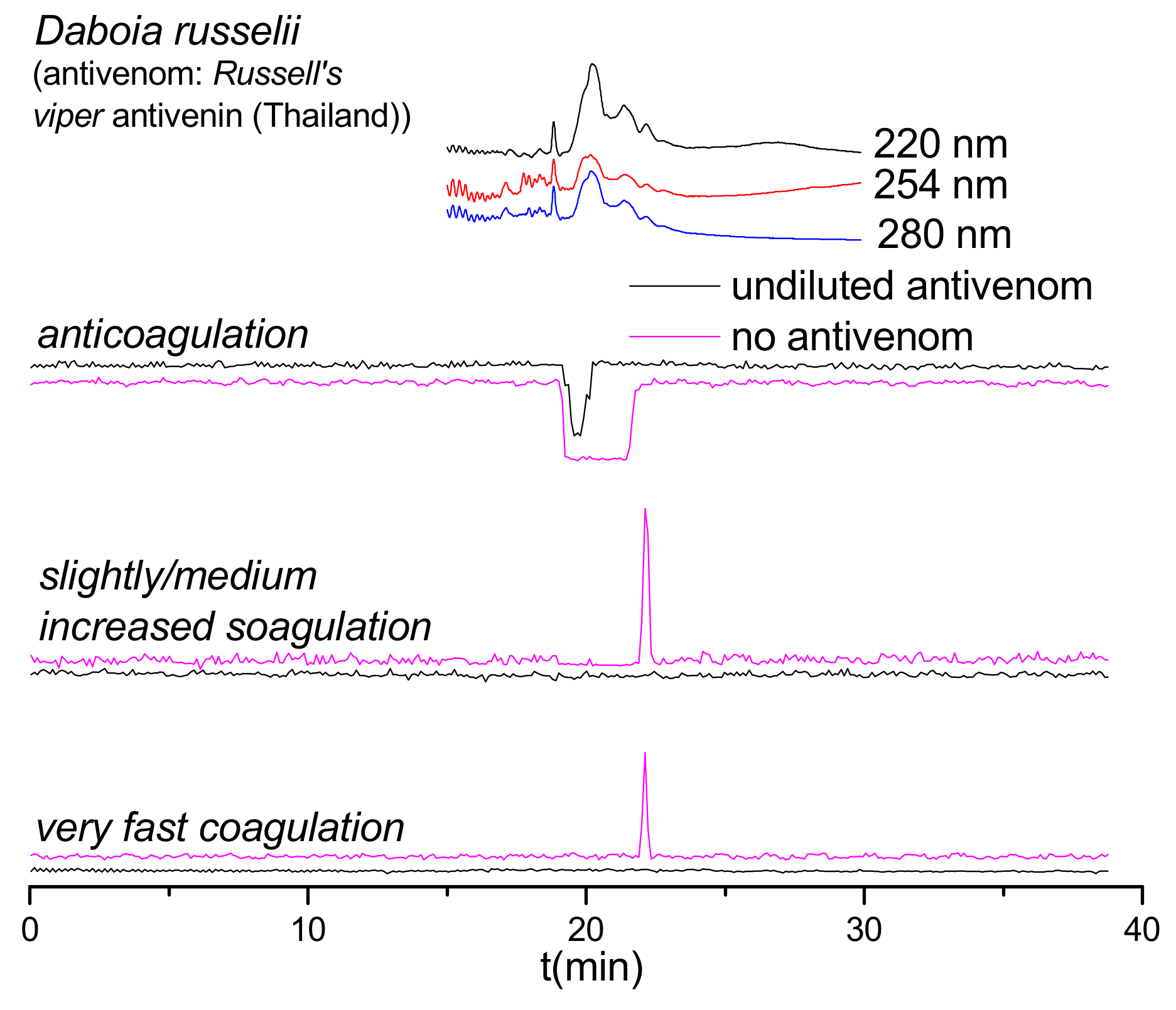
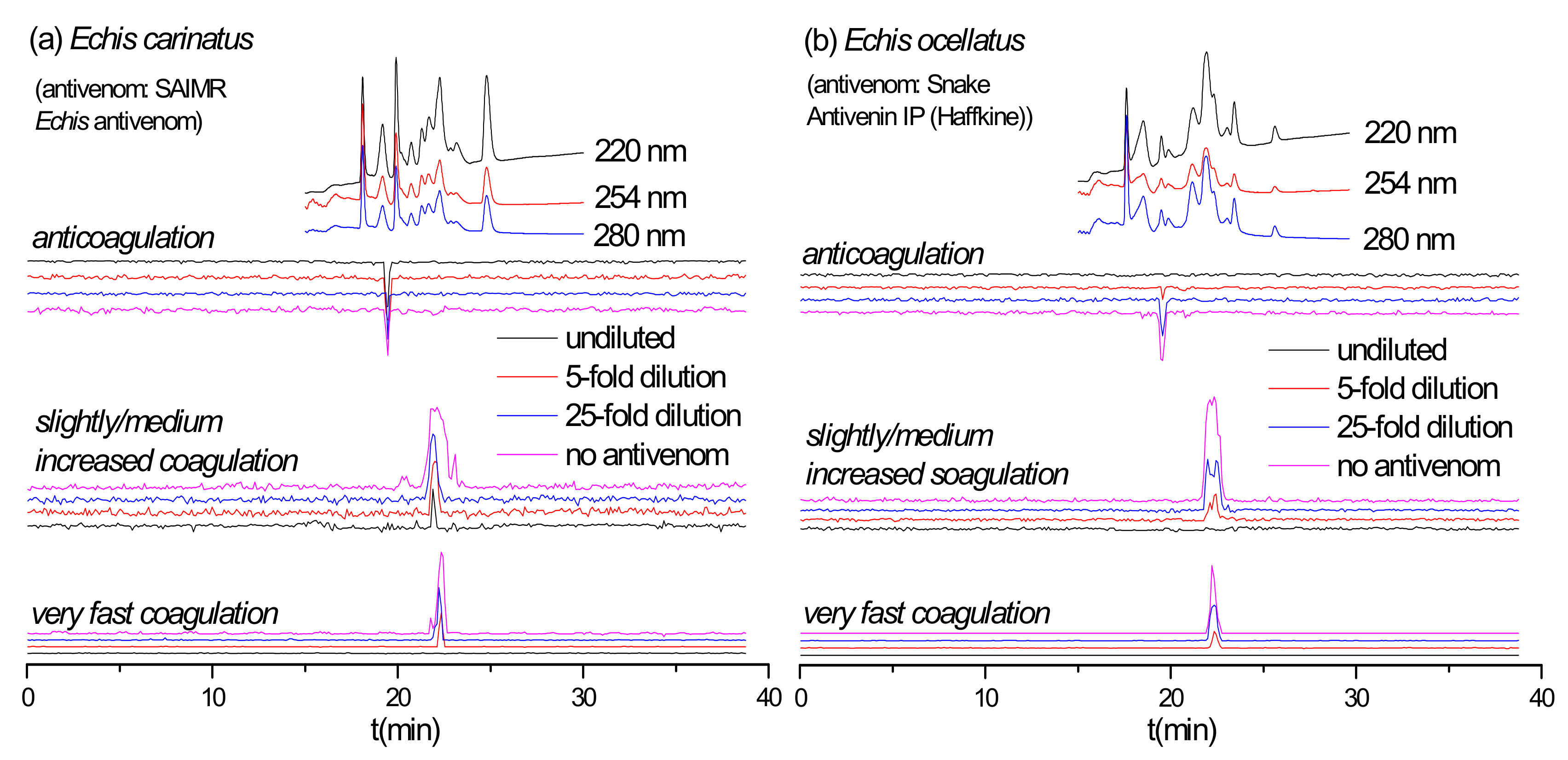
2.3. Antivenom Neutralization Potency
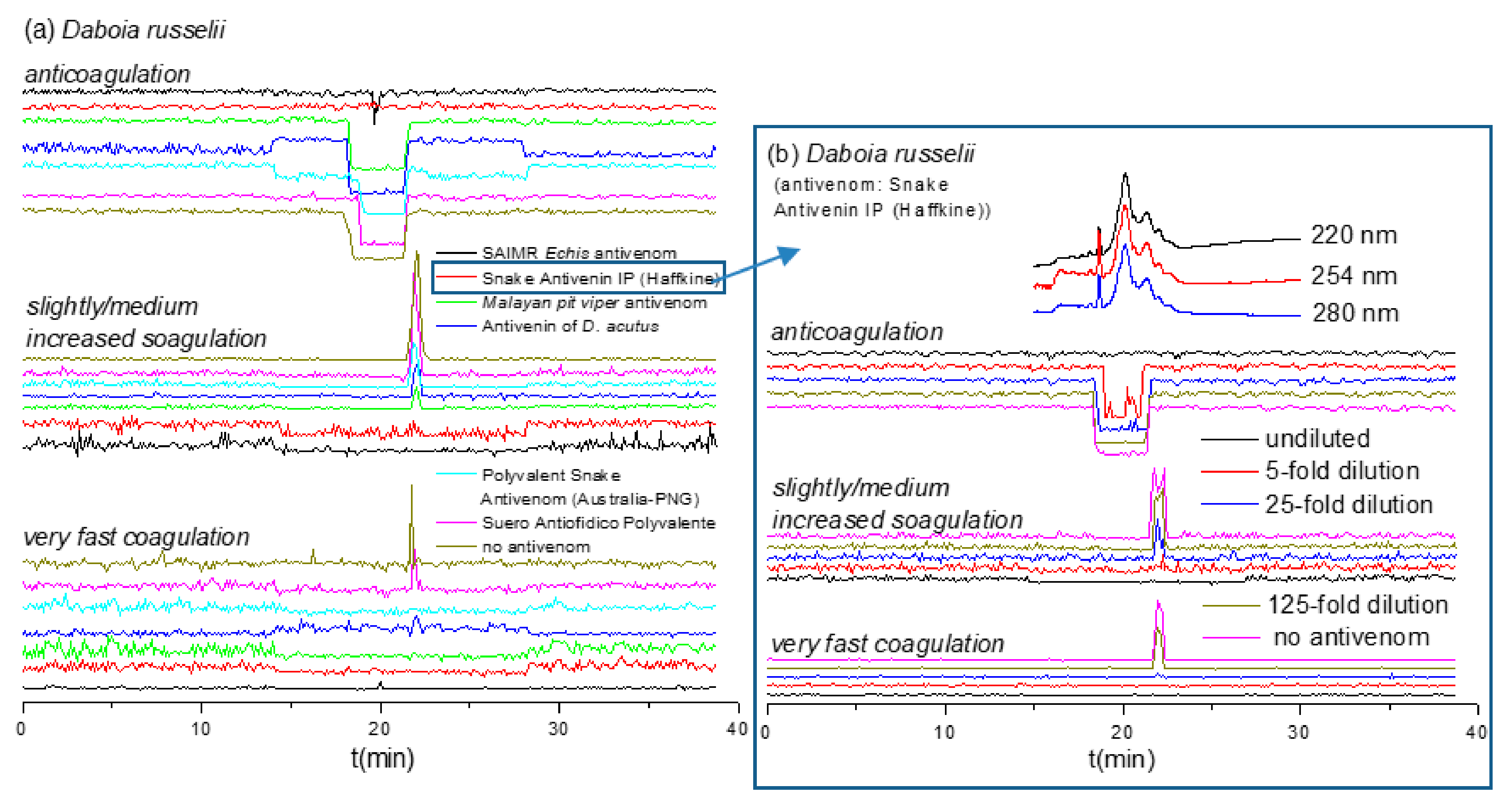
2.4. Antivenom Cross-Reactivity
2.5. Limitations
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals
5.2. List of Venoms and Antivenoms Included in This Study
5.3. LC-MS Nanofractionation
5.4. Plasma Coagulation Activity Assay
5.5. Correlation of Coagulation Data with MS and Proteomics Data
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lomonte, B. Snake venoms: From research to treatment. Acta Méd. Costarric. 2012, 54, 86–96. [Google Scholar]
- Pandey, R. Snake Bite: A Neglected Tropical Condition. Indian Pediatr. 2015, 52, 571–572. [Google Scholar]
- Kasturiratne, A.; Wickremasinghe, A.R.; de Silva, N.; Gunawardena, N.K.; Pathmeswaran, A.; Premaratna, R.; Savioli, L.; Lalloo, D.G.; de Silva, H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008, 5, e218. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.; Gutiérrez, J.M.; Harrison, R.; Warrell, D.A.; White, J.; Winkel, K.D.; Gopalakrishnakone, P. The Global Snake Bite Initiative: An antidote for snake bite. Lancet 2010, 375, 89–91. [Google Scholar] [CrossRef]
- Habib, A.G.; Kuznik, A.; Hamza, M.; Abdullahi, M.I.; Chedi, B.A.; Chippaux, J.-P.; Warrell, D.A. Snakebite is under appreciated: Appraisal of burden from West Africa. PLoS Negl. Trop. Dis. 2015, 9, e0004088. [Google Scholar] [CrossRef]
- Means, R.; Cabrera, J.; Moreno, X.; Amini, R. Remote South American Snakebite with Extensive Myonecrosis. Clin. Pract. Cases Emerg. Med. 2017, 1, 47. [Google Scholar]
- Spyres, M.B.; Ruha, A.-M.; Seifert, S.; Onisko, N.; Padilla-Jones, A.; Smith, E.A. Occupational snake bites: A prospective case series of patients reported to the ToxIC North American Snakebite Registry. J. Med. Toxicol. 2016, 12, 365–369. [Google Scholar] [CrossRef]
- Ainsworth, S.; Slagboom, J.; Alomran, N.; Pla, D.; Alhamdi, Y.; King, S.I.; Bolton, F.M.; Gutiérrez, J.M.; Vonk, F.J.; Toh, C.-H. The paraspecific neutralisation of snake venom induced coagulopathy by antivenoms. Commun. Biol. 2018, 1, 34. [Google Scholar] [CrossRef]
- Maduwage, K.; Isbister, G.K. Current treatment for venom-induced consumption coagulopathy resulting from snakebite. PLoS Negl. Trop. Dis. 2014, 8, e3220. [Google Scholar] [CrossRef]
- Still, K.; Nandlal, R.S.; Slagboom, J.; Somsen, G.W.; Casewell, N.R.; Kool, J. Multipurpose HTS Coagulation Analysis: Assay Development and Assessment of Coagulopathic Snake Venoms. Toxins 2017, 9, 382. [Google Scholar] [CrossRef]
- Sartim, M.A.; Cezarette, G.N.; Jacob-Ferreira, A.L.; Frantz, F.G.; Faccioli, L.H.; Sampaio, S.V. Disseminated intravascular coagulation caused by moojenactivase, a procoagulant snake venom metalloprotease. Int. J. Biol. Macromol. 2017, 103, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Slagboom, J.; Kool, J.; Harrison, R.A.; Casewell, N.R. Haemotoxic snake venoms: Their functional activity, impact on snakebite victims and pharmaceutical promise. Br. J. Haematol. 2017, 177, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Flores, M.P.; Faria, F.; de Andrade, S.A.; Chudzinski-Tavassi, A.M. Snake Venom Components Affecting the Coagulation System. Snake Venoms 2015, 1–20. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Escalante, T.; Rucavado, A.; Herrera, C. Hemorrhage caused by snake venom metalloproteinases: A journey of discovery and understanding. Toxins 2016, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M.; Koh, C.Y. Metalloproteases affecting blood coagulation, fibrinolysis and platelet aggregation from snake venoms: Definition and nomenclature of interaction sites. Toxins 2016, 8, 284. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, I.S.; Abubakar, S.B.; Habib, A.G.; Nasidi, A.; Durfa, N.; Yusuf, P.O.; Larnyang, S.; Garnvwa, J.; Sokomba, E.; Salako, L. Randomised controlled double-blind non-inferiority trial of two antivenoms for saw-scaled or carpet viper (Echis ocellatus) envenoming in Nigeria. PLoS Negl. Trop. Dis. 2010, 4, e767. [Google Scholar] [CrossRef]
- Roncolato, E.C.; Campos, L.B.; Pessenda, G.; e Silva, L.C.; Furtado, G.P.; Barbosa, J.E. Phage display as a novel promising antivenom therapy: A review. Toxicon 2015, 93, 79–84. [Google Scholar] [CrossRef]
- Deshpande, R.P.; Motghare, V.M.; Padwal, S.L.; Pore, R.R.; Bhamare, C.G.; Deshmukh, V.S.; Pise, H.N. Adverse drug reaction profile of anti-snake venom in a rural tertiary care teaching hospital. J. Young Pharm. 2013, 5, 41–45. [Google Scholar] [CrossRef]
- Hoogenboom, H.R. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 2005, 23, 1105. [Google Scholar] [CrossRef]
- Harrison, R.A.; Cook, D.A.; Renjifo, C.; Casewell, N.R.; Currier, R.B.; Wagstaff, S.C. Research strategies to improve snakebite treatment: Challenges and progress. J. Proteom. 2011, 74, 1768–1780. [Google Scholar] [CrossRef]
- Calvete, J.J.; Sanz, L.; Angulo, Y.; Lomonte, B.; Gutiérrez, J.M. Venoms, venomics, antivenomics. FEBS Lett. 2009, 583, 1736–1743. [Google Scholar] [CrossRef]
- Zietek, B.M.; Still, K.B.; Jaschusch, K.; Bruyneel, B.; Ariese, F.; Brouwer, T.J.; Luger, M.; Limburg, R.J.; Rosier, J.C.; v Iperen, D.J. Bioactivity profiling of small-volume samples by nano liquid chromatography coupled to microarray bioassaying using high-resolution fractionation. Anal. Chem. 2019, 91, 10458–10466. [Google Scholar] [CrossRef] [PubMed]
- Zietek, B.M.; Mayar, M.; Slagboom, J.; Bruyneel, B.; Vonk, F.J.; Somsen, G.W.; Casewell, N.R.; Kool, J. Liquid chromatographic nanofractionation with parallel mass spectrometric detection for the screening of plasmin inhibitors and (metallo) proteinases in snake venoms. Anal. Bioanal. Chem. 2018, 410, 5751–5763. [Google Scholar] [CrossRef] [PubMed]
- Slagboom, J.; Mladić, M.; Xie, C.; Vonk, F.; Somsen, G.W.; Casewell, N.R.; Kool, J. High throughput screening and identification of coagulopathic snake venom proteins and peptides using nanofractionation and proteomics approaches. bioRxiv 2019. [Google Scholar] [CrossRef]
- Cardoso, F.C.; Ferraz, C.R.; Arrahman, A.; Xie, C.; Casewell, N.R.; Lewis, R.J.; Kool, J. Multifunctional toxins in snake venoms and therapeutic implications: From pain to hemorrhage and necrosis. Front. Ecol. Evol. 2019, 7, 218. [Google Scholar]
- Mounier, C.M.; Bon, C.; Kini, R.M. Anticoagulant venom and mammalian secreted phospholipases A2: Protein-versus phospholipid-dependent mechanism of action. Pathophysiol. Haemost. Thromb. 2001, 31, 279–287. [Google Scholar] [CrossRef]
- Savanur, A.; Ali, S.A.; Munir, I.; Abbasi, A.; Alam, M.; Shaikh, H.A. Pharmacological and biochemical studies on the venom of a clinically important viper snake (Echis carinatus) of Pakistan. Toxicon 2014, 80, 47–57. [Google Scholar] [CrossRef]
- Ciscotto, P.H.; Rates, B.; Silva, D.A.; Richardson, M.; Silva, L.P.; Andrade, H.; Donato, M.F.; Cotta, G.A.; Maria, W.S.; Rodrigues, R.J. Venomic analysis and evaluation of antivenom cross-reactivity of South American Micrurus species. J. Proteom. 2011, 74, 1810–1825. [Google Scholar] [CrossRef]
- Leong, P.K.; Sim, S.M.; Fung, S.Y.; Sumana, K.; Sitprija, V.; Tan, N.H. Cross neutralization of Afro-Asian cobra and Asian krait venoms by a Thai polyvalent snake antivenom (Neuro Polyvalent Snake Antivenom). PLoS Negl. Trop. Dis. 2012, 6, e1672. [Google Scholar] [CrossRef]
- Howes, J.-M.; Kamiguti, A.; Theakston, R.D.G.; Wilkinson, M.; Laing, G. Effects of three novel metalloproteinases from the venom of the West African saw-scaled viper, Echis ocellatus on blood coagulation and platelets. Biochim. Biophys. Acta BBA Gen. Subj. 2005, 1724, 194–202. [Google Scholar] [CrossRef]
- Casewell, N.R.; Harrison, R.A.; Wüster, W.; Wagstaff, S.C. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genom. 2009, 10, 564. [Google Scholar] [CrossRef] [PubMed]
- Rogalski, A.; Soerensen, C.; Op den Brouw, B.; Lister, C.; Dashevsky, D.; Arbuckle, K.; Gloria, A.; Zdenek, C.N.; Casewell, N.R.; Gutiérrez, J.M. Differential procoagulant effects of saw-scaled viper (Serpentes: Viperidae: Echis) snake venoms on human plasma and the narrow taxonomic ranges of antivenom efficacies. Toxicol. Lett. 2017, 280, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Casewell, N.R.; Cook, D.A.N.; Wagstaff, S.C.; Nasidi, A.; Durfa, N.; Wüster, W.; Harrison, R.A.; Williams, D.J. Pre-Clinical Assays Predict Pan-African Echis Viper Efficacy for a Species-Specific Antivenom. PLoS Negl. Trop. Dis. 2010, 4, e851. [Google Scholar] [CrossRef] [PubMed]
- Visser, L.E.; Kyei-Faried, S.; Belcher, D.W.; Geelhoed, D.W.; Van Leeuwen, J.S.; Van Roosmalen, J. Failure of a new antivenom to treat Echis ocellatus snake bite in rural Ghana: The importance of quality surveillance. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Tsai, I.H.; Wang, Y.M.; Au, L.C.; Ko, T.P.; Chen, Y.H.; Chu, Y.F. Phospholipases A2 from Callosellasma rhodostoma venom gland: Cloning and sequencing of 10 of the cDNAs, three-dimensional modelling and chemical modification of the major isozyme. Eur. J. Biochem. 2000, 267, 6684–6691. [Google Scholar] [CrossRef] [PubMed]
- Faure, G.; Gowda, V.T.; Maroun, R.C. Characterization of a human coagulation factor Xa-binding site on Viperidae snake venom phospholipases A 2 by affinity binding studies and molecular bioinformatics. BMC Struct. Biol. 2007, 7, 82. [Google Scholar] [CrossRef]
- Kemparaju, K.; Krishnakanth, T.; Gowda, T.V. Purification and characterization of a platelet aggregation inhibitor acidic phospholipase A2 from Indian saw-scaled viper (Echis carinatus) venom. Toxicon 1999, 37, 1659–1671. [Google Scholar] [CrossRef]
- Jonker, W.; Ballesteros-Gómez, A.; Hamers, T.; Somsen, G.W.; Lamoree, M.H.; Kool, J. Highly selective screening of estrogenic compounds in consumer-electronics plastics by liquid chromatography in parallel combined with nanofractionation-bioactivity detection and mass spectrometry. Environ. Sci. Technol. 2016, 50, 12385–12393. [Google Scholar] [CrossRef]
- Otvos, R.A.; Mladic, M.; Arias-Alpizar, G.; Niessen, W.M.; Somsen, G.W.; Smit, A.B.; Kool, J. At-line cellular screening methodology for bioactives in mixtures targeting the α7-nicotinic acetylcholine receptor. J. Biomol. Screen. 2016, 21, 459–467. [Google Scholar] [CrossRef]
- Zietek, B.M.; Mladic, M.; Bruyneel, B.; Niessen, W.M.; Honing, M.; Somsen, G.W.; Kool, J. Nanofractionation platform with parallel mass spectrometry for identification of CYP1A2 inhibitors in metabolic mixtures. SLAS Discov. Adv. Life Sci. R D 2018, 23, 283–293. [Google Scholar] [CrossRef]
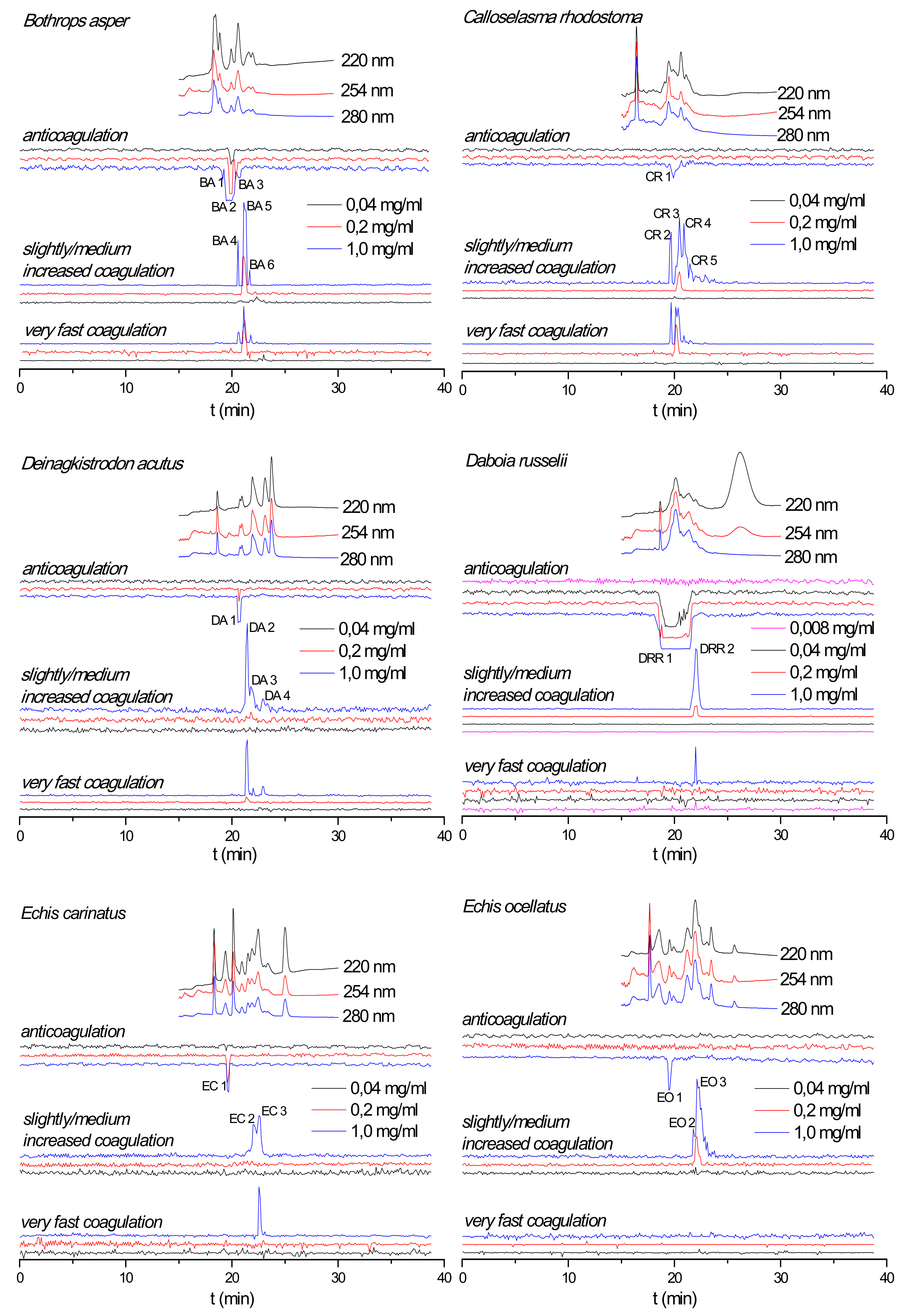
| Species | Peak Number | Retention Time (min) | Mascot Protein Hits | Exact Mass From MS Data | Exact Mass Calculated From Mascot Data | Toxin Class |
|---|---|---|---|---|---|---|
| B. asper | BA 1 | 19.1 | PA2H2_BOTAS | 13,714.5646 | 13,714.56817 | PLA2 |
| BA 2 | 19.4–20.2 | PA2HA_BOTAS | 13,912.4649 | 13,896.51308 | PLA2 | |
| BA 2 | 19.4–20.2 | PA2H3_BOTAS | 13,765.5812 | 13,765.58896 | PLA2 | |
| BA 3 | 20.5–20.8 | PA2B3_BOTAS | 13,957.5333 | 13,957.48720 | PLA2 | |
| BA 3 | 20.5–20.8 | VM2_BOTAS | - | 53,564 | SVMP | |
| BA 3 | 20.5–20.8 | PA2A2_BOTAS | - | 14,194 | PLA2 | |
| BA 4 | 20.6 | VSPL_BOTAS | - | 28,019 | SVSP | |
| BA 4 | 20.6 | VM1B1_BOTAS | - | 45,936 | SVMP | |
| BA 5 | 21.1–21.3 | SLA_BOTAS | - | 7084 | CTL | |
| BA 6 | 21.7 | - | - | - | - | |
| C. rhodostoma | CR 1 | 19.7–20.5 | PA2BD_CALRH | 13,665.0848 | 13,665.0237 | PLA2 |
| CR 1 | 19.7–20.5 | VSPF1_CALRH | - | 26,570 | SVSP | |
| CR 1 | 19.7–20.5 | SLEA_CALRH | - | 15,962 | CTL | |
| CR 1 | 19.7–20.5 | SLEB_CALRH | - | 15,190 | CTL | |
| CR 1 | 19.7–20.5 | PA2AB_CALRH | - | 14,352 | PLA2 | |
| CR 2 | 19.6–19.7 | VSPF2_CALRH | - | 29,145 | SVSP | |
| CR 3 | 20.1–20.7 | VSPF2_CALRH | - | 29,145 | SVSP | |
| CR 4 | 20.8–21.2 | VSPF2_CALRH | - | 29,145 | SVSP | |
| CR 4 | 20.8–21.2 | SLYA_CALRH | - | 15,796 | CTL | |
| CR 5 | 21.5–21.7 | SLYA_CALRH | - | 15,796 | CTL | |
| CR 5 | 21.5–21.7 | SLYB_CALRH | - | 16,770 | CTL | |
| D. acutus | DA 1 | 20.6–20.9 | PA2A_DEIAC | - | 14,820 | PLA2 |
| DA 1 | 20.6–20.9 | SL_DEIAC | - | 18,332 | CTL | |
| DA 2 | 21.3–21.7 | SLCB_DEIAC | - | 17,133 | CTL | |
| DA 2 | 21.3–21.7 | VSP1_DEIAC | - | 29,480 | SVSP | |
| DA 2 | 21.3–21.7 | VSPA_DEIAC | - | 26,132 | SVSP | |
| DA 2 | 21.3–21.7 | VM1AC_DEIAC | - | 47,690 | SVMP | |
| DA 2 | 21.3–21.7 | VM11_DEIAC | - | 47,845 | SVMP | |
| DA 2 | 21.3–21.7 | VM1H5_DEIAC | - | 46,518 | SVMP | |
| DA 2 | 21.3–21.7 | VM3AK_DEIAC | - | 69,752 | SVMP | |
| DA 3 | 21.8–22.1 | VM11_DEIAC | 47,845 | SVSP | ||
| DA 3 | 21.8–22.1 | VM1H5_DEIAC | 46,518 | SVSP | ||
| DA 4 | 22.8–23.1 | VM3A2_DEIAC | 27,151 | SVMP | ||
| DA 4 | 22.8–23.1 | VM3AH_DEIAC | 70,721 | SVMP | ||
| D. russelii | DRR 1 | 18.3–21.7 | PA2B8_DABRR | 13,587.2248 | 13,587.2027 | PLA2 |
| DRR 1 | 18.3–21.7 | PA2B5_DABRR | 13,587 | PLA2 | ||
| DRR 1 | 18.3–21.7 | PA2B3_DABRR | 13,687 | PLA2 | ||
| DRR 2 | 21.6–22.4 | - | - | - | - | |
| E. carinatus | EC 1 | 19.6–19.7 | PA2A1_ECHCA | - | 16,310 | PLA2 |
| EC 2 | 21.9–22.3 | - | - | - | - | |
| EC 3 | 22.3–22.9 | - | - | - | - | |
| E. ocellatus (Nigeria) | EO 1 | 19.4–19.8 | PA2A5_ECHOC | 13,856.1382 | 13,856.0665 | PLA2 |
| EO 2 | 21.8–21.9 | VM3E2_ECHOC | - | 69,426 | SVMP | |
| EO 2 | 21.8–21.9 | VM3E6_ECHOC | - | 57,658 | SVMP | |
| EO 2 | 21.8–21.9 | SL1_ECHOC | - | 16,601 | CTL | |
| EO 2 | 21.8–21.9 | SL124_ECHOC | - | 16,882 | CTL | |
| EO 3 | 22.0–23.1 | VM3E6_ECHOC | - | 57,658 | SVMP | |
| EO 3 | 22.0–23.1 | SL1_ECHOC | - | 16,601 | CTL | |
| EO 3 | 22.0–23.1 | SL124_ECHOC | - | 16,882 | CTL |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, C.; Slagboom, J.; Albulescu, L.-O.; Bruyneel, B.; Still, K.B.M.; Vonk, F.J.; Somsen, G.W.; Casewell, N.R.; Kool, J. Antivenom Neutralization of Coagulopathic Snake Venom Toxins Assessed by Bioactivity Profiling Using Nanofractionation Analytics. Toxins 2020, 12, 53. https://doi.org/10.3390/toxins12010053
Xie C, Slagboom J, Albulescu L-O, Bruyneel B, Still KBM, Vonk FJ, Somsen GW, Casewell NR, Kool J. Antivenom Neutralization of Coagulopathic Snake Venom Toxins Assessed by Bioactivity Profiling Using Nanofractionation Analytics. Toxins. 2020; 12(1):53. https://doi.org/10.3390/toxins12010053
Chicago/Turabian StyleXie, Chunfang, Julien Slagboom, Laura-Oana Albulescu, Ben Bruyneel, Kristina B. M. Still, Freek J. Vonk, Govert W. Somsen, Nicholas R. Casewell, and Jeroen Kool. 2020. "Antivenom Neutralization of Coagulopathic Snake Venom Toxins Assessed by Bioactivity Profiling Using Nanofractionation Analytics" Toxins 12, no. 1: 53. https://doi.org/10.3390/toxins12010053
APA StyleXie, C., Slagboom, J., Albulescu, L.-O., Bruyneel, B., Still, K. B. M., Vonk, F. J., Somsen, G. W., Casewell, N. R., & Kool, J. (2020). Antivenom Neutralization of Coagulopathic Snake Venom Toxins Assessed by Bioactivity Profiling Using Nanofractionation Analytics. Toxins, 12(1), 53. https://doi.org/10.3390/toxins12010053






