LC-HRMS Screening and Identification of Novel Peptide Markers of Ricin Based on Multiple Protease Digestion Strategies
Abstract
1. Introduction
2. Results and Discussion
2.1. Amino Acid Sequence Difference between Ricin and RCA120
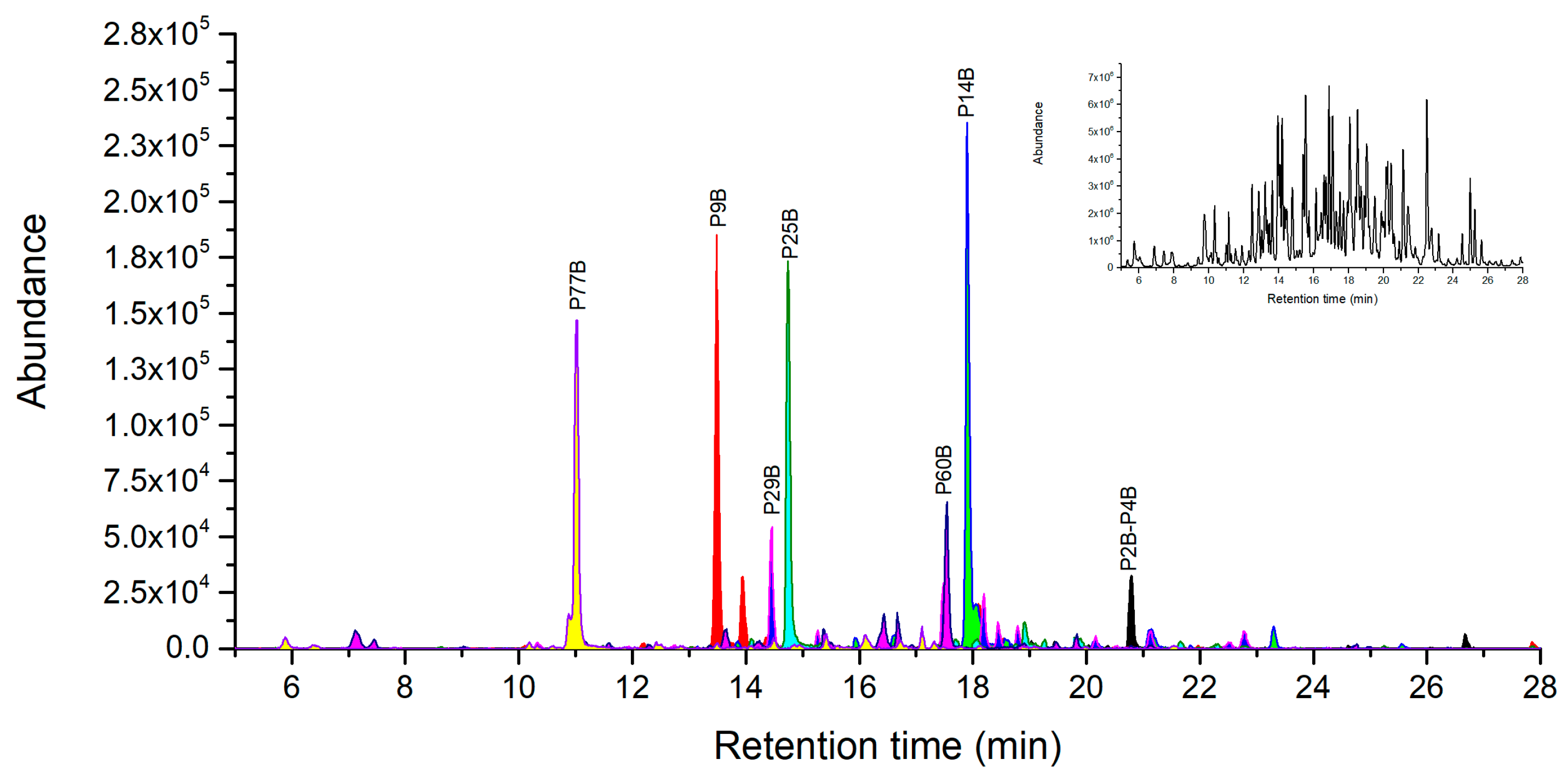
2.2. Ricin Peptide Markers from Trypsin Digestion
2.3. Novel Peptide Markers of Ricin from Tandem Digestion with Endoproteinase Glu-C after Trypsin
2.4. Novel Peptide Markers of Ricin from Chymotrypsin Digestion
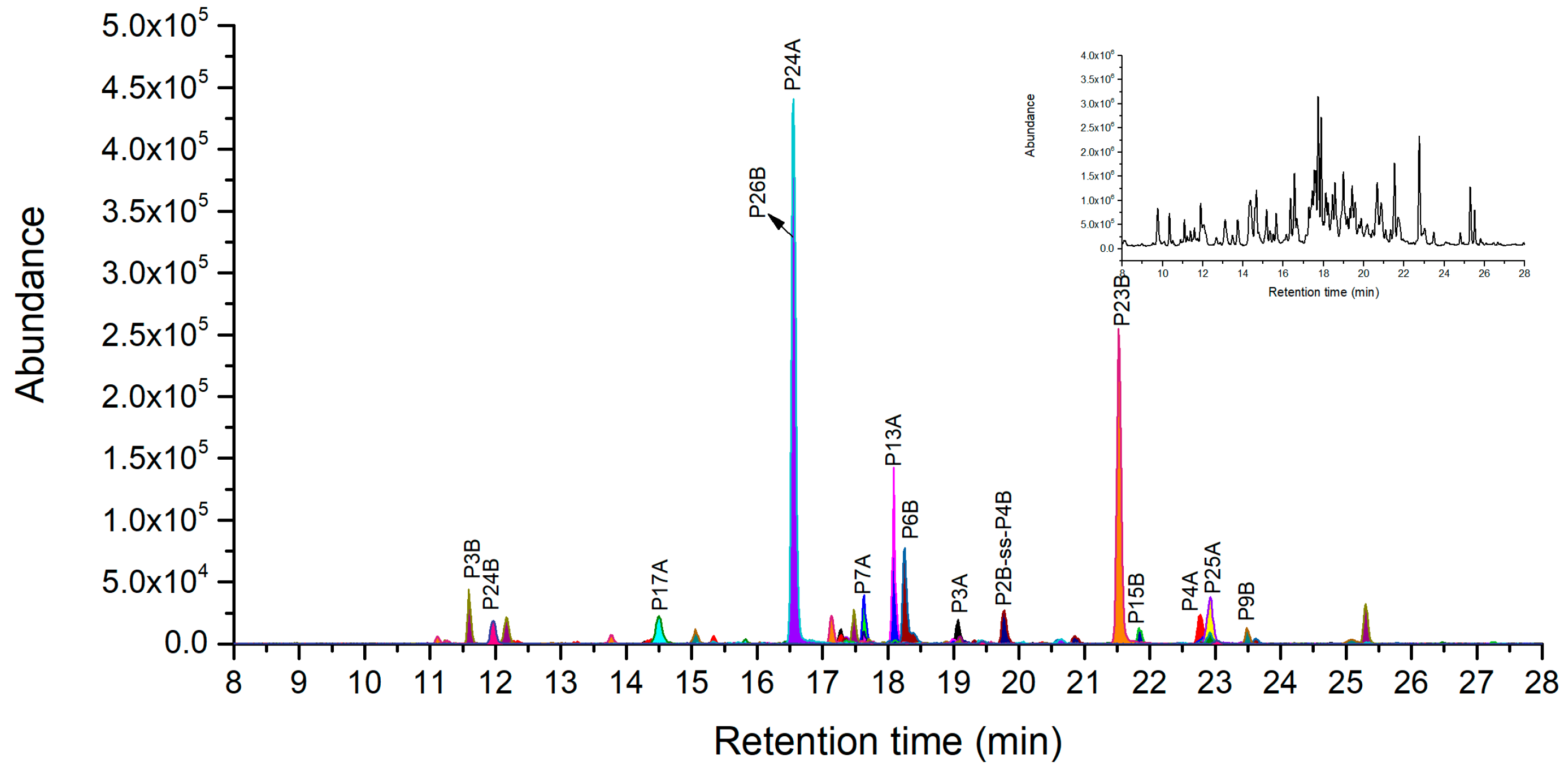
2.5. Novel Peptide Markers of Ricin from Other Proteinase Digestion
2.5.1. Novel Peptide Markers of Ricin from Pepsin Digestion
2.5.2. Novel Peptide Markers of Ricin from Proteinase K Digestion
3. Materials and Methods
3.1. Sequence Alignment and Identification of Uniqueness of Digested Peptides
3.2. Digestion of Ricin Using Multiple Proteinases
3.2.1. Trypsin Digestion of Ricin with Denaturation and Reduction
3.2.2. Solvent-Assisted Direct Digestion of Ricin
3.2.3. Tandem Digestion with Glu-C after Trypsin
3.2.4. Chymotrypsin Digestion with Denaturation and Reduction
3.2.5. Solvent-Assisted Chymotrypsin Digestion of Ricin
3.2.6. Pepsin Digestion
3.2.7. Proteinase K Digestion
3.3. Accurate Mass LC-MS Qualitative Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fabbrini, M.S.; Katayama, M.; Nakase, I.; Vago, R. Plant Ribosome-Inactivating Proteins: Progesses, Challenges and Biotechnological Applications (and a Few Digressions). Toxins 2017, 9, 314. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.; Hao, Q.; Chen, Y.; Barre, A.; Vandenbussche, F.; Desmyter, S.; Rougé, P.; Peumans, W.J. Ribosome-Inactivating Proteins: A Family of Plant Proteins That Do More Than Inactivate Ribosomes. Crit. Rev. Plant Sci. 2010, 20, 395–465. [Google Scholar] [CrossRef]
- Audi, J.; Belson, M.; Patel, M.; Schier, J.; Osterloh, J. Ricin Poisoning: A Comprehensive Review. JAMA 2005, 294, 2342–2451. [Google Scholar] [CrossRef] [PubMed]
- Worbs, S.; Kohler, K.; Pauly, D.; Avondet, M.A.; Schaer, M.; Dorner, M.B.; Dorner, B.G. Ricinus communis intoxications in human and veterinary medicine-a summary of real cases. Toxins 2011, 3, 1332–1372. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, F.; Battelli, M.G. Ribosome-inactivating proteins: Progress and problems. Cell. Mol. Life Sci. 2006, 63, 1850–1866. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Baudys, J.; Barr, J.R.; Kalb, S.R. Improved Sensitivity for the Qualitative and Quantitative Analysis of Active Ricin by MALDI-TOF Mass Spectrometry. Anal. Chem. 2016, 88, 6867–6872. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.J.; Sun, J.F.; Lui, R.; Zhang, Z.M.; Liu, J.F.; Xie, J.W. New Surface-Enhanced Raman Sensing Chip Designed for On-Site Detection of Active Ricin in Complex Matrices Based on Specific Depurination. ACS Appl. Mater. Interfaces 2016, 8, 2449–2455. [Google Scholar] [CrossRef]
- Falach, R.; Sapoznikov, A.; Gal, Y.; Israeli, O.; Leitner, M.; Seliger, N.; Ehrlich, S.; Kronman, C.; Sabo, T. Quantitative profiling of the in vivo enzymatic activity of ricin reveals disparate depurination of different pulmonary cell types. Toxicol. Lett. 2016, 258, 11–19. [Google Scholar] [CrossRef]
- Simon, S.; Worbs, S.; Avondet, M.A.; Tracz, D.M.; Dano, J.; Schmidt, L.; Volland, H.; Dorner, B.G.; Corbett, C.R. Recommended Immunological Assays to Screen for Ricin-Containing Samples. Toxins 2015, 7, 4967–4986. [Google Scholar] [CrossRef]
- Gaylord, S.T.; Dinh, T.L.; Goldman, E.R.; Anderson, G.P.; Ngan, K.C.; Walt, D.R. Ultrasensitive Detection of Ricin Toxin in Multiple Sample Matrixes Using Single-Domain Antibodies. Anal. Chem. 2015, 87, 6570–6577. [Google Scholar] [CrossRef]
- Worbs, S.; Skiba, M.; Soderstrom, M.; Rapinoja, M.L.; Zeleny, R.; Russmann, H.; Schimmel, H.; Vanninen, P.; Fredriksson, S.A.; Dorner, B.G. Characterization of Ricin and R. communis Agglutinin Reference Materials. Toxins 2015, 7, 4906–4934. [Google Scholar] [CrossRef] [PubMed]
- Ostin, A.; Bergstrom, T.; Fredriksson, S.A.; Nilsson, C. Solvent-Assisted Trypsin Digestion of Ricin for Forensic Identification by LC-ESI MS/MS. Anal. Chem. 2007, 79, 6271–6278. [Google Scholar] [CrossRef] [PubMed]
- McGrath, S.C.; Schieltz, D.M.; McWilliams, L.G.; Pirkle, J.L.; Barr, J.R. Detection and quantification of ricin in beverages using isotope dilution tandem mass spectrometry. Anal. Chem. 2011, 83, 2897–2905. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Tang, J.; Li, C.; Liu, Q.; Chen, J.; Li, H.; Guo, L.; Xie, J. Identification and quantification of ricin in biomedical samples by magnetic immunocapture enrichment and liquid chromatography electrospray ionization tandem mass spectrometry. Anal. Bioanal. Chem. 2014, 406, 5147–5155. [Google Scholar] [CrossRef] [PubMed]
- Schieltz, D.M.; McWilliams, L.G.; Kuklenyik, Z.; Prezioso, S.M.; Carter, A.J.; Williamson, Y.M.; McGrath, S.C.; Morse, S.A.; Barr, J.R. Quantification of ricin, RCA and comparison of enzymatic activity in 18 Ricinus communis cultivars by isotope dilution mass spectrometry. Toxicon 2015, 95, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Worbs, S.; Skiba, M.; Bender, J.; Zeleny, R.; Schimmel, H.; Luginbühl, W.; Dorner, B. An International Proficiency Test to Detect, Identify and Quantify Ricin in Complex Matrices. Toxins 2015, 7, 4987–5010. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, S.A.; Artursson, E.; Bergstrom, T.; Ostin, A.; Nilsson, C.; Astot, C. Identification of RIP-II toxins by affinity enrichment, enzymatic digestion and LC-MS. Anal. Chem. 2015, 87, 967–974. [Google Scholar] [CrossRef]
- Braun, A.V.; Taranchenko, V.F.; Tikhomirov, L.A.; Grechukhin, A.P.; Rybal’chenko, I.V. Detection of Ricin in Plant Extracts and Soil Using Liquid Chromatography–High-Resolution Mass Spectrometry. J. Anal. Chem. 2018, 73, 786–795. [Google Scholar] [CrossRef]
- Araki, T.; Funatsu, G. The complete amino acid sequence of the B-chain of ricin E isolated from small-grain castor bean seeds. Ricin E is a gene recombination product of ricin D and Ricinus communis agglutinin. Biochim. Biophys. Acta 1987, 911, 191–200. [Google Scholar] [CrossRef]
- Fredriksson, S.A.; Hulst, A.G.; Artursson, E.; De Jong, A.L.; Nilsson, C.; Van Baar, B.L. Forensic Identification of Neat Ricin and of Ricin from Crude Castor Bean Extracts by Mass Spectrometry. Anal. Chem. 2005, 77, 1545–1555. [Google Scholar] [CrossRef]
- Sweeney, P.J.; Walker, J.M. Proteolytic Enzymes for Peptide Production. Methods Mol. Biol. 1993, 16, 277–303. [Google Scholar] [CrossRef] [PubMed]

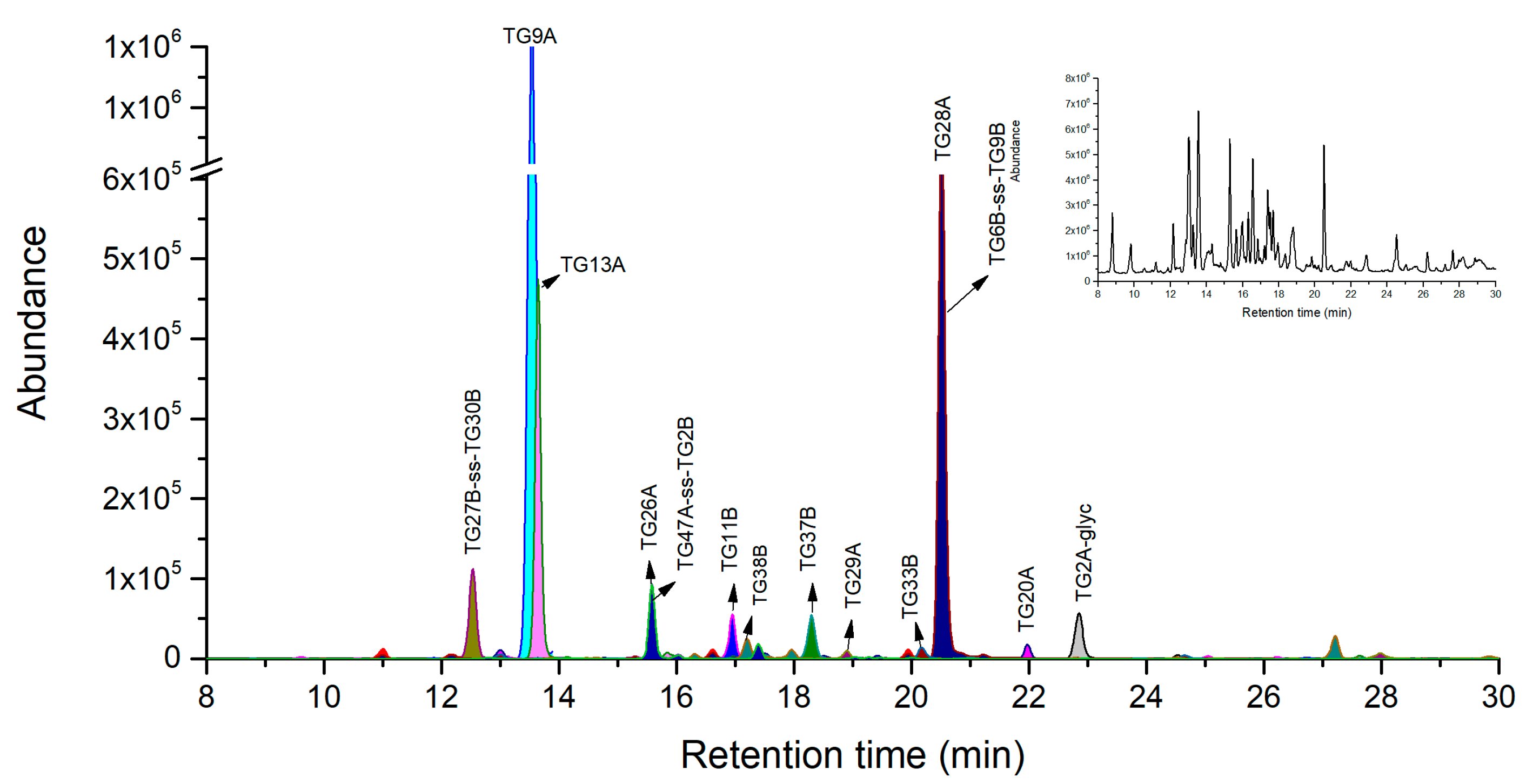
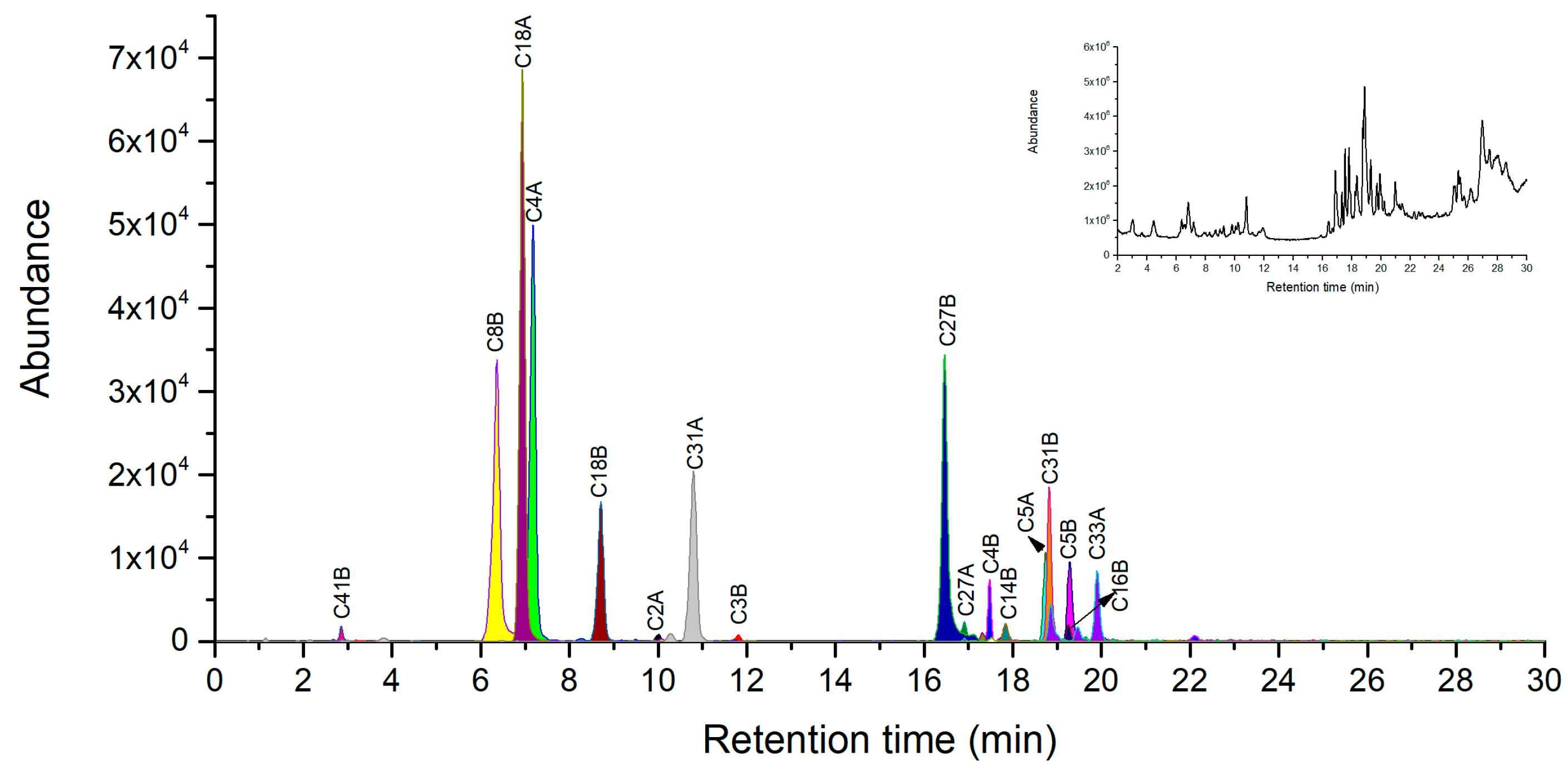
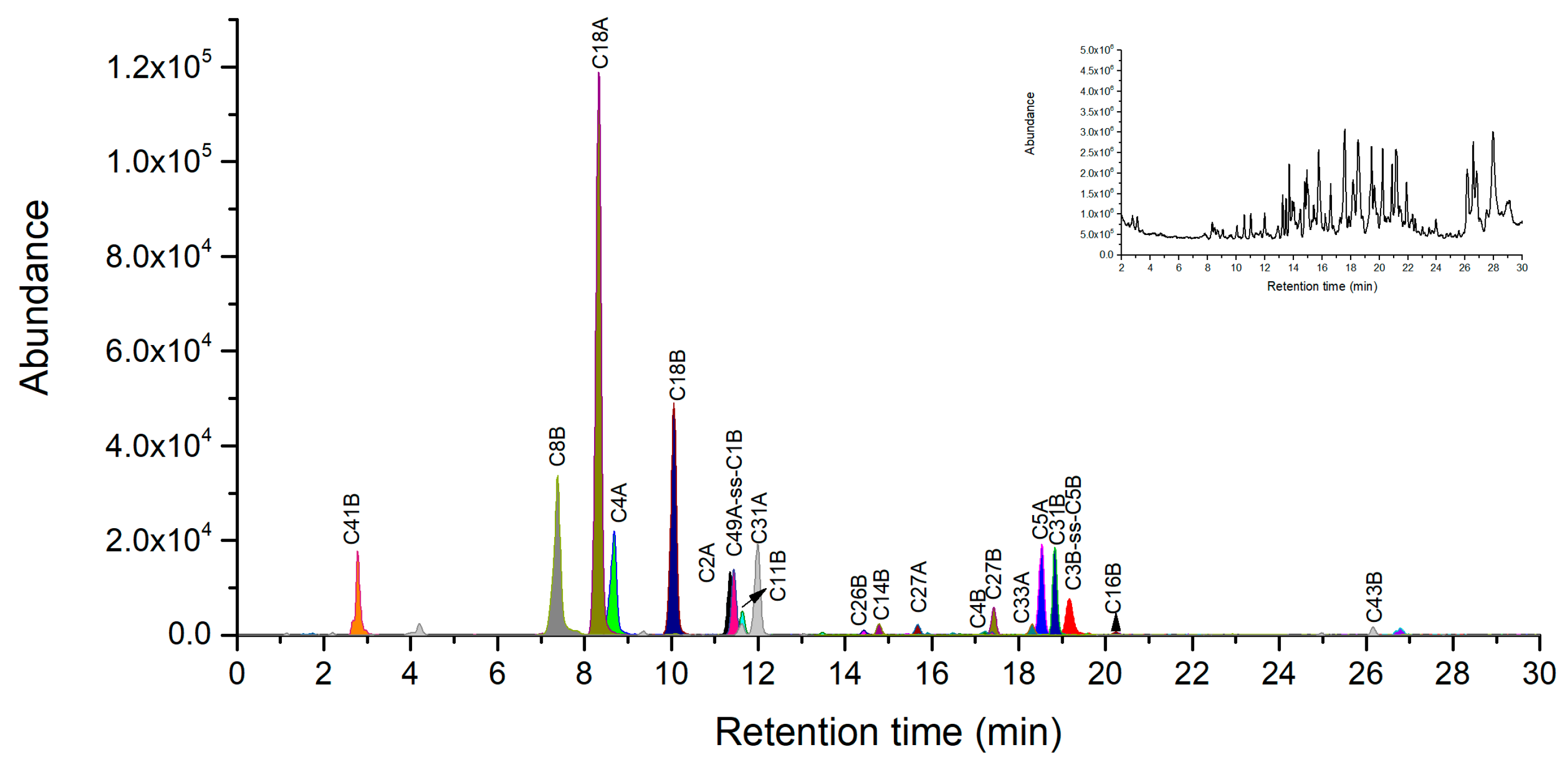
| TG#&Chain | Amino Acid Sequence | (M + H)+ | (M + 2H)2+ | (M + 3H)3+ | (M + 4H)4+ |
|---|---|---|---|---|---|
| TG2A-glyc * | QYPIINFTTAGATVQSYTNFIR | 3675.6946 | 1838.3509 | 1225.9031 | 919.6791 |
| TG9A | VGLPINQR | 896.5312 | 448.7692 | 299.5152 | - |
| TG13A | VTNAYVVGYR | 1141.6000 | 571.3036 | 381.2048 | - |
| TG20A | YTFAFGGNYD | 1154.4789 | 577.7431 | 385.4978 | - |
| TG26A | LGNGPLE | 699.3671 | 350.1872 | 233.7939 | - |
| TG28A | AISALYYYSTGGTQLPTLAR | 2146.1179 | 1073.5626 | 716.0441 | 537.2849 |
| TG29A | SFIICIQMISE | 1283.6374 | 642.3223 | 428.5506 | - |
| TG47A-ss-TG2B a | CAPPPSSQF-ss-VCMD | 1397.5535 | 699.2804 | 466.5226 | - |
| TG6B-ss-TG9B a | NGLCVD-ss-FHNGNAIQLWPCK | 2145.0003 | 1073.0038 | 715.7154 | 537.0555 |
| TG11B | ANQLWTLK | 973.5465 | 487.2769 | 325.1870 | - |
| TG15B-ss-TG16B a | CLTTYGYSPGVYVMIYD-ss-CNTAATD | 2637.1194 | 1319.0633 | 879.7113 | 660.0353 |
| TG20B | SSLVLAATSGNSGTTLTVQTNIYAVSQGWLPTNNTQPFVTTIVGLYGLCLQANSGQVWIED | 6284.1859 | 3142.5966 | 2095.4002 | 1571.8024 |
| TG27B-ss-TG30B a | NCLTSD-ss-ILSCGPASSGQR | 1824.7458 | 912.9094 | 608.9639 | 456.9912 |
| TG33B | GTILNLYSGLVLD | 1377.7624 | 689.3848 | 459.9256 | - |
| TG37B | QIILYPLHGD | 1168.6360 | 584.8217 | 390.2169 | - |
| TG38B | PNQIWLPLF | 1127.6248 | 564.3160 | 376.5464 | - |
| C# and Chain | Amino Acid Sequence | (M + H)+ | (M + 2H)2+ | (M + 3H)3+ | (M + 4H)4+ |
|---|---|---|---|---|---|
| C2A | TTAGATVQSY | 998.4789 | 499.7431 | 333.4978 | - |
| C4A | IRAVRGRL | 940.6163 | 470.8118 | 314.2103 | - |
| C5A | TTGADVRHEIPVLPNRVGLP INQRF | 2799.5376 | 1400.2724 | 933.8507 | 700.6418 |
| C18A | TDVQNRY | 895.4268 | 448.2170 | 299.1471 | - |
| C27A | EEAISAL | 732.3774 | 366.6923 | 265.1449 | - |
| C31A | STGGTQLPTL | 974.5153 | 487.7613 | 325.5099 | - |
| C33A | IICIQM | 720.3783 | 360.6928 | 240.7976 | - |
| C2B | DPEPIVRIVGRNGL | 1534.8699 | 767.9386 | 512.2948 | 384.4734 |
| C3B | CVDVRDGRF | 1066.5098 | 533.7585 | 356.1748 | - |
| C4B | HNGNAIQL | 866.4479 | 433.7276 | 289.4875 | - |
| C5B | WPCKSNTDANQL | 1376.6263 | 688.8168 | 459.5469 | 344.9125 |
| C8B | KRDNTIRSNGKCL | 1504.8012 | 752.9043 | 502.2719 | 376.9562 |
| C14B | DCNTAATDATRW | 1324.5586 | 643.8386 | 442.1911 | 331.8956 |
| C16B | DNGTIINPRSSL | 1286.6699 | 643.8386 | 429.5615 | 322.4234 |
| C18B | AATSGNSGTTL | 979.4691 | 490.2382 | 327.1612 | - |
| C26B | QANSGQVW | 889.4162 | 445.2118 | 297.1436 | - |
| C27B | IEDCSSEKAEQQW | 1552.6584 | 776.8328 | 518.2243 | 388.9205 |
| C31B | TSDSNIRETVVKIL | 1574.8748 | 787.9410 | 525.6298 | 394.4746 |
| C40B | DVRASDPSL | 959.4792 | 480.2432 | 320.4979 | - |
| C41B | KQIIL | 614.4235 | 307.7154 | 205.4794 | - |
| C43B | HGDPNQIW | 966.4428 | 483.7250 | 322.8191 | - |
| K#&Chain | Amino Acid Sequence | (M + H)+ | (M + 2H)2+ | (M + 3H)3+ | (M + 4H)4+ |
|---|---|---|---|---|---|
| K71A | STGGTQL | 663.3308 | 332.1690 | 221.7818 | - |
| K15B | PCKSNTDA | 835.3614 | 418.1843 | 279.1253 | - |
| K41B | TSGNSGTTL | 837.3948 | 419.2011 | 279.8031 | - |
| K61B | EDCSSEKA | 868.3353 | 434.6713 | 290.1166 | - |
| K92B | HGDPNQI | 780.3635 | 390.6854 | 260.7927 | - |
| Peptide Marker | Amino Acid Sequence | (M + H)+ | (M + 2H)2+ | (M + 3H)3+ | (M + 4H)4+ |
|---|---|---|---|---|---|
| T7A | VGLPINQR | 896.5312 | 448.7692 | 299.5152 | - |
| T10A | YTFAFGGNYDR | 1310.5800 | 655.7936 | 437.5315 | - |
| T11A | LEQLAGNLR | 1013.5738 | 507.2905 | 338.5294 | - |
| T6B | SNTDANQLWTLK | 1390.6961 | 695.8517 | 464.2369 | - |
| T20B | QIILYPLHGDPNQIWLPLF | 2277.2430 | 1139.1251 | 759.7525 | 570.0667 |
| C2A | TTAGATVQSY | 998.4789 | 499.7431 | 333.4978 | - |
| C18A | TDVQNRY | 895.4268 | 448.2170 | 299.1471 | - |
| VL-C18B | VLAATSGNSGTTL | ||||
| TG13A | VTNAYVVGYR | 1141.6000 | 571.3036 | 381.2048 | - |
| TG26A | LGNGPLE | 699.3671 | 350.1872 | 233.7939 | - |
| TG28A | AISALYYYSTGGTQLPTLAR | 2146.1179 | 1073.5626 | 716.0441 | 537.2849 |
| TG6B-ss-TG9B a | NGLCVD-ss-FHNGNAIQLWPCK | 2145.0003 | 1073.0038 | 715.7154 | 537.0555 |
| TG11B | ANQLWTLK | 973.5465 | 487.2769 | 325.1870 | - |
| TG27B-ss-TG30B a | NCLTSD-ss-ILSCGPASSGQR | 1824.7458 | 912.9094 | 608.9639 | 456.9912 |
| TG37B | QIILYPLHGD | 1168.6360 | 584.8217 | 390.2169 | - |
| C4A | IRAVRGRL | 940.6163 | 470.8118 | 314.2103 | - |
| C31A | STGGTQLPTL | 974.5153 | 487.7613 | 325.5099 | - |
| C4B | HNGNAIQL | 866.4479 | 433.7276 | 289.4875 | - |
| C8B | KRDNTIRSNGKCL | 1504.8012 | 752.9043 | 502.2719 | 376.9562 |
| C18B | AATSGNSGTTL | 979.4691 | 490.2382 | 327.1612 | - |
| C27B | IEDCSSEKAEQQW | 1552.6584 | 776.8328 | 518.2243 | 388.9205 |
| C31B | TSDSNIRETVVKIL | 1574.8748 | 787.9410 | 525.6298 | 394.4746 |
| P7A | PINQRF | 774.4257 | 387.7165 | 258.8134 | - |
| P13A | DVTNAYVVGYRAGNSAYF | 1966.9293 | 983.9683 | 656.3146 | 492.4883 |
| P24A | EEAISAL | 732.3774 | 366.6923 | 244.7973 | - |
| P6B | KRDNTIRSNGKCL | 1504.8012 | 752.9043 | 502.2719 | 376.9562 |
| P23B | DVRASDPSL | 959.4792 | 480.2432 | 320.4979 | - |
| P26B | HGDPNQIWL | 1079.5268 | 540.2671 | 360.5138 | - |
| K71A | STGGTQL | 663.3308 | 332.1690 | 221.7818 | - |
| K15B | PCKSNTDA | 835.3614 | 418.1843 | 279.1253 | - |
| K41B | TSGNSGTTL | 837.3948 | 419.2011 | 279.8031 | - |
| K61B | EDCSSEKA | 868.3353 | 434.6713 | 290.1166 | - |
| K92B | HGDPNQI | 780.3635 | 390.6854 | 260.7927 | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, L.-H.; Liu, C.-C.; Chen, B.; Yan, L.; Yu, H.-L.; Yang, Y.; Wu, J.-N.; Li, X.-S.; Liu, S.-L. LC-HRMS Screening and Identification of Novel Peptide Markers of Ricin Based on Multiple Protease Digestion Strategies. Toxins 2019, 11, 393. https://doi.org/10.3390/toxins11070393
Liang L-H, Liu C-C, Chen B, Yan L, Yu H-L, Yang Y, Wu J-N, Li X-S, Liu S-L. LC-HRMS Screening and Identification of Novel Peptide Markers of Ricin Based on Multiple Protease Digestion Strategies. Toxins. 2019; 11(7):393. https://doi.org/10.3390/toxins11070393
Chicago/Turabian StyleLiang, Long-Hui, Chang-Cai Liu, Bo Chen, Long Yan, Hui-Lan Yu, Yang Yang, Ji-Na Wu, Xiao-Sen Li, and Shi-Lei Liu. 2019. "LC-HRMS Screening and Identification of Novel Peptide Markers of Ricin Based on Multiple Protease Digestion Strategies" Toxins 11, no. 7: 393. https://doi.org/10.3390/toxins11070393
APA StyleLiang, L.-H., Liu, C.-C., Chen, B., Yan, L., Yu, H.-L., Yang, Y., Wu, J.-N., Li, X.-S., & Liu, S.-L. (2019). LC-HRMS Screening and Identification of Novel Peptide Markers of Ricin Based on Multiple Protease Digestion Strategies. Toxins, 11(7), 393. https://doi.org/10.3390/toxins11070393




