1. Introduction
Prebiotic definitions vary among different scientific and political arenas across the world [
1]. Depending on the local definition, nearly all prebiotics can be classified as dietary fiber, but not all fibers are considered prebiotics [
2]. The most recent definition describes a prebiotic as “a substrate that is selectively utilized by host microorganisms conferring a health benefit” [
3]. Functional characteristics of prebiotics include the ability to: resist the low pH of the stomach, resist hydrolysis by mammalian enzymes, resist absorption in the upper gastrointestinal tract, the ability to be fermented by intestinal microbiota and selectively stimulate the growth and/or activity of intestinal bacteria associated with host health and overall well-being [
4,
5]. Inulin, beta-glucans, and xylooligosaccharides all provide health benefits to consumers that are related to the fermentation of these compounds in the distal gastrointestinal tract, and are also considered functional fibers with many other benefits [
6]. As the definition of “prebiotic” broadens to include the overall impact from the metabolism from these compounds, the category of prebiotics will expand [
7]. The importance of displaying direct health benefits due to bacterial fermentation is still the driving mechanism for all prebiotics.
As our awareness and understanding of the importance of the gut microbiome and gut microbiota increases, it is imperative for consumers to understand the key differences between different forms of prebiotics, and where they can be found in various foods and food products. XOS is an emerging prebiotic with well-displayed, consistent health benefits [
8] and is composed of sugar oligomers composed of xylose units [
9], found naturally in fruits, vegetables, milk, honey and bamboo shoots. XOS is commonly produced from xylan containing lignocellulosic materials through various chemical methods, direct enzymatic hydrolysis, or a combination of both treatments [
10,
11,
12,
13,
14]. Inulin is a heterogeneous blend of fructose polymers (degree of polymerization, DP < 10) [
15] which occurs naturally in thousands of plant species, including wheat, onion, bananas, garlic and chicory [
16]. Beta-glucan is a polysaccharide composed of
d-glucose monomers with beta-glycosidic linkages, present in either linear chains in grains, such as oat and barley (up to 7%), or in branched structures in fungi, yeast and certain bacteria [
17]. These prebiotics, or prebiotic mixtures, each provide a unique carbon source for selective stimulation of different bacterial taxa and are important microbiota-shaping compounds.
Because no analytical method currently exists to measure the prebiotic capacity of foods in terms of their influence on gastrointestinal taxa, this field relies heavily on fecalbiotics (living or once living fecal microbial populations) to quantify the effects of these compounds. In vitro fermentation models allow for quantitative analysis of specific materials and are semi-representative models of colonic fermentation [
18]. Although not a complete substitute for human studies, when paired with in vivo models, in vitro analysis can be an accurate systematic approach to analyzing different parameters and end points in colonic fermentation [
19].
With the recent release of the International Scientific Association for Probiotics and Prebiotics consensus statement, XOS has been categorized as a prebiotic or prebiotic candidate [
3]. The prebiotic effects of XOS have previously been summarized by Broekaert et al. [
20]. While there is less evidence supporting the prebiotic effects of XOS compared to other types of prebiotics, studies have shown that XOS supplementation in humans can increase SCFA and bifidobacteria, as well as improve stool consistency and frequency [
21,
22,
23]. This paper compares the fermentation effects of XOS to previously established prebiotics (inulin and beta-glucans) in a controlled in vitro model. To the authors’ knowledge, this is the first controlled in vitro study comparing the effects of XOS to these known prebiotics. The objective of this project was to compare currently available prebiotics by their ability to change specific taxa as well as compare differences in the production of gas and common short chain fatty acids (SCFA) between these products. Inulin, XOS and beta-glucan based products were chosen for this experiment because they are established and emerging prebiotics that are commonly consumed, and offer well-demonstrated health benefits to their consumers.
2. Materials and Methods
2.1. Prebiotic Dietary Fibers Analyzed
Five common prebiotic dietary fibers were chosen for this study (
Table 1), including different types of beta-glucans, inulin and xylooligosaccharide supplements.
2.2. Donor Information
Fecal samples were collected from three healthy volunteers (2 males, 1 female) under anaerobic conditions. Donors included individuals (ages 22–28) consuming non-specific Western diets, who do not consume any supplements, including fiber supplements. Donors were non-smokers, did not receive any antibiotic treatments in the last year, and were not affected by any known gastrointestinal diseases (
Table 2).
2.3. Fecal Collection
Fecal samples were anaerobically collected within 5 min of the start of the fermentation (Medline Specimen Collection Kit, Medline, Inc., Rogers, MN, USA), and homogenized immediately upon collection. All data and samples collected were done in accordance with University of Minnesota policies and procedures.
2.4. Fermentation
The fiber samples were fermented using in vitro methods to mimic the environment of the distal colon. These methods have been used in previous in vitro studies, including Koecher et al., who found complementary results between these in vitro methods and a human intervention study of the same fibers [
24]. Fiber samples (0.5 g) were hydrated in 40 mL of prepared sterile trypticase peptone fermentation media in 100 mL serum bottles, capped, and incubated for 12 h at 4 °C to limit possibility of microbial growth [
25]. Following incubation, serum bottles were transferred to a circulating water bath at 37 °C for 2 h to allow the samples to reach body temperature. Post-collection, fecal samples were mixed using a 6:1 ratio of phosphate buffer solution to fecal sample. After mixing, obtained fecal slurry was combined with prepared reducing solution (2.52 g cysteine hydrochloride, 16 mL 1 N NaOH, 2.56 g sodium sulfide nonanhydride, 380 mL DD H
2O) at a 2:15 ratio. 10 mL of the prepared fecal inoculum was added to each of the serum bottles, 0.8 mL Oxyrase
® was added, flushed with CO
2, sealed, and then immediately placed in a 37 °C circulating water bath. Fecal inoculum control samples with no fiber added were prepared for SCFA and gas production comparison. Baseline pH of the fermentation media was measured, with a mean of 6.83 ± 0.04, to mimic the environment of the distal colon. Samples were prepared in triplicate and analyzed at 0, 12 and 24 h. Upon removal at each time point, total gas volume was measured. Then samples were divided into aliquots for analysis and 1 mL of copper sulfate (200 g/L) was added to cease fermentation. All samples were immediately frozen and stored at −80 °C for further analysis.
2.5. SCFA Analysis
SCFA samples were extracted according to Schneider et al. [
26] with minor modifications, and analyzed with previously described methods [
27].
2.6. DNA Extractions
Fecal bacteria DNA from the in vitro system were extracted using a PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) following the provided operating instruction, including bead beating for 20 min.
2.6.1. Primary/Secondary Amplification
The V1-V3 region of the 16S rRNA was amplified using a two-step PCR protocol. The primary amplification was done using an ABI7900 qPCR machine (Applied Biosystems, Foster City, CA, USA). The following recipe was used: 3 μL template DNA, 0.48 μL nuclease-free water, 1.2 μL 5× KAPA HiFi buffer (Kapa Biosystems, Woburn, MA, USA), 0.18 μL 10 mM dNTPs (Kapa Biosystems, Woburn, MA, USA), 0.3 μL DMSO (Fisher Scientific, Waltham, MA, USA), 0.12 μL ROX (25 μM) (Life Technologies, Carlsbad, CA, USA), 0.003 μL 1000× SYBR Green, 0.12 μL KAPA HiFi Polymerase (Kapa Biosystems, Woburn, MA, USA), 0.3 μL forward primer (10 μM), 0.3 μL reverse primer (10 μM). Cycling conditions were: 95 °C for 5 min, followed by 20 cycles of 98 °C for 20 s, 55 °C for 15 s, and 72 °C for 1 min. The primers for the primary amplification contained both 16S-specific primers (V1_27F and V3_V34R), as well as adapter tails for adding indices and Illumina flow cell adapters in a secondary amplification. The following primers were used (16S-specific sequences in bold): Meta_V1_27F (TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGAGTTTGATCMTGGCTCAG) and Meta_V3_534R (GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGATTACCGCGGCTGCTGG).
The amplicons from the primary PCR were diluted 1:100 in sterile, nuclease-free water, and a second PCR reaction was set up to add the Illumina flow cell adapters (Illumina Inc., San Diego, CA, USA) and indices. The secondary amplification was done on a fixed block BioRad Tetrad PCR machine (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using the following recipe: 5 μL template DNA, 1 μL nuclease-free water, 2 μL 5× KAPA HiFi buffer (Kapa Biosystems, Woburn, MA, USA), 0.3 μL 10 mM dNTPs (Kapa Biosystems, Woburn, MA, USA), 0.5 μL DMSO (Fisher Scientific, Waltham, MA, USA) 0.2 μL KAPA HiFi Polymerase (Kapa Biosystems, Woburn, MA, USA), 0.5 μL forward primer (10 μM), 0.5 μL reverse primer (10 μM). Cycling conditions were: 95 °C for 5 min, followed by 10 cycles of 98 °C for 20 s, 55 °C for 15 s, 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. The following indexing primers were used (X indicates the positions of the 8 bp indices): Forward indexing primer: AATGATACGGCGACCACCGAGATCTACACXXXXXXXXTCGTCGGCAGCGTC and Reverse indexing primer: CAAGCAGAAGACGGCATACGAGATXXXXXXXXGTCTCGTGGGCTCGG.
2.6.2. Normalization and Sequencing
The samples were normalized using a SequalPrep capture-resin bead plate (Life Technologies, Carlsbad, CA, USA) and pooled using equal volume. The final pools were quantified via PicoGreen dsDNA assay (Life Technologies, Carlsbad, CA, USA) and diluted to 2 nM. 10 μL of the 2 nM pool was denatured with 10 μL of 0.2 N NaOH, diluted to 8 pM in Illumina’s HT1 buffer, spiked with 15% phiX, heat denatured at 96 °C for 2 min, and sequenced using a MiSeq 600 cycle v3 kit (Illumina, San Diego, CA, USA).
2.6.3. Sequence Processing and Analysis
Generated sequence data was processed and analyzed using QIIME [
28]. Fastq sequence data was processed with the University of Minnesota’s gopher-pipeline for metagenomics [
29]. Sequence data had adapters removed and sliding quality trimming window by Trimmomatic [
30]; primers removed and overlapping reads merged by Pandaseq [
31]. Within QIIME, chimera checking done by chimera slayer, Open reference OTU picking completed with Usearch61, taxonomic identification using GreenGenes (Version 13.8) reference database, rarefied to 14,393 sequences per sample. Analysis was performed using
R (
R Development Core Team, Vienna, Austria, 2012).
2.7. Statistical Analysis
All statistical analysis was performed using R software (Version 3.2.2, R Development Core Team, Vienna, Austria, 2012). Differences in means were determined using the Kruskal-Wallis ANOVA test, testing the null hypothesis that the location parameter of the groups of abundancies for a given OTU is the same. Multiple comparisons were corrected using the Benjamini-Hochberg FDR (false discovery rate) procedure for multiple comparisons. For gas and SCFA data, ANOVA with Tukey HSD was used to compare means. Significance was set for p-values < 0.05 for all statistical tests.
2.8. Consent Ethics Approval Code
Voluntary informed consent was obtained from all fecal donors prior to this study according to University of Minnesota policies and procedures.
4. Discussion
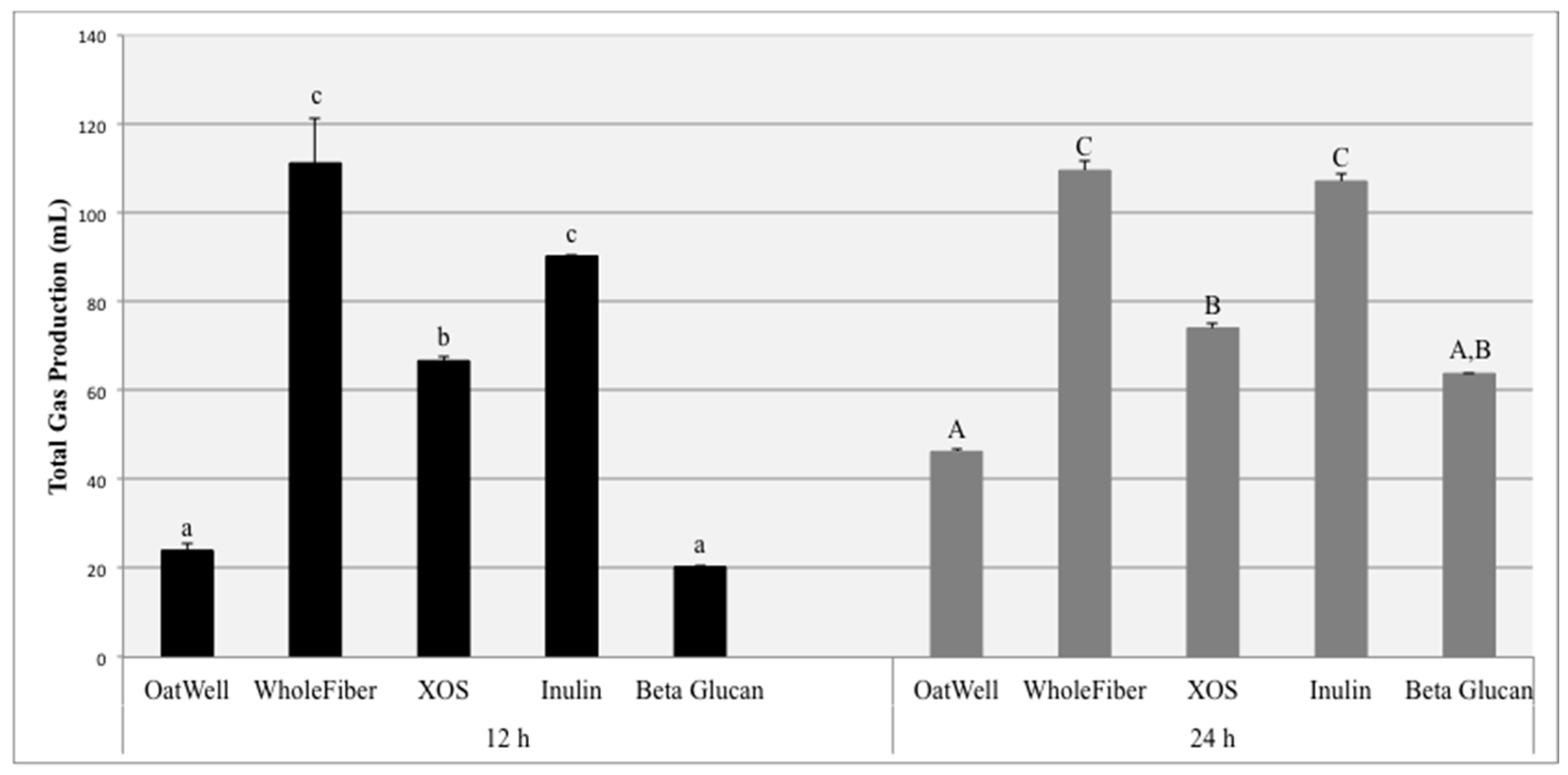
The aim of this study was to investigate the beneficial effects of commonly consumed prebiotic dietary fibers, including their ability to influence the growth of identified bacterial populations, form beneficial SCFAs, and the amount of gas they produce due to fermentation. Total gas production due to fiber fermentation depends on a wide range of factors. The inulin samples and the WholeFiber samples (mixture of dried chicory root inulin, pectin and hemi/cellulose) resulted in the highest gas production at both 12 and 24 h. These results are consistent with results from both clinical feeding studies and other in vitro experiments, in which fermentation of inulin products resulted in high amounts of gas production, sometimes resulting in mild negative gastrointestinal symptoms, depending on the dosage [
32,
33]. Similar in vitro studies have found inulin to be much more fermentable than beta-glucan products, for both barley and oat-derived beta-glucans [
34]. XOS fermentation results in less gas production than the inulin products, and more gas than beta-glucan products. Because of these findings, previous studies based on digestive tolerance and parameters have established a tolerated daily dosage for XOS of approximately 12 g/day [
35].
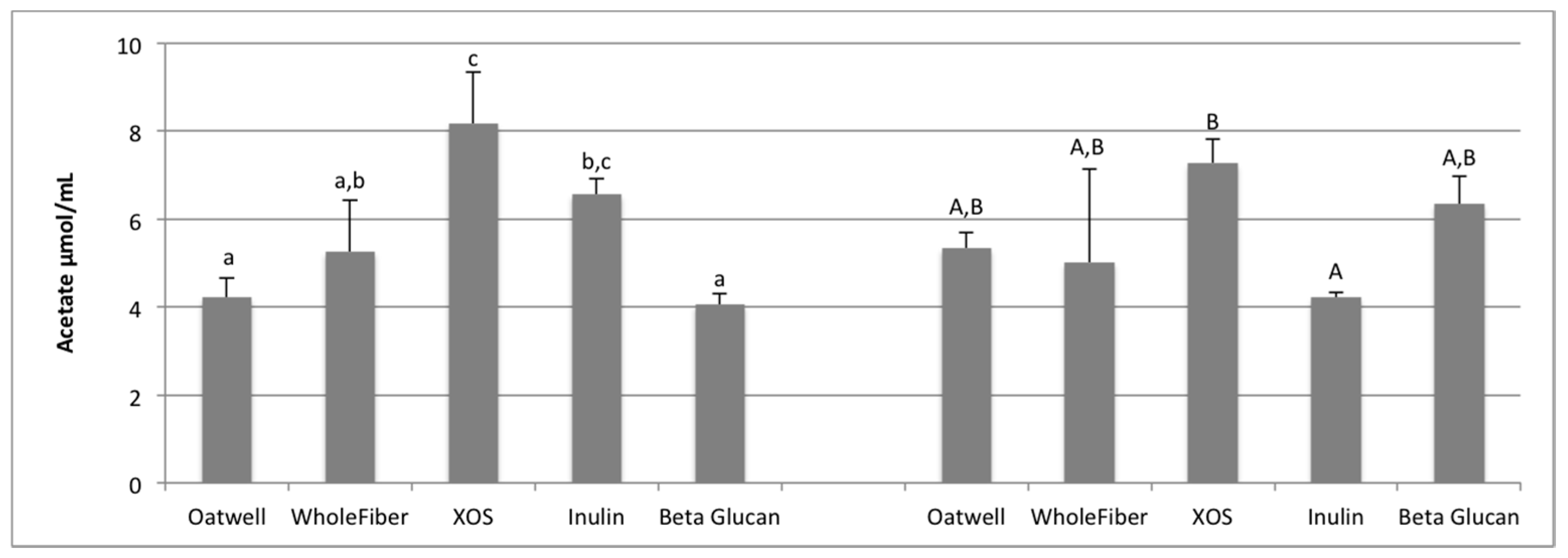
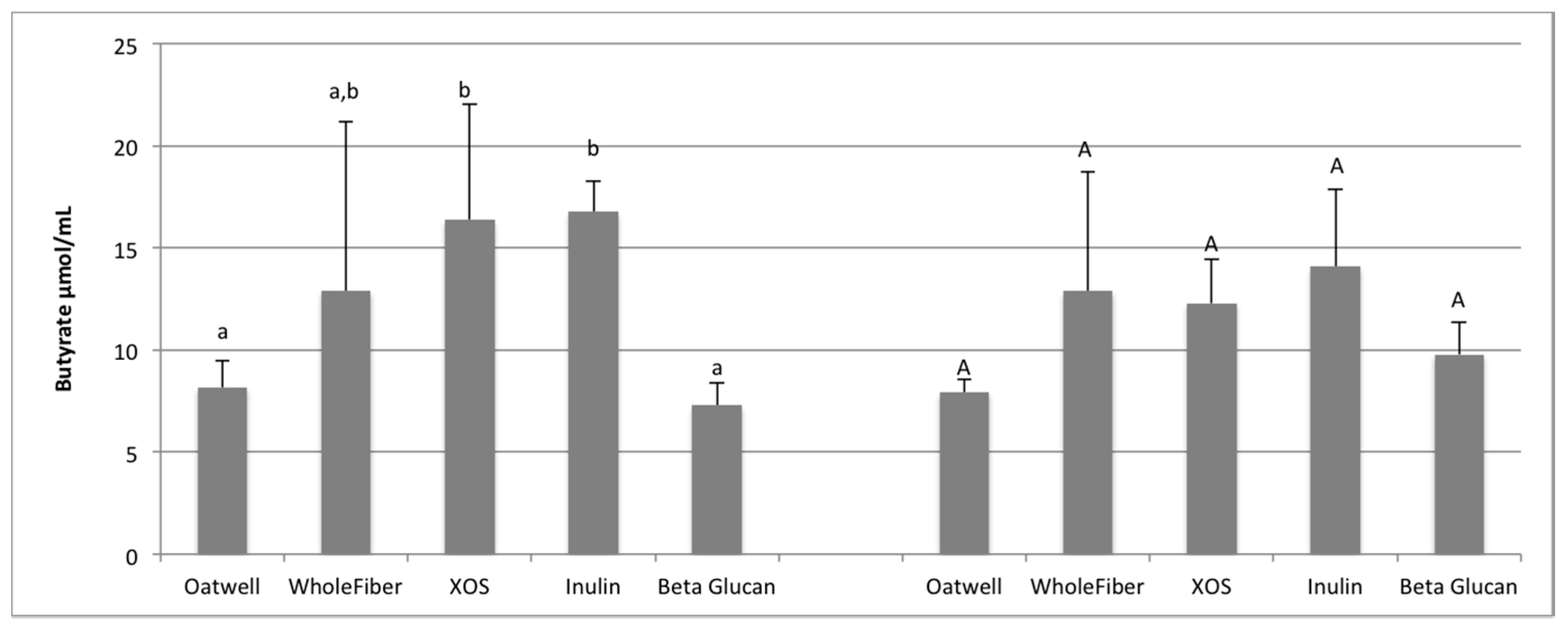
SCFA production due to the fermentation of prebiotic dietary fibers promotes many beneficial health outcomes to the host. SCFA production may contribute to up to 10% of the host’s metabolizable energy daily, with production of total SCFAs usually between 100–200 mmol/day, but is highly dependent on the donor and availability of substrates for fermentation [
36,
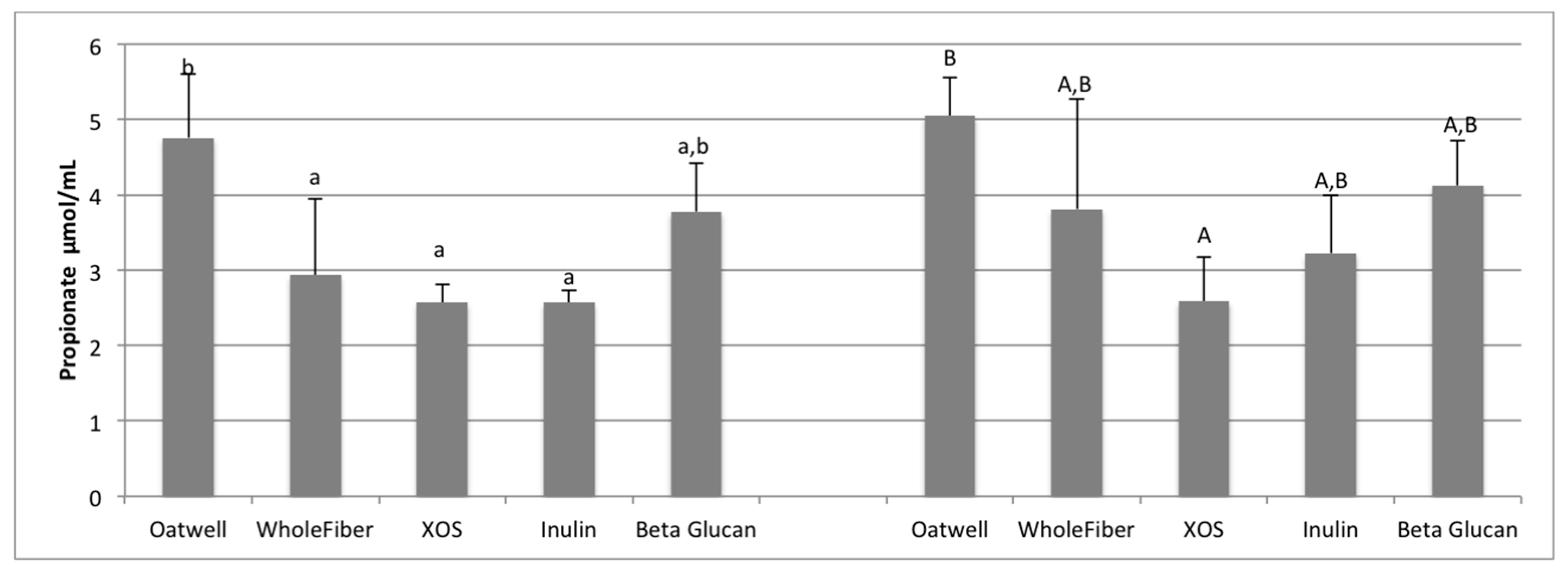
37]. At 12 h of fermentation, the OatWell and beta-glucan samples had significantly higher concentrations of propionate, and the highest mean concentration at 24 h, compared to the other prebiotic dietary fibers analyzed. Similar in vitro studies with beta-glucan based products have also shown similar preference for these products to result in propionate formation [
34]. Although no mechanism has been identified, and studies show conflicting results [
38], elevated serum propionate concentrations have been shown to have a hypocholesterolaemic effect [
39]. Propionate may also play an influential role in satiety, although mechanisms still remain unclear [
40,
41]. Cholesterol-lowering properties of beta-glucans may be limited to effects from the upper-GI, although many propionate-producing bacteria have a preference to fermenting various types of beta-glucans (
Bacteroides,
Prevotella,
Clostridium) based on the presence of genes responsible for endo-β-glucanase enzyme production [
42].
Microbial diversity among fecal donors complicates the identification of trends among the five treatment groups (
Supplemental Figures S8 and S9). In terms of taxonomic shift, the inulin-based products were fermented nearly identically by all three fecal donors. Both pure inulin and WholeFiber promoted the growth of
Collinsella comparing the 24 h samples to the 0 h samples. Inulin-type fructans have been shown in clinical studies to promote substantial growth of
Collinsella, paralleled with increased urinary hippurate levels [
43]. Hippurate is a metabolite derived from various fermentation processes in the gut that has been found in decreased concentrations in obese individuals compared to lean individuals, and also between diabetics and non-diabetics [
44,
45,
46]. The genus
Collinsella has been found in lower concentrations in individuals with IBD compared to healthy controls [
47], while
Collinsella aerofaciens has been associated with low risk of colorectal cancer [
48]. Increases in
Collinsella and increased urinary hippurate levels are considered a beneficial effect of inulin consumption due to its prebiotic capacity [
43]. In vivo studies with inulin, scFOS and resistant starch supplementation have found decreases in the SMB53 genus, consistent with the OatWell treatment in the present study [
30,
31].
A significant increase in the genus
Bifidobacterium was observed only with the XOS treatment. Rycroft et al. found a similar affinity of
Bifidobacterium toward XOS [
49]. However, previous studies have shown inulin to also stimulate the growth of
Bifidobacterium [
49,
50,
51,
52]. While there was an overall rise in
Bifidobacterium with the inulin treatment group between 0 h and 24 h (1.33, 5.44 OTUs respectively), this study did not find that increase to be significant (
p = 0.304). The small sample size and individual microbiome variability likely played a role in this result. Increases in
Bifidobacterium have been heavily studied and reviewed, and are considered a beneficial effect due to their correlation with many positive health outcomes [
1].
Bifidobacteria reside naturally in the gastrointestinal tract of healthy human adults and have a strong affinity to ferment oligosaccharides, making them a common marker for prebiotic capacity.
Bifidobacterium is a unique genus of bacteria in that no gas is formed as an end product of metabolism [
53]. Like
Lactobacillus, these bacteria are saccharolytic, often considered a beneficial trait [
54].
Bifidobacteria also do not produce any known carcinogenic substances in vivo.
Bifidobacteria concentrations have been negatively associated with obesity and weight gain [
55,
56,
57,
58]. Increases in
Bifidobacteria have also been correlated with a decrease in blood lipopolysaccharides (LPS), inflammatory reagents that play a role in the development of inflammatory metabolic disorders and conditions, and are primarily found in gram-negative bacteria [
59]. LPS induce the activation of Toll-like receptor 4 (TLR4), which leads to inflammation due to release of pro-inflammatory cytokines and chemokines [
60].
In vitro fermentations are semi-representative models of colonic fermentation, but have limitations [
18]. This study did not include an in vitro digestion process, which would remove digestible contents from the samples prior to fermentation, and would be a more representative model. However, because the test substrates are primarily fiber, which is non-digestible, this should have minimal impact on the results of this study. In vivo, formed gases are continually absorbed and colonic absorption is rapid. Because SCFAs are rapidly absorbed and difficult to measure, in vitro models help to understand the kinetics of colonic fermentation. However, in vitro models must be paired with similar in vivo models to better understand the full mechanisms of action resulting from colonic fermentation of prebiotic dietary fibers. Because the SCFA are not absorbed in in vitro models, the SCFA produced can alter the pH of the fermentation media. While the fermentation media was designed to mimic the pH of the distal colon at baseline, the pH of the media in this study was not further controlled throughout the experiment. This is another limitation of this in vitro model. An additional limitation of this study was the small sample size. The present study was conducted using fecal inoculum from only three donors. Due to the individual variability between the donor’s microbiota (
Figure S7), a larger sample size may be needed to achieve a more representative view of the effects of each of the fibers.