One-Week Elderberry Juice Intervention Promotes Metabolic Flexibility in the Transcriptome of Overweight Adults During a Meal Challenge
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Clinical Trial
2.2. RNA Sequencing
2.3. Read Processing and Alignment
2.4. Differential Gene Expression Analysis
2.5. Pathway-Based Filtering of Differentially Expressed Genes
2.6. Gene Ontology Enrichment Analysis
2.7. Gene Set Enrichment Analysis
3. Results
3.1. DEG Main Effects and Contrasts
3.2. Metabolic Flexibility Gene Expression
3.3. Gene Ontology Enrichment Analysis
3.4. Gene Set Enrichment Analysis
4. Discussion
4.1. Systemic Absorption and Bioavailability of Anthocyanins from EBJ
4.2. Elderberry Juice and Blood Glucose Homeostasis
4.3. Lipid Metabolism, FAO Suppression, and Mitochondrial Function
4.4. Fatty Acid Oxidation and Mitochondrial Modulation
4.5. Insulin and Nutrient-Sensing Signaling
4.6. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPK | AMP-activated protein kinase |
| BP | Biological Process (Gene Ontology category) |
| CC | Cellular Component (Gene Ontology category) |
| C3GE | Cyanidin-3-glucoside equivalents |
| DEG | Differentially expressed gene |
| EBJ | Elderberry juice |
| FAO | Fatty acid oxidation |
| FC | Fold change |
| FDR | False discovery rate |
| FoxO | Forkhead box O |
| GSEA | Gene set enrichment analysis |
| GO | Gene Ontology |
| HK2 | Hexokinase 2 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | Mitogen-activated protein kinase |
| MF | Molecular Function (Gene Ontology category) |
| mTOR | Mechanistic target of rapamycin |
| MTT | Meal tolerance test |
| NES | Normalized enrichment score |
| PBMC | Peripheral blood mononuclear cell |
| PDK2 | Pyruvate dehydrogenase kinase 2 |
| PDK4 | Pyruvate dehydrogenase kinase 4 |
| PI3K–AKT | Phosphoinositide 3-kinase–protein kinase B |
| PL | Placebo |
| PPARGC1B | Peroxisome proliferator-activated receptor gamma coactivator 1-beta |
| RQ | Respiratory quotient |
| TCA | Tricarboxylic acid |
| TOR | Target of rapamycin |
| VST | Variance-stabilizing transformation |
References
- Galgani, J.E.; Bergouignan, A.; Rieusset, J.; Moro, C.; Nazare, J.A. Editorial: Metabolic Flexibility. Front. Nutr. 2022, 9, 946300. [Google Scholar] [CrossRef] [PubMed]
- Palmer, B.F.; Clegg, D.J. Metabolic Flexibility and Its Impact on Health Outcomes. Mayo Clin. Proc. 2022, 97, 761–776. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.L.; Soeters, M.R.; Wüst, R.C.I.; Houtkooper, R.H. Metabolic Flexibility as an Adaptation to Energy Resources and Requirements in Health and Disease. Endocr. Rev. 2018, 39, 489–517. [Google Scholar] [CrossRef] [PubMed]
- Goodpaster, B.H.; Sparks, L.M. Metabolic flexibility in health and disease. Cell Metab. 2017, 25, 1027–1036. [Google Scholar] [CrossRef]
- Tareen, S.H.; Kutmon, M.; Arts, I.C.; De Kok, T.M.; Evelo, C.T.; Adriaens, M.E. Logical modelling reveals the PDC-PDK interaction as the regulatory switch driving metabolic flexibility at the cellular level. Genes Nutr. 2019, 14, 27. [Google Scholar] [CrossRef]
- Nistor, M.; Pop, R.; Daescu, A.; Pintea, A.; Socaciu, C.; Rugina, D. Anthocyanins as Key Phytochemicals Acting for the Prevention of Metabolic Diseases: An Overview. Molecules 2022, 27, 4254. [Google Scholar] [CrossRef]
- Riordan, J.; Solverson, P. Berry Anthocyanins in Rodent and Human Obesity and Diabetes: A Review of the Evidence. BioMed 2022, 2, 210–237. [Google Scholar] [CrossRef]
- Sharma, S.; Pandita, G.; Bhosale, Y.K. Anthocyanin: Potential tool for diabetes management and different delivery aspects. Trends Food Sci. Technol. 2023, 140, 104170. [Google Scholar] [CrossRef]
- Osman, A.G.; Avula, B.; Katragunta, K.; Ali, Z.; Chittiboyina, A.G.; Khan, I.A. Elderberry Extracts: Characterization of the Polyphenolic Chemical Composition, Quality Consistency, Safety, Adulteration, and Attenuation of Oxidative Stress- and Inflammation-Induced Health Disorders. Molecules 2023, 28, 3148. [Google Scholar] [CrossRef]
- Teets, C.; Ghanem, N.; Ma, G.; Minj, J.; Perkins-Veazie, P.; Johnson, S.A.; Etter, A.J.; Carbonero, F.G.; Solverson, P.M. A One-Week Elderberry Juice Intervention Augments the Fecal Microbiota and Suggests Improvement in Glucose Tolerance and Fat Oxidation in a Randomized Controlled Trial. Nutrients 2024, 16, 3555. [Google Scholar] [CrossRef]
- Abueg, L.A.L.; Afgan, E.; Allart, O.; Awan, A.H.; Bacon, W.A.; Baker, D.; Bassetti, M.; Batut, B.; Bernt, M.; Blankenberg, D.; et al. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Carlson, M. org.Hs.eg.db: Genome Wide Annotation for Human. Available online: https://bioconductor.org/packages/org.Hs.eg.db/ (accessed on 7 August 2025).
- Lim, C.-Y.; In, J. Considerations for crossover design in clinical study. Korean J. Anesthesiol. 2021, 74, 293–299. [Google Scholar] [CrossRef]
- Duda, J.C.; Drenda, C.; Kästel, H.; Rahnenführer, J.; Kappenberg, F. Benefit of using interaction effects for the analysis of high-dimensional time-response or dose-response data for two-group comparisons. Sci. Rep. 2023, 13, 20804. [Google Scholar] [CrossRef]
- Vestal, B.E.; Wynn, E.; Moore, C.M. lmerSeq: An R package for analyzing transformed RNA-Seq data with linear mixed effects models. BMC Bioinform. 2022, 23, 489. [Google Scholar] [CrossRef]
- Laurent, G.S.; Shtokalo, D.; Tackett, M.R.; Yang, Z.; Vyatkin, Y.; Milos, P.M.; Seilheimer, B.; McCaffrey, T.A.; Kapranov, P. On the importance of small changes in RNA expression. Methods 2013, 63, 18–24. [Google Scholar] [CrossRef]
- Costa, A.; van der Stelt, I.; Reynés, B.; Konieczna, J.; Fiol, M.; Keijer, J.; Palou, A.; Romaguera, D.; van Schothorst, E.M.; Oliver, P. Whole-Genome Transcriptomics of PBMC to Identify Biomarkers of Early Metabolic Risk in Apparently Healthy People with Overweight-Obesity and in Normal-Weight Subjects. Mol. Nutr. Food Res. 2023, 67, 2200503. [Google Scholar] [CrossRef] [PubMed]
- Tareen, S.H.K.; Adriaens, M.E.; Arts, I.C.W.; De Kok, T.M.; Vink, R.G.; Roumans, N.J.T.; Van Baak, M.A.; Mariman, E.C.M.; Evelo, C.T.; Kutmon, M. Profiling Cellular Processes in Adipose Tissue During Weight Loss Using Time Series Gene Expression. Genes 2018, 9, 525. [Google Scholar] [CrossRef] [PubMed]
- Solverson, P. Anthocyanin Bioactivity in Obesity and Diabetes: The Essential Role of Glucose Transporters in the Gut and Periphery. Cells 2020, 9, 2515. [Google Scholar] [CrossRef] [PubMed]
- Tareen, S.H.K.; Kutmon, M.; Adriaens, M.E.; Mariman, E.C.M.; De Kok, T.M.; Arts, I.C.W.; Evelo, C.T. Exploring the cellular network of metabolic flexibility in the adipose tissue. Genes. Nutr. 2018, 13, 17. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Reynés, B.; Priego, T.; Cifre, M.; Oliver, P.; Palou, A. Peripheral blood cells, a transcriptomic tool in nutrigenomic and obesity studies: Current state of the art. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1006–1020. [Google Scholar] [CrossRef]
- O’Grada, C.M.; Morine, M.J.; Morris, C.; Ryan, M.; Dillon, E.T.; Walsh, M.; Gibney, E.R.; Brennan, L.; Gibney, M.J.; Roche, H.M. PBMCs reflect the immune component of the WAT transcriptome—implications as biomarkers of metabolic health in the postprandial state. Mol. Nutr. Food Res. 2014, 58, 808–820. [Google Scholar] [CrossRef]
- Inami, O.; Tamura, I.; Kikuzaki, H.; Nakatani, N. Stability of anthocyanins of Sambucus canadensis and Sambucus nigra. J. Agric. Food Chem. 1996, 44, 3090–3096. [Google Scholar] [CrossRef]
- Jokioja, J.; Yang, B.; Linderborg, K.M. Acylated anthocyanins: A review on their bioavailability and effects on postprandial carbohydrate metabolism and inflammation. Compr. Rev. Food Sci. Food Saf. 2021, 20, 5570–5615. [Google Scholar] [CrossRef]
- Pascariu, O.-E.; Israel-Roming, F. Bioactive Compounds from Elderberry: Extraction, Health Benefits, and Food Applications. Processes 2022, 10, 2288. [Google Scholar] [CrossRef]
- Lee, J.; Finn, C.E. Anthocyanins and other polyphenolics in American elderberry (Sambucus canadensis) and European elderberry (S. nigra) cultivars. J. Sci. Food Agric. 2007, 87, 2665–2675. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Kortesniemi, M.K.; Linderborg, K.M.; Yang, B. Anthocyanins as Promising Molecules Affecting Energy Homeostasis, Inflammation, and Gut Microbiota in Type 2 Diabetes with Special Reference to Impact of Acylation. J. Agric. Food Chem. 2023, 71, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Roma-Rodrigues, C.; Santos, A.; Veigas, B.; Brás, N.; Faria, A.; Calhau, C.; de Freitas, V.; Baptista, P.V.; Mateus, N. GLUT1 and GLUT3 involvement in anthocyanin gastric transport-Nanobased targeted approach. Sci. Rep. 2019, 9, 789. [Google Scholar] [CrossRef] [PubMed]
- Mueckler, M.; Thorens, B. The SLC2 (GLUT) family of membrane transporters. Mol. Asp. Med. 2013, 34, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, W.Y.; Guertin, D.A. De Novo Lipogenesis as a Source of Second Messengers in Adipocytes. Curr. Diab. Rep. 2019, 19, 138. [Google Scholar] [CrossRef]
- Jiang, S.; Liu, H.; Li, C. Dietary Regulation of Oxidative Stress in Chronic Metabolic Diseases. Foods 2021, 10, 1854. [Google Scholar] [CrossRef]
- Yang, W.; Jiang, W.; Guo, S. Regulation of macronutrients in insulin resistance and glucose homeostasis during type 2 diabetes mellitus. Nutrients 2023, 15, 4671. [Google Scholar] [CrossRef]
- Randeni, N.; Luo, J.; Xu, B. Critical Review on Anti-Obesity Effects of Anthocyanins Through PI3K/Akt Signaling Pathways. Nutrients 2025, 17, 1126. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierliński, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef]
- Smolders, L.; Mensink, R.P.; Boekschoten, M.V.; De Ridder, R.J.J.; Plat, J. The acute effects on duodenal gene expression in healthy men following consumption of a low-fat meal enriched with theobromine or fat. Sci. Rep. 2018, 8, 1700. [Google Scholar] [CrossRef]
- Sagaya, F.M.; Hurrell, R.F.; Vergères, G. Postprandial blood cell transcriptomics in response to the ingestion of dairy products by healthy individuals. J. Nutr. Biochem. 2012, 23, 1701–1715. [Google Scholar] [CrossRef]
- Davis, R.; Murgia, C.; Dordevic, A.L.; Bonham, M.P.; Huggins, C.E. Diurnal variation in gene expression of human peripheral blood mononuclear cells after eating a standard meal compared with a high protein meal: A cross-over study. Clin. Nutr. 2021, 40, 4349–4359. [Google Scholar] [CrossRef]
- Gui, H.; Sun, L.; Liu, R.; Si, X.; Li, D.; Wang, Y.; Shu, C.; Sun, X.; Jiang, Q.; Qiao, Y.; et al. Current knowledge of anthocyanin metabolism in the digestive tract: Absorption, distribution, degradation, and interconversion. Crit. Rev. Food Sci. Nutr. 2023, 63, 5953–5966. [Google Scholar] [CrossRef]
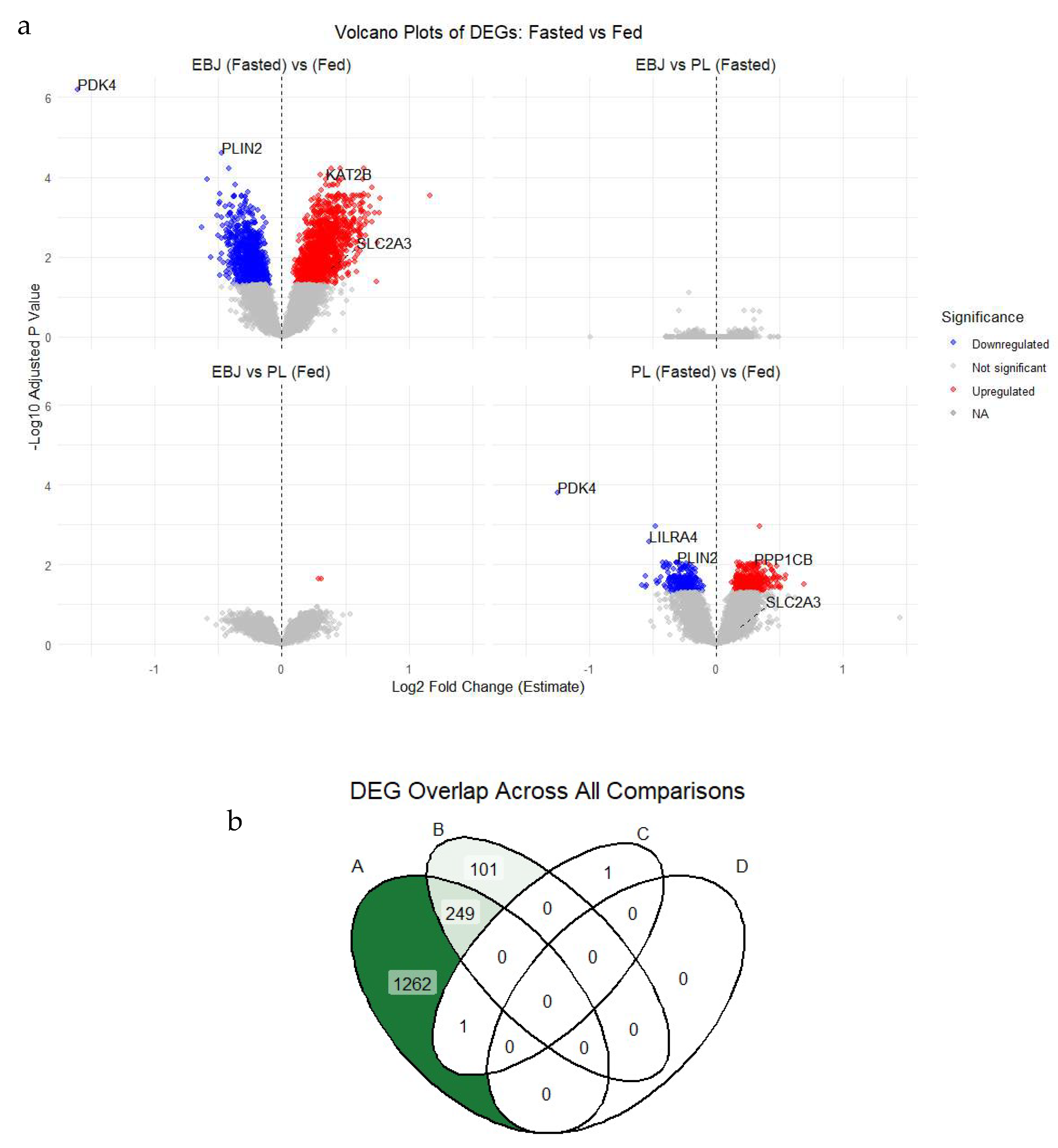
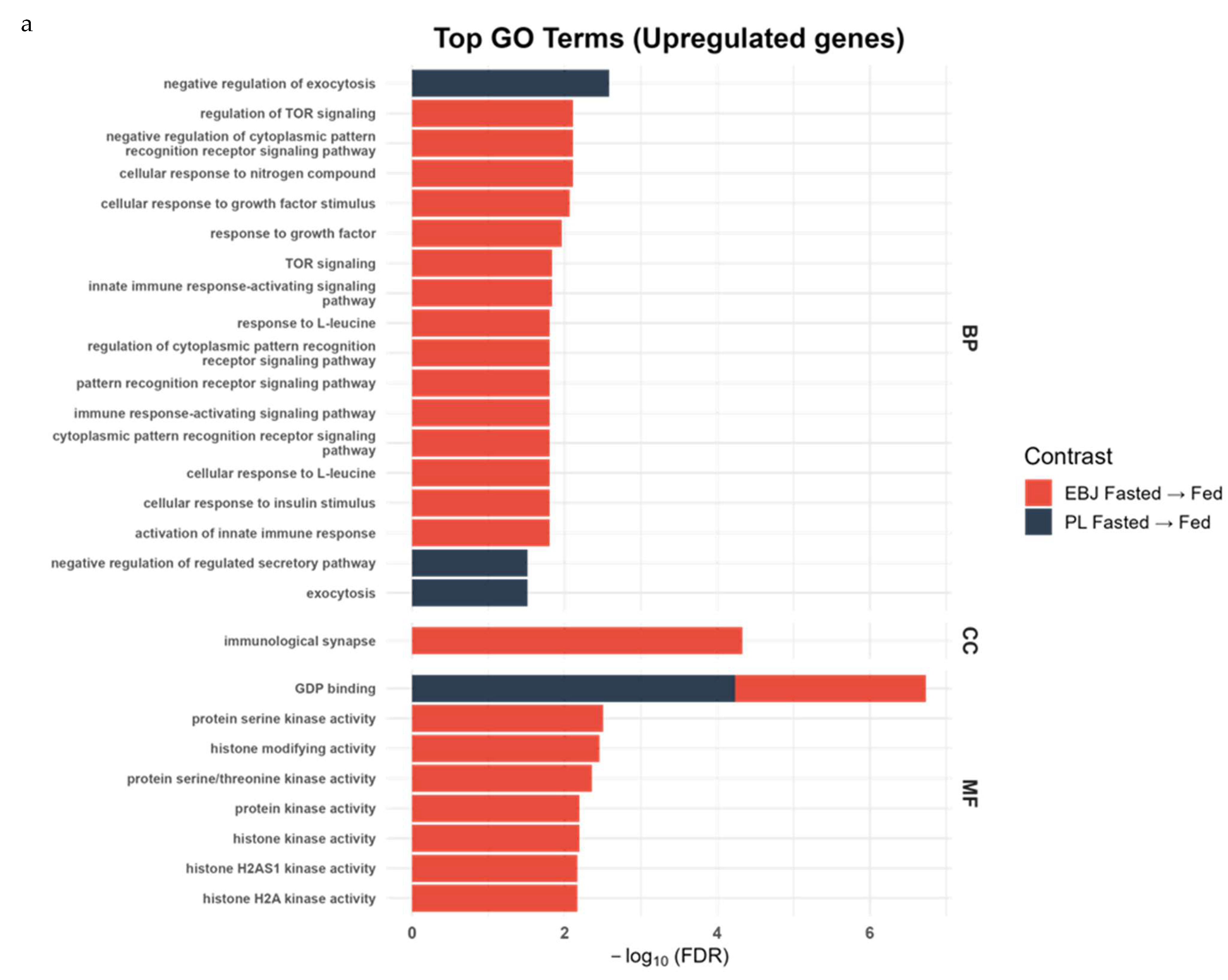

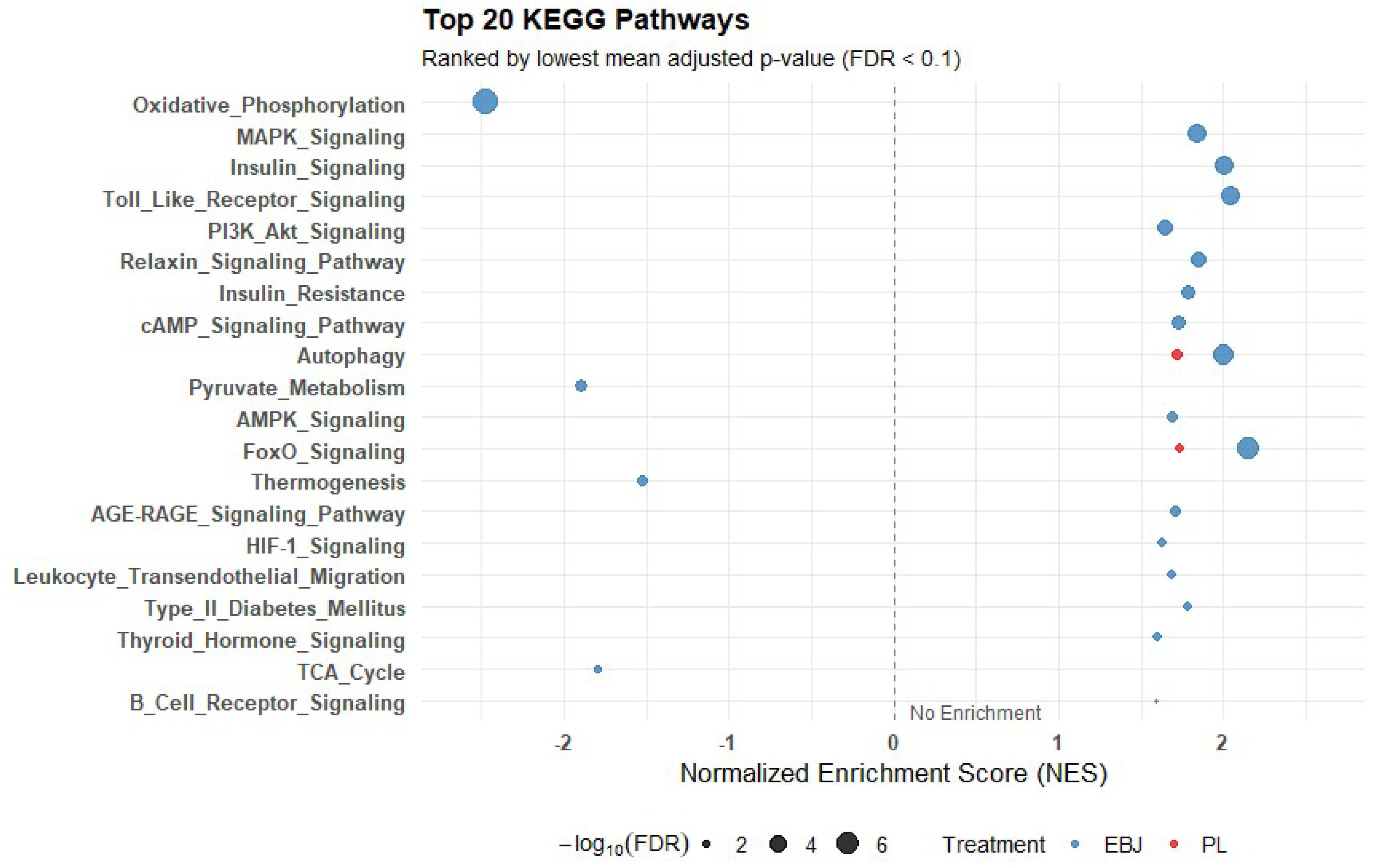
| Pathway | EBJ Count | PL Count | Shared Count | FDR |
|---|---|---|---|---|
| Insulin Signaling | 18 | 7 | 6 | 1.56E-05 |
| FoxO Signaling | 8 | 3 | 3 | <0.001 |
| Literature Curation | 4 | 2 | 2 | 0.001 |
| MAPK Signaling | 21 | 8 | 5 | 0.001 |
| Circadian Rhythm | 4 | 2 | 2 | 0.001 |
| PI3K–Akt Signaling | 29 | 9 | 5 | 0.009 |
| Autophagy | 17 | 5 | 3 | 0.013 |
| Cytokine–Cytokine Receptor Interaction | 19 | 5 | 3 | 0.015 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teets, C.; Etter, A.J.; Solverson, P.M. One-Week Elderberry Juice Intervention Promotes Metabolic Flexibility in the Transcriptome of Overweight Adults During a Meal Challenge. Nutrients 2025, 17, 3142. https://doi.org/10.3390/nu17193142
Teets C, Etter AJ, Solverson PM. One-Week Elderberry Juice Intervention Promotes Metabolic Flexibility in the Transcriptome of Overweight Adults During a Meal Challenge. Nutrients. 2025; 17(19):3142. https://doi.org/10.3390/nu17193142
Chicago/Turabian StyleTeets, Christy, Andrea J. Etter, and Patrick M. Solverson. 2025. "One-Week Elderberry Juice Intervention Promotes Metabolic Flexibility in the Transcriptome of Overweight Adults During a Meal Challenge" Nutrients 17, no. 19: 3142. https://doi.org/10.3390/nu17193142
APA StyleTeets, C., Etter, A. J., & Solverson, P. M. (2025). One-Week Elderberry Juice Intervention Promotes Metabolic Flexibility in the Transcriptome of Overweight Adults During a Meal Challenge. Nutrients, 17(19), 3142. https://doi.org/10.3390/nu17193142






