Effect of Alpha-1 Antitrypsin Deficiency on Zinc Homeostasis Gene Regulation and Interaction with Endoplasmic Reticulum Stress Response-Associated Genes
Highlights
- The expression of the Alpha-1 antitrypsin mutant Z may influence zinc metabolism in liver cells.
- Bayesian network analysis uncovered novel gene-to-gene interactions among zinc transporters, as well as between genes involved in zinc homeostasis and the unfolded protein response.
- Patients with Alpha-1 antitrypsin deficiency may have altered zinc metabolism.
- Genes involved in zinc homeostasis may serve as potential therapeutic targets for Alpha-1 antitrypsin deficiency treatment.
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Labile Zinc and Cell Viability Measurement
2.3. mRNA Quantitation by RT-PCR
2.4. Western Blot Analysis
2.5. GEO Data Analysis
2.6. Bayesian Network Analysis
2.7. Statistical Analysis
3. Results
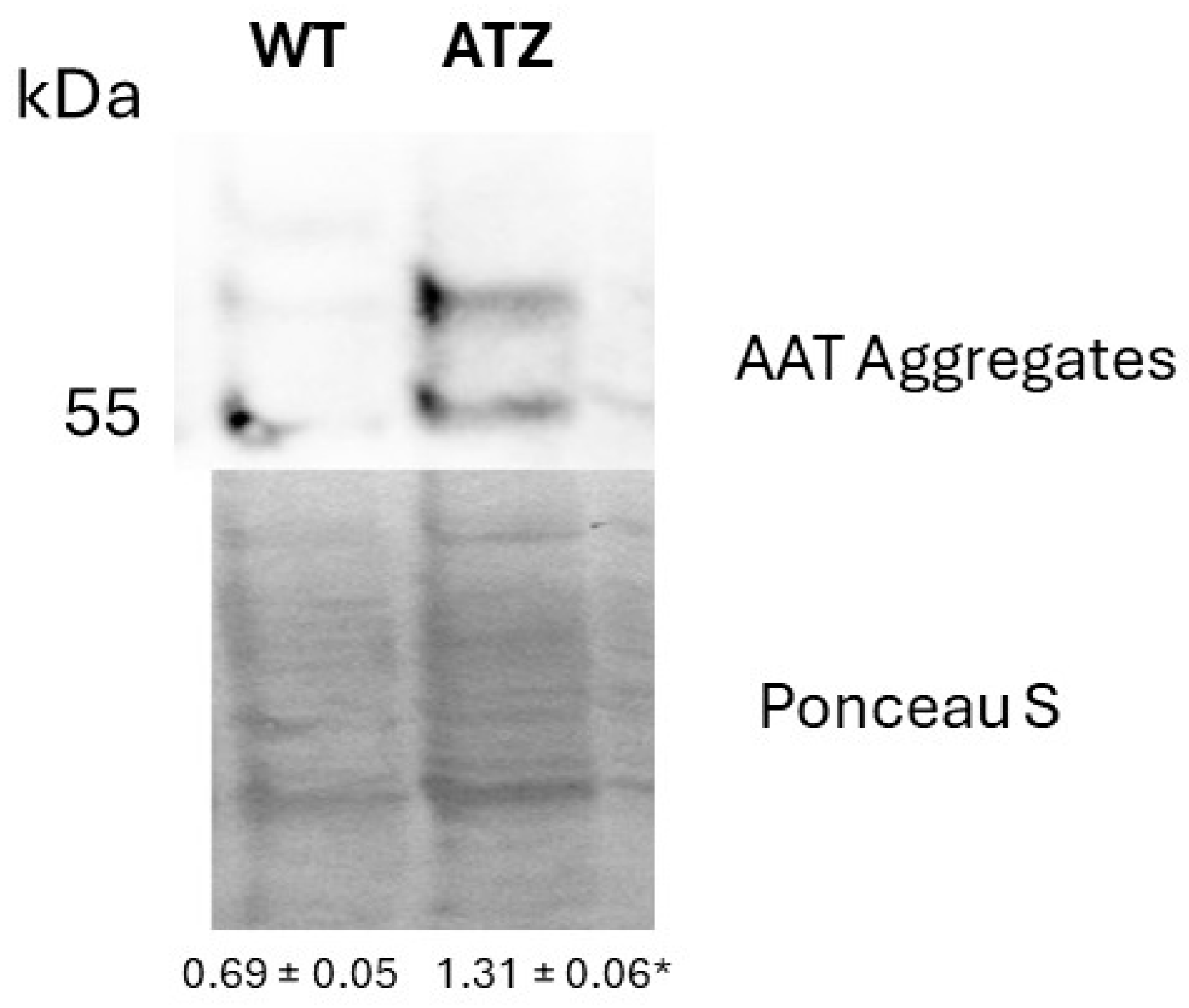
3.1. Expression of AAT
3.2. Labile Zinc
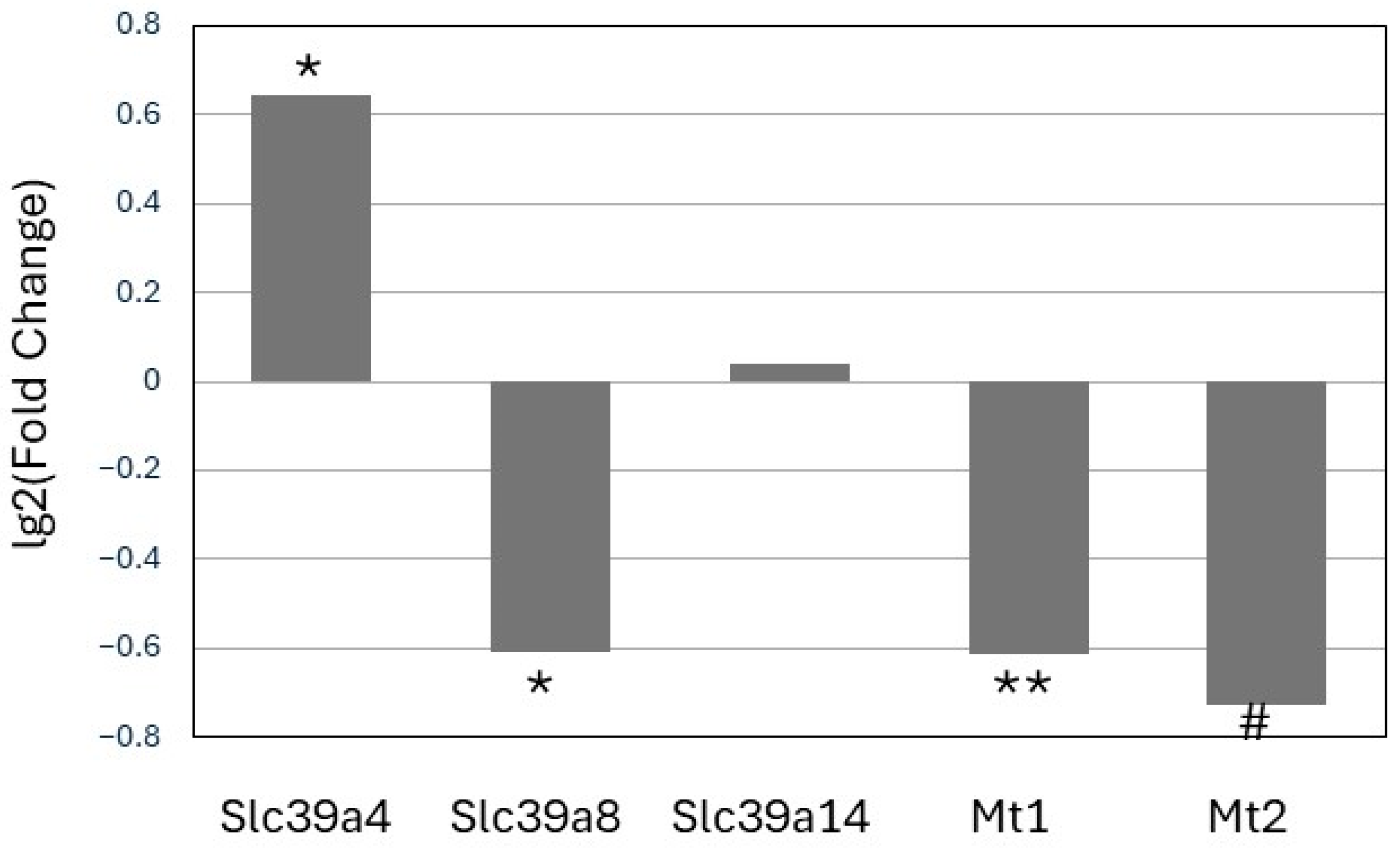
3.3. Expression of Zinc Transporters and Mt1 and Mt2 in Hepatocytes
3.4. Gene Expression of Genes Involved in Zinc Homeostasis and UPR in Liver Samples of PiZ and Wildtype Mice (GEO)
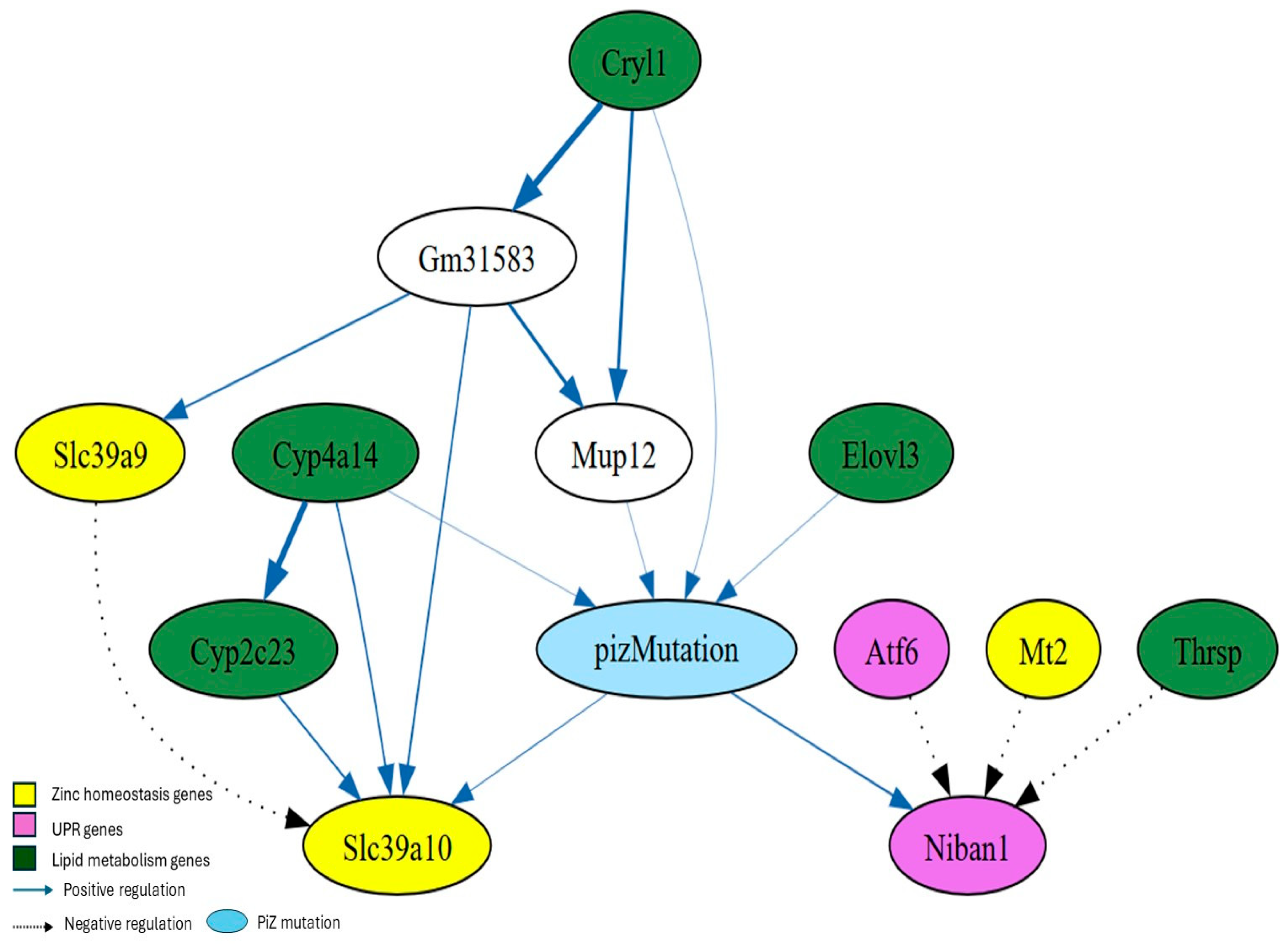
3.5. Bayesian Networks Analysis
3.6. GeNIe Simulations
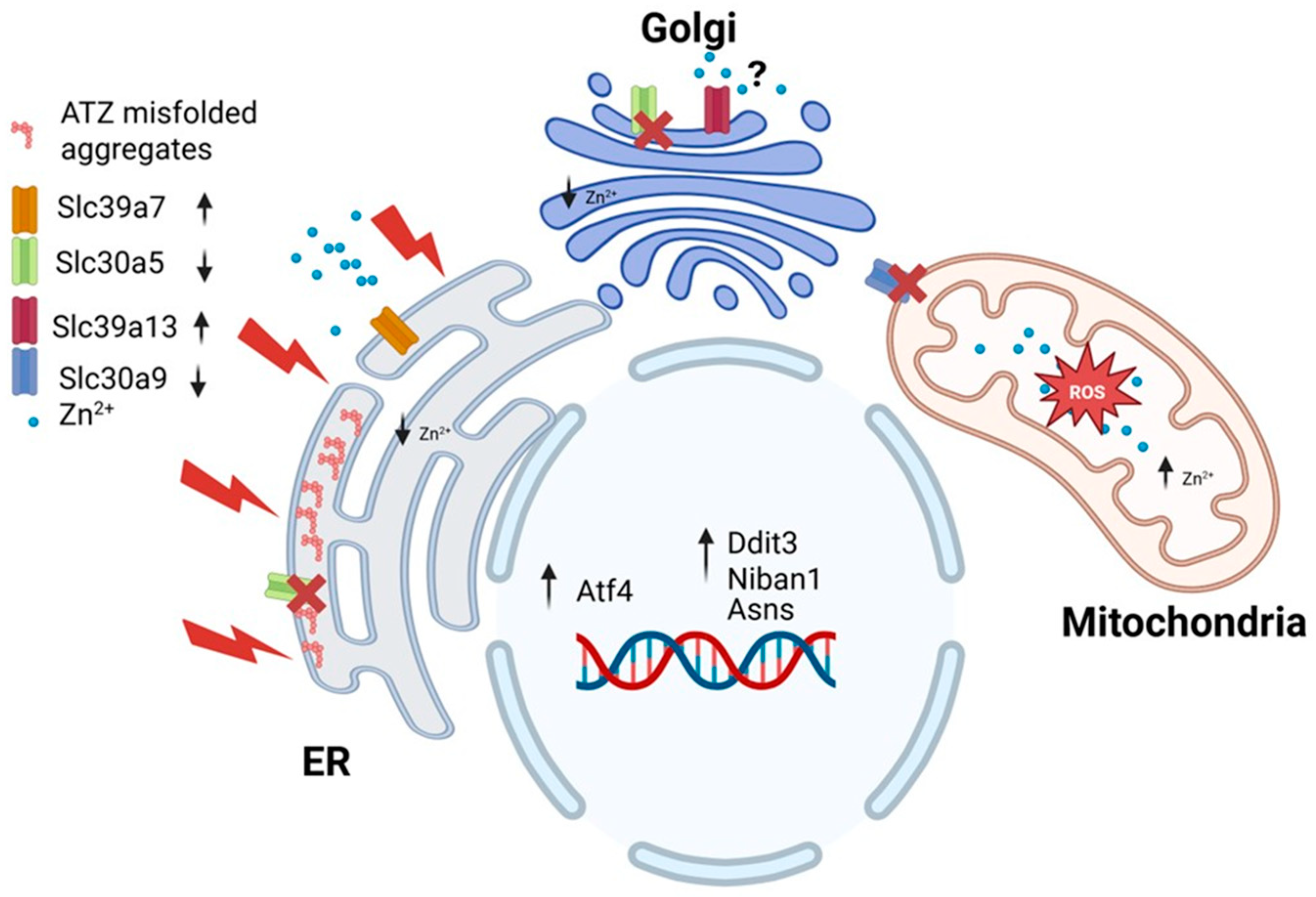
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| UPR | Unfolded protein response |
| MB | Markov blanket |
| ER | Endoplasmic reticulum |
| ROS | Reactive oxygen species |
| BN | Bayesian network |
| ERAD | ER-associated degradation |
| AAT | Alpha-1 antitrypsin |
| AATD | Alpha-1 antitrypsin |
| ATZ | Alpha-1 antitrypsin mutant Z |
| PiZ | Alpha-1 antitrypsin mutant Z |
| AI | Artificial intelligence |
References
- Seixas, S.; Marques, P.I. Known Mutations at the Cause of Alpha-1 Antitrypsin Deficiency an Updated Overview of SERPINA1 Variation Spectrum. Appl. Clin. Genet. 2021, 14, 173–194. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Teckman, J.H.; Perlmutter, D.H. The endoplasmic reticulum degradation pathway for mutant secretory proteins α1-antitrypsin Z and S is distinct from that for an unassembled membrane protein. J. Biol. Chem. 1996, 271, 13215–13220. [Google Scholar] [CrossRef] [PubMed]
- Rondanelli, M.; Gasparri, C.; Razza, C.; Ferraris, C.; Perna, S.; Ferrarotti, I.; Corsico, A.G. Practical dietary advices for subjects with alpha-1 antitrypsin deficiency. Biomed. Pharmacother. 2023, 163, 114753. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, B.; Haschka, D.; Finkenstedt, A.; Petersen, B.-S.; Theurl, I.; Henninger, B.; Janecke, A.R.; Wang, C.-Y.; Lin, H.Y.; Veits, L.; et al. Impaired hepcidin expression in alpha-1-antitrypsin deficiency associated with iron overload and progressive liver disease. Hum. Mol. Genet. 2015, 24, 6254–6263. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Elzouki, A.N.; Hultcrantz, R.; Stal, P.; Befrits, R.; Eriksson, S. Increased PiZ gene frequency for alpha 1 antitrypsin in patients with genetic haemochromatosis. Gut 1995, 36, 922–926. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rabinovitz, M.; Gavaler, J.S.; Kelly, R.H.; Van Thiel, D.H. Association between heterozygous β1–antitrypsin deficiency and genetic hemochromatosis. Hepatology 1992, 16, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Cousins, R.J. Regulatory aspects of zinc metabolism in liver and intestine. Nutr. Rev. 1979, 37, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Stamoulis, I.; Kouraklis, G.; Theocharis, S. Zinc and the liver: An active interaction. Dig. Dis. Sci. 2007, 52, 1595–1612. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhang, Z.; Liu, S.; Aluo, Z.; Zhang, L.; Yu, L.; Li, Y.; Song, Z.; Zhou, L. Zinc Supplementation Alleviates Lipid and Glucose Metabolic Disorders Induced by a High-Fat Diet. J. Agric. Food Chem. 2020, 68, 5189–5200. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Liu, J.; Song, Z.; McClain, C.J.; Kang, Y.J. Zinc supplementation inhibits hepatic apoptosis in mice subjected to a long-term ethanol exposure. Exp. Biol. Med. 2008, 233, 540–548. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhong, W.; Zhao, Y.; Sun, X.; Song, Z.; McClain, C.J.; Zhou, Z. Dietary zinc deficiency exaggerates ethanol-induced liver injury in mice: Involvement of intrahepatic and extrahepatic factors. PLoS ONE 2013, 8, e76522. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Szuster-Ciesielska, A.; Plewka, K.; Daniluk, J.; Kandefer-Szerszeń, M. Zinc supplementation attenuates ethanol- and acetaldehyde-induced liver stellate cell activation by inhibiting reactive oxygen species (ROS) production and by influencing intracellular signaling. Biochem. Pharmacol. 2009, 78, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, J.P.; Narayanan, V.; Doan, H.; Yoo, C. Effect of zinc intake on hepatic autophagy during acute alcohol intoxication. BioMetals 2018, 31, 217–232. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kambe, T.; Matsunaga, M.; Takeda, T.-A. Understanding the Contribution of Zinc Transporters in the Function of the Early Secretory Pathway. Int. J. Mol. Sci. 2017, 18, 2179. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vembar, S.S.; Brodsky, J.L. One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008, 9, 944–957. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Solovyov, A.; Gilbert, H.F. Zinc-dependent dimerization of the folding catalyst, protein disulfide isomerase. Protein Sci. 2004, 13, 1902–1907. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, M.-H.; Aydemir, T.B.; Cousins, R.J. Dietary Zinc Regulates Apoptosis through the Phosphorylated Eukaryotic Initiation Factor 2α/Activating Transcription Factor-4/C/EBP-Homologous Protein Pathway during Pharmacologically Induced Endoplasmic Reticulum Stress in Livers of Mice. J. Nutr. 2016, 146, 2180–2186. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cousins, R.J.; Liuzzi, J.P.; Lichten, L.A. Mammalian zinc transport, trafficking, and signals. J. Biol. Chem. 2006, 281, 24085–24089. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.K.; Taylor, K.M.; Ford, D.; Valentine, R.A. Differential subcellular localization of the splice variants of the zinc transporter ZnT5 is dictated by the different C-terminal regions. PLoS ONE 2011, 6, e23878. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, L.; Kirschke, C.P.; Gitschier, J. Functional characterization of a novel mammalian zinc transporter, ZnT6. J. Biol. Chem. 2002, 277, 26389–26395. [Google Scholar] [CrossRef] [PubMed]
- Kirschke, C.P.; Huang, L. ZnT7, a novel mammalian zinc transporter, accumulates zinc in the golgi apparatus. J. Biol. Chem. 2003, 278, 4096–4102. [Google Scholar] [CrossRef] [PubMed]
- Fukunaka, A.; Suzuki, T.; Kurokawa, Y.; Yamazaki, T.; Fujiwara, N.; Ishihara, K.; Migaki, H.; Okumura, K.; Masuda, S.; Yamaguchi-Iwai, Y.; et al. Demonstration and characterization of the heterodimerization of ZnT5 and ZnT6 in the early secretory pathway. J. Biol. Chem. 2009, 284, 30798–30806. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ishihara, K.; Yamazaki, T.; Ishida, Y.; Suzuki, T.; Oda, K.; Nagao, M.; Yamaguchi-Iwai, Y.; Kambe, T. Zinc transport complexes contribute to the homeostatic maintenance of secretory pathway function in vertebrate cells. J. Biol. Chem. 2006, 281, 17743–17750. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.M.; Morgan, H.E.; Johnson, A.; Nicholson, R.I. Structure-function analysis of HKE4, a member of the new LIV-1 subfamily of zinc transporters. Biochem. J. 2004, 377, 131–139. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bin, B.-H.; Bhin, J.; Seo, J.; Kim, S.-Y.; Lee, E.; Park, K.; Choi, D.-H.; Takagishi, T.; Hara, T.; Hwang, D.; et al. Requirement of Zinc Transporter SLC39A7/ZIP7 for Dermal Development to Fine-Tune Endoplasmic Reticulum Function by Regulating Protein Disulfide Isomerase. J. Investig. Dermatol. 2017, 137, 1682–1691. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-H.; Aydemir, T.B.; Kim, J.; Cousins, R.J. Hepatic ZIP14-mediated zinc transport is required for adaptation to endoplasmic reticulum stress. Proc. Natl. Acad. Sci. USA 2017, 114, E5805–E5814. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Teckman, J.H.; Perlmutter, D.H. Retention of mutant α1-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G961–G974. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, J.P.; Bobo, J.A.; Lichten, L.A.; Samuelson, D.A.; Cousins, R.J. Responsive transporter genes within the murine intestinal-pancreatic axis form a basis of zinc homeostasis. Proc. Natl. Acad. Sci. USA 2004, 101, 14355–14360. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Attanasiio, S.; Ferriero, R.; Gernoux, G.; De Cegli, R.; Carissimo, A.; Nusco, E.; Campione, S.; Teckman, J.; Mueller, C.; Piccolo, P.; et al. CHOP and c-JUN up-regulate the mutant Z α1-antitrypsin, exacerbating its aggregation and liver proteotoxicity. J. Biol. Chem. 2020, 295, 13213–13223. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Pastore, N.; Attanasio, S.; Granese, B.; Castello, R.; Teckman, J.; Wilson, A.A.; Ballabio, A.; Brunetti-Pierri, N. Activation of the c-Jun N-terminal kinase pathway aggravates proteotoxicity of hepatic mutant Z alpha1-antitrypsin. Hepatology 2017, 65, 1865–1874. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pearl, J. Probabilistic Reasoning in Intelligent Systems: Representation and Reasoning; Brachman, R.J., Ed.; Morgan Kaufmann: San Mateo, CA, USA, 1988. [Google Scholar]
- Heckerman, D. A Bayesian Approach to Learning Causal Networks; Morgan Kaufmann: San Francisco, CA, USA, 1995. [Google Scholar]
- Banjo: Bayesian Network Inference with Java Objects. Available online: https://users.cs.duke.edu/~amink/software/banjo/ (accessed on 1 May 2025).
- Tsukumo, Y.; Tomida, A.; Kitahara, O.; Nakamura, Y.; Asada, S.; Mori, K.; Tsuruo, T. Nucleobindin 1 controls the unfolded protein response by inhibiting ATF6 activation. J. Biol. Chem. 2007, 282, 29264–29272. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Wang, P.; Chen, W.; Wang, J.-R.; Ma, X.; Zhang, H.; Zhang, W.J.; Wei, C. Hepatic ELOVL3 is dispensable for lipid metabolism in mice. Biochem. Biophys. Res. Commun. 2023, 658, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Kuemmerle, N.B.; Kinlaw, W.B. THRSP (thyroid hormone responsive). Atlas Genet. Cytogenet. Oncol. Haematol. 2011, 15, 480–482. [Google Scholar] [CrossRef] [PubMed] [PubMed Central][Green Version]
- Sarparast, M.; Dattmore, D.; Alan, J.; Lee, K.S.S. Cytochrome P450 Metabolism of Polyunsaturated Fatty Acids and Neurodegeneration. Nutrients 2020, 12, 3523. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, X.; Li, S.; Zhou, Y.; Su, W.; Ruan, X.; Wang, B.; Zheng, F.; Warner, M.; Gustafsson, J.; Guan, Y. Ablation of cytochrome P450 omega-hydroxylase 4A14 gene attenuates hepatic steatosis and fibrosis. Proc. Natl. Acad. Sci. USA 2017, 114, 3181–3185, Erratum in Proc. Natl. Acad. Sci. USA 2023, 120, e2315591120. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Feng, L.; Ding, G.; Zhou, Y.; Zhu, H.; Jiang, H. Downregulation of Crystallin Lambda 1 is a New Independent Prognostic Marker in Clear Cell Renal Cell Carcinoma. Pharmgenomics Pers. Med. 2022, 15, 857–866. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Real, M.V.F.; Colvin, M.S.; Sheehan, M.J.; Moeller, A.H. Major urinary protein (Mup) gene family deletion drives sex-specific alterations in the house-mouse gut microbiota. bioRxiv [Preprint] 2023, 551491, Update in Microbiol. Spectr. 2024, 12, e0356623. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Quaife, C.; Hammer, R.E.; Mottet, N.; Palmiter, R.D. Glucocorticoid regulation of metallothionein during murine development. Dev. Biol. 1986, 118, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Wang, L.; Li, L.; Huang, Z.; Ye, L. Metallothionein 1: A New Spotlight on Inflammatory Diseases. Front. Immunol. 2021, 12, 739918. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Matsuura, W.; Yamazaki, T.; Yamaguchi-Iwai, Y.; Masuda, S.; Nagao, M.; Andrews, G.K.; Kambe, T. SLC39A9 (ZIP9) Regulates Zinc Homeostasis in the Secretory Pathway: Characterization of the ZIP Subfamily I Protein in Vertebrate Cells. Biosci. Biotechnol. Biochem. 2009, 73, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Pang, Y.; Dong, J.; Berg, A.H. Identification and characterization of membrane androgen receptors in the ZIP9 zinc transporter subfamily: II. Role of human ZIP9 in testosterone-induced prostate and breast cancer cell apoptosis. Endocrinology 2014, 155, 4250–4265. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lichten, L.A.; Ryu, M.-S.; Guo, L.; Embury, J.; Cousins, R.J. MTF-1-mediated repression of the zinc transporter Zip10 is alleviated by zinc restriction. PLoS ONE 2011, 6, e21526. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ohashi, W.; Kimura, S.; Iwanaga, T.; Furusawa, Y.; Irié, T.; Izumi, H.; Watanabe, T.; Hijikata, A.; Hara, T.; Ohara, O.; et al. Zinc Transporter SLC39A7/ZIP7 Promotes Intestinal Epithelial Self-Renewal by Resolving ER Stress. PLoS Genet. 2016, 12, e1006349. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Deng, H.; Qiao, X.; Xie, T.; Fu, W.; Li, H.; Zhao, Y.; Guo, M.; Feng, Y.; Chen, L.; Zhao, Y.; et al. SLC-30A9 is required for Zn2+ homeostasis, Zn2+ mobilization, and mitochondrial health. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, H.Y.; Gale, J.R.; Reynolds, I.J.; Weiss, J.H.; Aizenman, E. The Multifaceted Roles of Zinc in Neuronal Mitochondrial Dysfunction. Biomedicines 2021, 9, 489. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ma, P.; Wang, Z.; Wang, Y.; Hou, B.; Sun, J.; Tian, H.; Li, B.; Shui, G.; Yang, X.; Yang, X.; et al. Integration of Metabolomics and Transcriptomics Reveals Ketone Body and Lipid Metabolism Disturbance Related to ER Stress in the Liver. J. Proteome Res. 2021, 20, 3875–3888. [Google Scholar] [CrossRef] [PubMed]
- Fukada, T.; Civic, N.; Furuichi, T.; Shimoda, S.; Mishima, K.; Higashiyama, H.; Idaira, Y.; Asada, Y.; Kitamura, H.; Yamasaki, S.; et al. The zinc transporter SLC39A13/ZIP13 is required for connective tissue development; its involvement in BMP/TGF-β signaling pathways. PLoS ONE 2008, 3, e3642, Erratum in PLoS ONE 2008, 3. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jeong, J.; Walker, J.M.; Wang, F.; Park, J.G.; Palmer, A.E.; Giunta, C.; Rohrbach, M.; Steinmann, B.; Eide, D.J. Promotion of vesicular zinc efflux by ZIP13 and its implications for spondylocheiro dysplastic Ehlers–Danlos syndrome. Proc. Natl. Acad. Sci. USA 2012, 109, E3530–E3538. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]


| GSE93115 | GSE141593 | |
|---|---|---|
| Mt1 | 3.4 * | 1.7 * |
| Mt2 | 3.5 * | 3.6 ** |
| Mtf1 | −0.23 | 0.4 |
| Slc39a1 | −0.5 * | 0.3 |
| Slc39a3 | 0.6 * | −0.1 |
| Slc39a4 | 0.3 | 1.8 ** |
| Slc39a6 | 0.3 | 0.02 |
| Slc39a7 | 0.6 * | 1.3 ** |
| Slc39a8 | −0.6 * | −0.7 |
| Slc39a9 | 0.5 | −0.3 |
| Slc39a10 | NF | 0.3 |
| Slc39a11 | 0.3 | 1.4 ** |
| Sl39a13 | 0.4 | 0.7 |
| Slc39a14 | 0.3 | −0.1 |
| Slc30a1 | −0.4 | 0.1 |
| Slc30a4 | −0.1 | 0.4 |
| Slc30a5 | −0.3 | −0.02 |
| Slc30a6 | −0.4 | 0.1 |
| Slc30a7 | −0.02 | 0.3 |
| Slc30a9 | −0.5 * | −0.3 |
| GSE93115 | GSE141593 | |
|---|---|---|
| Atf3 | 5.3 ** | 3.5 ** |
| Atf4 | 0.9 * | 0.8 ** |
| Atf6 | −0.5 | 0.1 |
| Ern1 (IRE1) | 0.3 | 0.4 |
| Sdf2l1 | 0.34 | 2.9 ** |
| Xbp1 | −0.9 * | −0.1 |
| Eif2ak3 (Perk) | 0.3 | 0.6 |
| Derl3 | 1.8 * | 5.7 ** |
| Trib3 | 3.8 ** | 3.0 ** |
| Ddit3 (Chop) | 3.7 ** | 3.0 ** |
| Ddit4 | 0.5 * | 4.9 ** |
| Bhlha15 (Mist1) | 0.3 | 6.7 ** |
| Niban1 (Fam129a) | 1.05 * | 2.3 ** |
| Condition | |
|---|---|
| PiZ | Cyp4a14 [0], Elovl3 [0] |
| PiZ | Elovl3 [0], Mt2 [0], Niban1 [2] |
| PiZ | Atf6 [0], Cyp4a14 [0], Elovl3 [0] |
| PiZ | Cyp4a14 [0], Niban1 [2], Thrsp [0] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liuzzi, J.P.; Gonzales, S.; Barbieri, M.A.; Vidal, R.; Yoo, C. Effect of Alpha-1 Antitrypsin Deficiency on Zinc Homeostasis Gene Regulation and Interaction with Endoplasmic Reticulum Stress Response-Associated Genes. Nutrients 2025, 17, 1913. https://doi.org/10.3390/nu17111913
Liuzzi JP, Gonzales S, Barbieri MA, Vidal R, Yoo C. Effect of Alpha-1 Antitrypsin Deficiency on Zinc Homeostasis Gene Regulation and Interaction with Endoplasmic Reticulum Stress Response-Associated Genes. Nutrients. 2025; 17(11):1913. https://doi.org/10.3390/nu17111913
Chicago/Turabian StyleLiuzzi, Juan P., Samantha Gonzales, Manuel A. Barbieri, Rebecca Vidal, and Changwon Yoo. 2025. "Effect of Alpha-1 Antitrypsin Deficiency on Zinc Homeostasis Gene Regulation and Interaction with Endoplasmic Reticulum Stress Response-Associated Genes" Nutrients 17, no. 11: 1913. https://doi.org/10.3390/nu17111913
APA StyleLiuzzi, J. P., Gonzales, S., Barbieri, M. A., Vidal, R., & Yoo, C. (2025). Effect of Alpha-1 Antitrypsin Deficiency on Zinc Homeostasis Gene Regulation and Interaction with Endoplasmic Reticulum Stress Response-Associated Genes. Nutrients, 17(11), 1913. https://doi.org/10.3390/nu17111913




