Methodological Review of Classification Trees for Risk Stratification: An Application Example in the Obesity Paradox
Abstract
1. Introduction
1.1. Concept of a Classification Tree
1.2. Phases in the Construction of a Classification Tree
- Phase 1—Tree Development: From the root node, the most appropriate variable is identified to split the node into two child nodes by establishing an optimal cut-off point if the variable is continuous. Each child node is subsequently split following the same methodology. A supervised machine learning model is used, with all records including predictor variables and the outcome variable submitted to the algorithm.
- Phase 2—Tree Growth Stopping Criteria: Tree development can continue until terminal nodes contain only a single case, or when the value of the dependent variable is the same for all cases within a node. Additional criteria, such as a minimum number of cases per node, can be defined to prevent excessive branching.
- Phase 3—Tree Pruning: A CT developed using the aforementioned method tends to be overly complex and branched, which may lead to overfitting the training dataset. Removing superfluous branches results in a simpler tree with better generalizability. The pruning process uses predefined cost–complexity criteria to eliminate branches that add more complexity than effectiveness. Supervised learning aims to reduce classification error.
- Phase 4—Selection of the Optimal CT: Selecting the optimal CT requires an internal validation system. This can be achieved by randomly splitting the sample into a training set and a validation set, or by applying cross-validation techniques. Cross-validation divides the dataset into subsets—e.g., 10 partitions, using 9 for training and 1 for validation in a recursive process.
1.3. Use of Classification Trees in Medicine
1.4. Types of Classification Trees
| 1. Simple Classification Trees | ||||
|---|---|---|---|---|
| CART | CHAID | C4.5 | ctree | |
| Description | Classification and regression tree | Chi-square automatic interaction detection | Concept Learning Systems Version 4.5 | Conditional inference trees |
| Developer | Breiman (1984) [16] | Kass (1980) [17] | Quinlan (1993) [18] | Hothorm (2006) [19] |
| Primary Use | Many disciplines with few data | Applied statisticians | Data miners | Applied statisticians |
| Splitting Method | Entropy Gini index | Chi-square tests F test | Gain ratio | Asymptotic approximations |
| Branch Limitations | Best binary split | Number of values of the input | Best binary split | Bonferroni-adjusted p-values |
| Pruning | Cross-validation | Best binary split p-value | Misclassification rates | No pruning |
| Software * | AnswerTree WEKA R-Python | AnswerTree R-Python | WEKA R-Python | R-Python |
| 2. Ensembled Classification Trees | ||||
| Random Forest | AdaBoost | XG-Boost | ||
| Description | Uncorrelated forest | Adaptive boosting | Extreme gradient boosting | |
| Developer | Breiman and Cutler (2001) [24] | Freund and Schapire (1995) [25] | Chen and Guestrin (2016) [26] | |
| Ensembled Method | Parallel bagging | Adaptive boosting | Boosting gradient descent | |
| Software * | R/Python/Java | R/Python/Java | R/Python/Java | |
| 3. Hybrid Models | ||||
| Fuzzy Random Forest | Random Forest Neural Network | |||
| Other method | Fuzzy logic | Neural network | ||
| Developer | Olaru (2003) [15] | Khozeimeh (2022) [14] | ||
| Software * | C language | Python | ||
1.5. Advantages and Disadvantages of Classification Trees
1.5.1. Advantages
- Non-parametric models.
- Can handle all variable types (continuous, ordinal, categorical).
- Easy to interpret, with clinically meaningful classification rules.
- No additional calculations required to determine individual patient risk.
- Perform variable selection and establish variable hierarchy.
- Identify optimal cut-off points for continuous variables.
- Detect relationships among variables without assuming independence.
- Less affected by outliers or missing values.
1.5.2. Disadvantages
- Risk of overfitting and limited generalizability.
- High sensitivity to data, leading to model instability.
- Complex trees may lose interpretability.
- Require specific software and development methodology.
- Many CT types exist, and the most suitable one for a specific problem may not be obvious in advance.
2. ENPIC Study and the Obesity Paradox
2.1. The ENPIC Study
2.2. The Obesity Paradox
3. Use of Classification Trees to Determine Cut-Off Points
4. Use of Classification Trees to Identify Relationships Between Variables
5. Multivariable Risk Models Using Classification Trees
6. Ensemble Classification Tree Models
7. Model Evaluation
8. Discussion
9. Conclusions
- How CTs can be used to establish cut-off points for continuous variables.
- How they can identify interactions between variables that might go unnoticed in traditional regression models.
- How multivariable CT models generate decision rules and stratify patient risk based on the most influential predictors.
- How ensemble methods such as random forest and XGBoost improve predictive accuracy.
- And finally, how explanatory tools like SHAP values can provide insight into the structure and predictions of complex models.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AdaBoost | adaptive boosting |
| APACHE II | Acute Physiology and Chronic Health disease Classification System II |
| AUC ROC | area under the ROC curve |
| BMI | body mass index |
| C4.5 | Concept Learning Systems Version 4.5 |
| CART | classification and regression tree |
| CHAID | chi-square automatic interaction detection tree |
| CRUISE | Classification Rule with Unbiased Interaction Selection and Estimation |
| CT | classification tree |
| ctree | conditional inference trees |
| ENPIC | Evaluation of Practical Nutrition Practices in the Critical Care Patient |
| FACT | Fast and Accurate Classification Tree |
| GUIDE | Generalized Unbiased Interaction Detection and Estimation |
| ICU | intensive care unit |
| LR | logistic regression |
| Python | Python software |
| QUEST | Quick Unbiased and Efficient Statistical Tree |
| R | The R Project for Statistical Computing |
| SHAP | SHapley Additive exPlanations |
| TNF-α | Tumor Necrosis Factor-alpha |
| TRIPOD | Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis |
| WEKA | Waikato Environment for Knowledge Analysis |
| XGBoost | extreme gradient boosting |
References
- Masic, I. Medical Decision Making—An Overview. Acta Inform. Med. 2022, 30, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Podgorelec, V.; Kokol, P.; Stiglic, B.; Rozman, I. Decision trees: An overview and their use in medicine. J. Med. Syst. 2002, 26, 445–463. [Google Scholar] [CrossRef]
- Wei-Yin, L. Fifty Years of Classification and Regression Trees. Int. Stat. Rev. 2014, 82, 329–348. [Google Scholar] [CrossRef]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Trujillano, J.; Sarria-Santamera, A.; Esquerda, A.; Badia, M.; Palma, M.; March, J. Approach to the methodology of classification and regression trees. Gac. Sanit. 2008, 22, 65–72. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mitchell, T. (Ed.) Decision tree learning. In Machine Learning; McGraw Hill: New York, NY, USA, 1997. [Google Scholar]
- Lemon, S.C.; Roy, J.; Clark, M.A.; Friedmann, P.D.; Rakowski, W. Classification and regression tree analysis in public health: Methodological review and comparison with logistic regression. Ann. Behav. Med. 2003, 26, 172–181. [Google Scholar] [CrossRef]
- Shang, H.; Ji, Y.; Cao, W.; Yi, J. A predictive model for depression in elderly people with arthritis based on the TRIPOD guidelines. Geriatr. Nurs. 2025, 29, 85–93. [Google Scholar] [CrossRef]
- Speiser, J.L.; Callahan, K.E.; Houston, D.K.; Fanning, J.; Gill, T.M.; Guralnik, J.M.; Newman, A.B.; Pahor, M.; Rejeski, W.J.; Miller, M.E. Machine Learning in Aging: An Example of Developing Prediction Models for Serious Fall Injury in Older Adults. J. Gerontol. A Biol. Sci. Med. Sci. 2021, 76, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Trujillano, J.; Badia, M.; Serviá, L.; March, J.; Rodriguez-Pozo, A. Stratification of the severity of critically ill patients with classification trees. BMC Med. Res. Methodol. 2009, 9, 83. [Google Scholar] [CrossRef]
- Yin, L.; Lin, X.; Liu, J.; Li, N.; He, X.; Zhang, M.; Guo, J.; Yang, J.; Deng, L.; Wang, Y.; et al. Investigation on Nutrition Status and Clinical Outcome of Common Cancers (INSCOC) Group. Classification Tree-Based Machine Learning to Visualize and Validate a Decision Tool for Identifying Malnutrition in Cancer Patients. JPEN J. Parenter. Enteral Nutr. 2021, 45, 736–1748. [Google Scholar] [CrossRef]
- Tay, E.; Barnett, D.; Rowland, M.; Kerse, N.; Edlin, R.; Waters, D.L.; Connolly, M.; Pillai, A.; Tupou, E.; The, R. Sociodemographic and Health Indicators of Diet Quality in Pre-Frail Older Adults in New Zealand. Nutrients. 2023, 15, 4416. [Google Scholar] [CrossRef] [PubMed]
- Marchitelli, S.; Mazza, C.; Ricci, E.; Faia, V.; Biondi, S.; Colasanti, M.; Cardinale, A.; Roma, P.; Tambelli, R. Identification of Psychological Treatment Dropout Predictors Using Machine Learning Models on Italian Patients Living with Overweight and Obesity Ineligible for Bariatric Surgery. Nutrients 2024, 16, 2605. [Google Scholar] [CrossRef]
- Khozeimeh, F.; Sharifrazi, D.; Izadi, N.H.; Joloudari, J.H.; Shoeibi, A.; Alizadehsani, R.; Tartibi, M.; Hussain, S.; Sani, Z.A.; Khodatars, M.; et al. RF-CNN-F: Random forest with convolutional neural network features for coronary artery disease diagnosis based on cardiac magnetic resonance. Sci. Rep. 2022, 12, 11178. [Google Scholar] [CrossRef] [PubMed]
- Olaru, C.; Wehenkel, L. A complete fuzzy decision tree technique. Fuzzy Sets Syst. 2003, 138, 221–254. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Wadsworth Publishing Co.: Belmont, CA, USA, 1984. [Google Scholar]
- Kass, G.V. An exploratory for investigating large quantities of categorical data. Ann. Appl. Stat. 1980, 29, 119–127. [Google Scholar] [CrossRef]
- Quinlan, J.R. C4.5: Programs for Machine Learning; Morgan Kaufmann: San Mateo, CA, USA, 1993. [Google Scholar]
- Hothorn, T.; Hornik, K.; Zeileis, A. Unbiased Recursive Partitioning: A Conditional Inference Framework. J. Comput. Graph. Stat. 2006, 15, 651–674. [Google Scholar] [CrossRef]
- SPSS Inc. AnswerTree User’s Guide; SPSS Inc.: Chicago, IL, USA, 2001. [Google Scholar]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software. ACM SIGKDD Explor. Newslett. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 1 January 2023).
- Breiman, L. Random Forest. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD ‘16), Association for Computing Machinery, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Lopez-Delgado, J.C.; Sanchez-Ales, L.; Flordelis-Lasierra, J.L.; Mor-Marco, E.; Bordeje-Laguna, M.L.; Portugal-Rodriguez, E.; Lorencio-Cardenas, C.; Vera-Artazcoz, P.; Aldunate-Calvo, S.; Llorente-Ruiz, B.; et al. Nutrition Therapy in Critically Ill Patients with Obesity: An Observational Study. Nutrients 2025, 17, 732. [Google Scholar] [CrossRef]
- Yébenes, J.C.; Bordeje-Laguna, M.L.; Lopez-Delgado, J.C.; Lorencio-Cardenas, C.; Martinez De Lagran Zurbano, I.; Navas-Moya, E.; Servia-Goixart, L. Smartfeeding: A Dynamic Strategy to Increase Nutritional Efficiency in Critically Ill Patients-Positioning Document of the Metabolism and Nutrition Working Group and the Early Mobilization Working Group of the Catalan Society of Intensive and Critical Care Medicine (SOCMiC). Nutrients 2024, 16, 1157. [Google Scholar] [CrossRef]
- Servia-Goixart, L.; Lopez-Delgado, J.C.; Grau-Carmona, T.; Trujillano-Cabello, J.; Bordeje-Laguna, M.L.; Mor-Marco, E.; Portugal-Rodriguez, E.; Lorencio-Cardenas, C.; Montejo-Gonzalez, J.C.; Vera-Artazcoz, P.; et al. Evaluation of Nutritional Practices in the Critical Care patient (The ENPIC study): Does nutrition really affect ICU mortality? Clin. Nutr. ESPEN 2022, 47, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Cahill, N.E.; Heyland, D.K. Bridging the guideline-practice gap in critical care nutrition: A review of guideline implementation studies. J. Parenter Enteral Nutr. 2010, 34, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Gruberg, L.; Weissman, N.J.; Waksman, R.; Fuchs, S.; Deible, R.; Pinnow, E.E.; Ahmed, L.M.; Kent, K.M.; Pichard, A.D.; Suddath, W.O.; et al. The impact of obesity on the short-term and long-term outcomes after percutaneous coronary intervention: The obesity paradox? J. Am. Coll. Cardiol. 2002, 39, 578–584. [Google Scholar] [CrossRef]
- Oreopoulos, A.; Padwal, R.; Kalantar-Zadeh, K.; Fonarow, G.C.; Norris, C.M.; McAlister, F.A. Body mass index and mortality in heart failure: A meta-analysis. Am. Heart J. 2008, 156, 13–22. [Google Scholar] [CrossRef]
- Robinson, M.K.; Mogensen, K.M.; Casey, J.D.; McKane, C.K.; Moromizato, T.; Rawn, J.D.; Christopher, K.B. The relationship among obesity, nutritional status, and mortality in the critically ill. Crit. Care Med. 2015, 43, 87–100. [Google Scholar] [CrossRef]
- Zhou, D.; Wang, C.; Lin, Q.; Li, T. The obesity paradox for survivors of critically ill patients. Crit. Care 2022, 26, 198. [Google Scholar] [CrossRef]
- Dickerson, R.N. The obesity paradox in the ICU: Real or not? Crit. Care 2013, 17, 154. [Google Scholar] [CrossRef]
- Ripoll, J.G.; Bittner, E.A. Obesity and Critical Illness-Associated Mortality: Paradox, Persistence and Progress. Crit. Care Med. 2023, 51, 551–554. [Google Scholar] [CrossRef]
- Knaus, W.A.; Zimmerman, J.E.; Wagner, D.P.; Draper, E.A.; Lawrence, D.E. APACHE-acute physiology and chronic health evaluation: A physiologically based classification system. Crit. Care Med. 1981, 9, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Porcel, J.M.; Trujillano, J.; Porcel, L.; Esquerda, A.; Bielsa, S. Development and validation of a diagnostic prediction model for heart failure-related pleural effusions: The BANCA score. ERJ Open Res. 2025, in press. [CrossRef]
- Díaz-Prieto, L.E.; Gómez-Martínez, S.; Vicente-Castro, I.; Heredia, C.; González-Romero, E.A.; Martín-Ridaura, M.D.C.; Ceinos, M.; Picón, M.J.; Marcos, A.; Nova, E. Effects of Moringa oleifera Lam. Supplementation on Inflammatory and Cardiometabolic Markers in Subjects with Prediabetes. Nutrients 2022, 14, 1937. [Google Scholar] [CrossRef]
- Cawthon, P.M.; Patel, S.M.; Kritchevsky, S.B.; Newman, A.B.; Santanasto, A.; Kiel, D.P.; Travison, T.G.; Lane, N.; Cummings, S.R.; Orwoll, E.S.; et al. What Cut-Point in Gait Speed Best Discriminates Community-Dwelling Older Adults with Mobility Complaints from Those Without? A Pooled Analysis from the Sarcopenia Definitions and Outcomes Consortium. J. Gerontol. A Biol. Sci. Med. Sci. 2021, 76, e321–e327. [Google Scholar] [CrossRef] [PubMed]
- Tamanna, T.; Mahmud, S.; Salma, N.; Hossain, M.M.; Karim, M.R. Identifying determinants of malnutrition in under-five children in Bangladesh: Insights from the BDHS-2022 cross-sectional study. Sci. Rep. 2025, 15, 14336. [Google Scholar] [CrossRef]
- Turjo, E.A.; Rahman, M.H. Assessing risk factors for malnutrition among women in Bangladesh and forecasting malnutrition using machine learning approaches. BMC Nutr. 2024, 10, 22. [Google Scholar] [CrossRef]
- Reusken, M.; Coffey, C.; Cruijssen, F.; Melenberg, B.; van Wanrooij, C. Identification of factors associated with acute malnutrition in children under 5 years and forecasting future prevalence: Assessing the potential of statistical and machine learning methods. BMJ Public Health 2025, 3, e001460. [Google Scholar] [CrossRef]
- Li, N.; Peng, E.; Liu, F. Prediction of lymph node metastasis in cervical cancer patients using AdaBoost machine learning model: Analysis of risk factors. Am. J. Cancer Res. 2025, 5, 1158–1173. [Google Scholar] [CrossRef]
- Wong, J.E.; Yamaguchi, M.; Nishi, N.; Araki, M.; Wee, L.H. Predicting Overweight and Obesity Status Among Malaysian Working Adults with Machine Learning or Logistic Regression: Retrospective Comparison Study. JMIR Form. Res. 2022, 6, e40404. [Google Scholar] [CrossRef]
- Lian, R.; Tang, H.; Chen, Z.; Chen, X.; Luo, S.; Jiang, W.; Jiang, J.; Yang, M. Development and multi-center cross-setting validation of an explainable prediction model for sarcopenic obesity: A machine learning approach based on readily available clinical features. Aging Clin. Exp. Res. 2025, 37, 63. [Google Scholar] [CrossRef]
- Gu, Y.; Su, S.; Wang, X.; Mao, J.; Ni, X.; Li, A.; Liang, Y.; Zeng, X. Comparative study of XGBoost and logistic regression for predicting sarcopenia in postsurgical gastric cancer patients. Sci. Rep. 2025, 5, 12808. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD statement. Ann. Intern. Med. 2015, 62, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Van Dort, B.A.; Engelsma, T.; Medlock, S.; Dusseljee-Peute, L. User-Centered Methods in Explainable AI Development for Hospital Clinical Decision Support: A Scoping Review. Stud. Health Technol. Inform. 2025, 326, 17–21. [Google Scholar] [CrossRef]
- Moser, D.; Sariyar, M. Explainable Versus Interpretable AI in Healthcare: How to Achieve Understanding. Stud. Health Technol. Inform. 2025, 327, 1433–1437. [Google Scholar] [CrossRef] [PubMed]
- Alkhanbouli, R.; Matar Abdulla Almadhaani, H.; Alhosani, F.; Simsekler, M.C.E. The role of explainable artificial intelligence in disease prediction: A systematic literature review and future research directions. BMC Med. Inform. Decis. Mak. 2025, 25, 110. [Google Scholar] [CrossRef]
- Sandri, E.; Cerdá Olmedo, G.; Piredda, M.; Werner, L.U.; Dentamaro, V. Explanatory AI Predicts the Diet Adopted Based on Nutritional and Lifestyle Habits in the Spanish Population. Eur. J. Investig. Health Psychol. Educ. 2025, 15, 11. [Google Scholar] [CrossRef]
- Harper, P.R. A review and comparison of classification algorithms for medical decision making. Health Policy. 2005, 71, 315–331. [Google Scholar] [CrossRef]
- El-Latif, E.I.A.; El-Dosuky, M.; Darwish, A.; Hassanien, A.E. A deep learning approach for ovarian cancer detection and classification based on fuzzy deep learning. Sci. Rep. 2024, 14, 26463. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Nair, B.; Vavilala, M.S.; Horibe, M.; Eisses, M.J.; Adams, T.; Liston, D.E.; Low, D.K.; Newman, S.F.; Kim, J.; et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2018, 2, 749–760. [Google Scholar] [CrossRef]

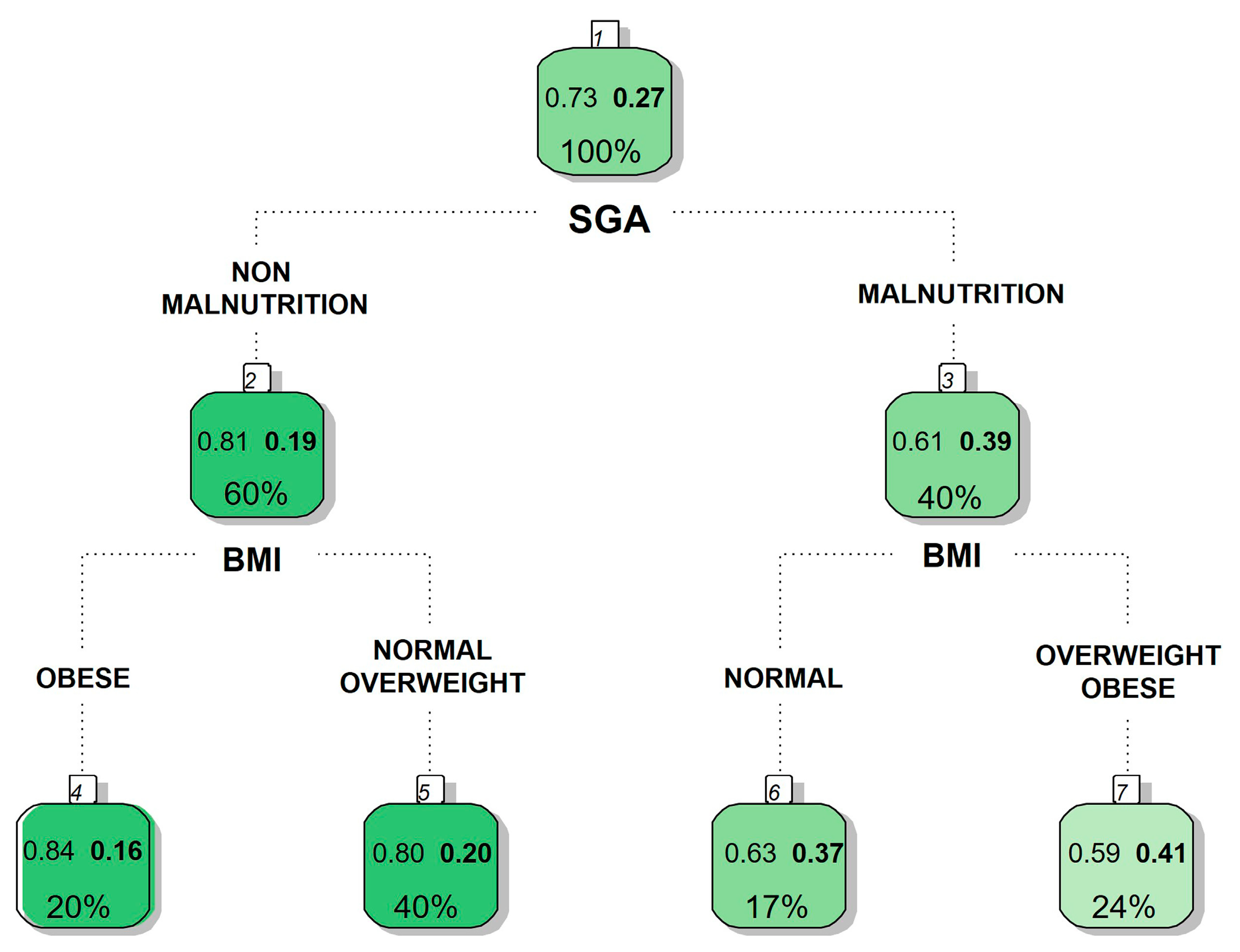



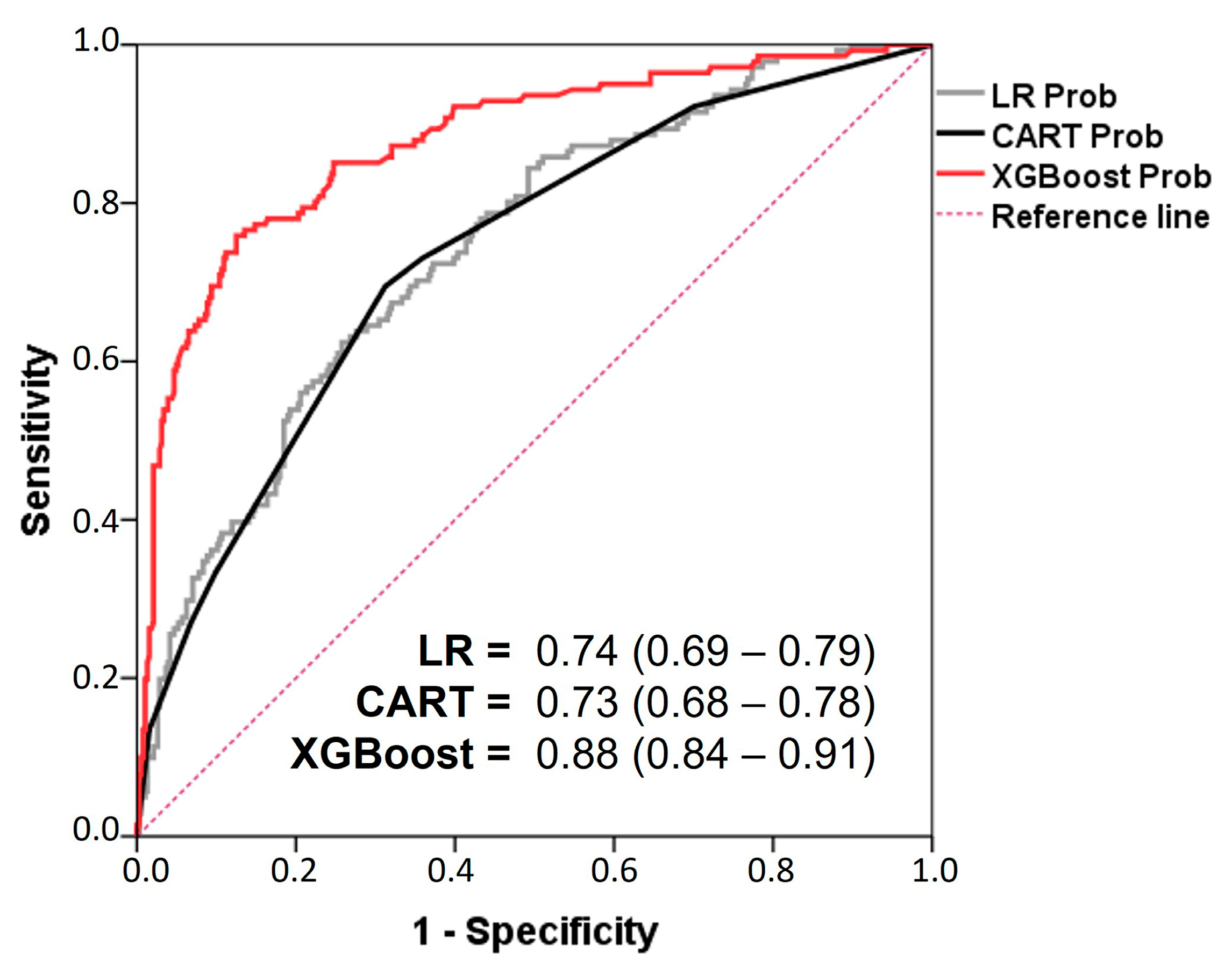
| All Patients n = 525 | Normal n = 165 | Overweight n = 210 | Obese n = 150 | p-Value | ||
|---|---|---|---|---|---|---|
| Baseline characteristics and comorbidities | ||||||
| Age, years, mean ± SD | 61.5 ± 15 | 58.8 ± 16.5 | 62.8 ± 14.7 | 62.7 ± 13.5 | 0.05 | |
| Sex, male patients, n (%) | 67.2% (353) | 64.8% (107) | 74.8% (157) | 59.3% (89) | 0.003 B | |
| Hypertension, n (%) | 43.6% (229) | 33.9% (56) | 41.9% (88) | 56.7% (85) | 0.01 A,B | |
| Diabetes mellitus, n (%) | 25% (131) | 21.2% (35) | 20% (42) | 36% (54) | 0.001 A,B | |
| AMI, n (%) | 14.1% (74) | 8.5% (14) | 16.7% (35) | 16.7% (25) | 0.04 B | |
| Neoplasia, n (%) | 20.6% (108) | 24.2% (40) | 19.5% (41) | 18% (27) | 0.11 | |
| Type of patient | Medical, n (%) | 63.8% (335) | 65.5% (108) | 62.9% (132) | 63.3% (95) | 0.81 |
| Trauma, n (%) | 12.6% (66) | 10.9% (18) | 15.2% (32) | 10.7% (16) | 0.75 | |
| Surgery, n (%) | 23.6% (124) | 23.6% (39) | 21.9% (46) | 26% (39) | 0.67 | |
| Prognosis ICU scores and nutrition status on ICU admission | ||||||
| APACHE II, mean ± SD | 20.3 ± 7.9 | 19.7 ± 7.6 | 20.1 ± 7.5 | 21.2 ± 8.5 | 0.18 | |
| Malnutrition (based on SGA), n (%) | 41% (215) | 52.7% (87) | 37.1% (78) | 33.3% (50) | 0.01 B | |
| Characteristics of Medical Nutrition Therapy | ||||||
| Early nutrition, <48 h, n (%) | 74.9% (393) | 77.6% (128) | 75.2% (158) | 71.3% (107) | 0.43 | |
| Kcal/kg/day, mean ± SD | 19 ± 5.6 | 23.1 ± 6 | 18.6 ± 3.7 | 15.27 ± 4.24 | 0.001 A | |
| Protein, g/kg/day, mean ± SD | 1 ± 0.4 | 1.2 ± 0.4 | 1 ± 0.3 | 0.8 ± 0.2 | 0.01 A,B | |
| EN | 63.2% (332) | 59.4% (98) | 64.3% (135) | 66% (99) | 0.34 | |
| PN | 15.4% (81) | 13.3% (22) | 16.2% (34) | 16.7% (25) | 0.85 | |
| EN-PN | 7.8% (41) | 8.5% (14) | 7.6% (16) | 7.3% (11) | 0.92 | |
| PN-EN | 13.5% (71) | 18.8% (31) | 11.9% (25) | 10% (15) | 0.27 | |
| Outcomes | ||||||
| Mechanical ventilation, n (%) | 92.8% (487) | 89.1% (147) | 93.8% (197) | 95.3% (143) | 0.08 | |
| Vasoactive drug support, n (%) | 77% (404) | 73.9% (122) | 79.5% (167) | 76.7% (115) | 0.44 | |
| Renal replacement therapy, n (%) | 16.6% (87) | 16.4% (27) | 12.9% (27) | 22% (33) | 0.07 | |
| ICU stay, days, mean ± SD | 20.3 ± 18 | 18.2 ± 13.8 | 21.1 ± 17.1 | 21.6 ± 22.5 | 0.08 | |
| 28-day mortality, n (%) | 26.7% (140) | 29.1% (48) | 27.1% (57) | 23.3% (35) | 0.51 | |
| Variable | OR (95% CI) | p-Value |
|---|---|---|
| Age (years) | ||
| <50 | 1 | |
| 50–75 | 3.3 (1.7–6.5) | 0.001 |
| >75 | 7.0 (3.3–14.9) | <0.001 |
| Sex (Male) | 1.0 (0.6–1.6) | 0.998 |
| APACHE II score | ||
| ≤25 | 1 | |
| >25 | 2.2 (1.4–3.5) | <0.001 |
| BMI groups | ||
| Normal | 1 | |
| Overweight | 0.9 (0.5–1.5) | 0.623 |
| Obese | 0.7 (0.4–1.4) | 0.651 |
| SGA | ||
| Non-malnutrition | 1 | |
| Malnutrition | 2.6 (1.74.0) | <0.001 |
| Median Kcal/Kg/day | 1.1 (0.9–1.1) | 0.315 |
| Median g protein/Kg/day | 0.3 (0.1–0.9) | 0.022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trujillano, J.; Serviá, L.; Badia, M.; Serrano, J.C.E.; Bordejé-Laguna, M.L.; Lorencio, C.; Vaquerizo, C.; Flordelis-Lasierra, J.L.; Martínez de Lagrán, I.; Portugal-Rodríguez, E.; et al. Methodological Review of Classification Trees for Risk Stratification: An Application Example in the Obesity Paradox. Nutrients 2025, 17, 1903. https://doi.org/10.3390/nu17111903
Trujillano J, Serviá L, Badia M, Serrano JCE, Bordejé-Laguna ML, Lorencio C, Vaquerizo C, Flordelis-Lasierra JL, Martínez de Lagrán I, Portugal-Rodríguez E, et al. Methodological Review of Classification Trees for Risk Stratification: An Application Example in the Obesity Paradox. Nutrients. 2025; 17(11):1903. https://doi.org/10.3390/nu17111903
Chicago/Turabian StyleTrujillano, Javier, Luis Serviá, Mariona Badia, José C. E. Serrano, María Luisa Bordejé-Laguna, Carol Lorencio, Clara Vaquerizo, José Luis Flordelis-Lasierra, Itziar Martínez de Lagrán, Esther Portugal-Rodríguez, and et al. 2025. "Methodological Review of Classification Trees for Risk Stratification: An Application Example in the Obesity Paradox" Nutrients 17, no. 11: 1903. https://doi.org/10.3390/nu17111903
APA StyleTrujillano, J., Serviá, L., Badia, M., Serrano, J. C. E., Bordejé-Laguna, M. L., Lorencio, C., Vaquerizo, C., Flordelis-Lasierra, J. L., Martínez de Lagrán, I., Portugal-Rodríguez, E., & López-Delgado, J. C. (2025). Methodological Review of Classification Trees for Risk Stratification: An Application Example in the Obesity Paradox. Nutrients, 17(11), 1903. https://doi.org/10.3390/nu17111903







