Selenium May Be Involved in Esophageal Squamous Cancer Prevention by Affecting GPx3 and FABP1 Expression: A Case-Control Study Based on Bioinformatic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformation Analysis
2.2. The Case-Control Study
2.2.1. Population-Sample Collection
2.2.2. Laboratory Measurements
2.2.3. Statistical Analysis
3. Results
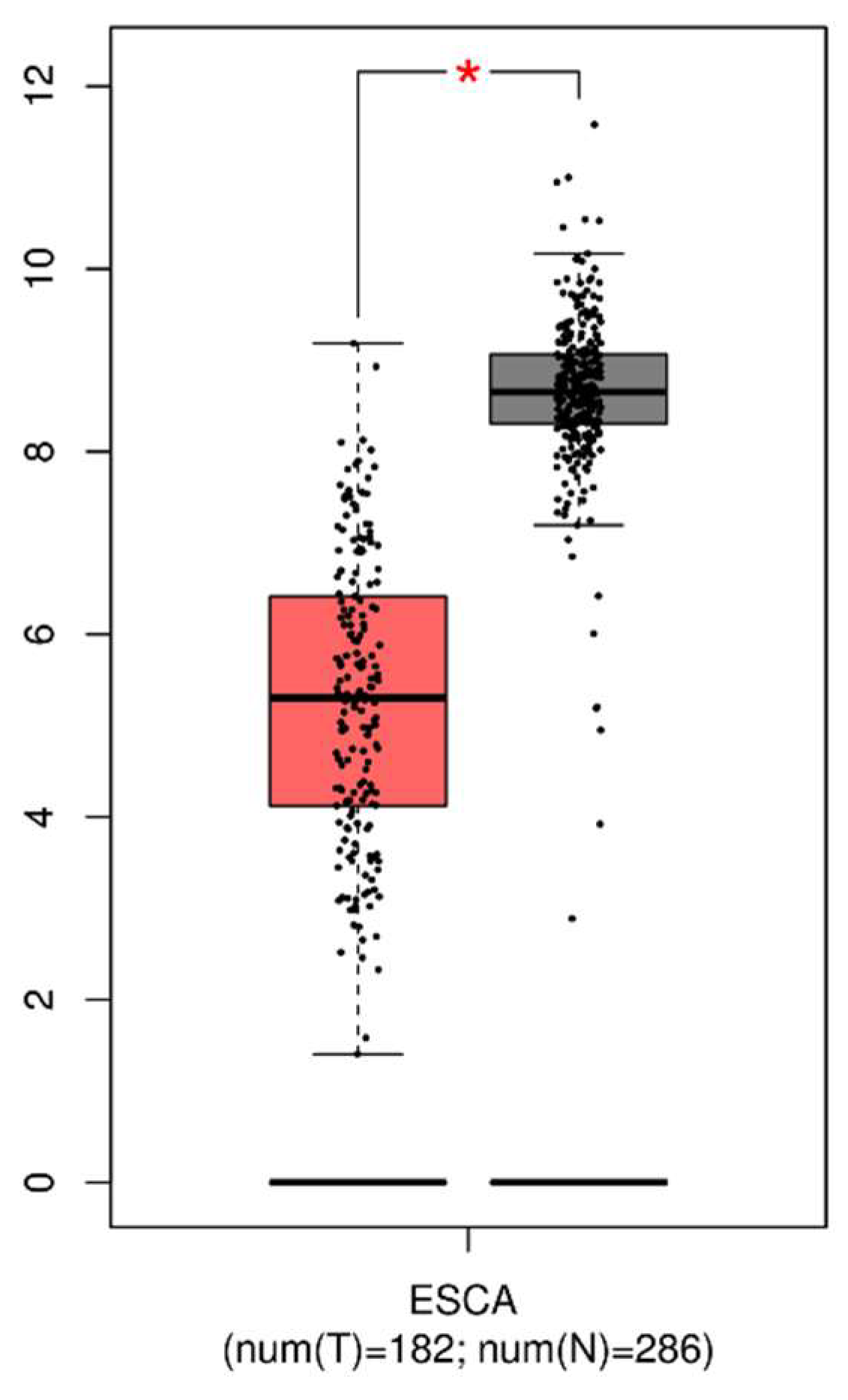
3.1. Expression of GPx3 in Tumor and Normal Tissues
3.2. Visualization of DEGs in the Tumor Tissues with Low Expressed GPx3
3.3. Selenium-Related Variables of the Subjects
3.4. Correlation between Selenium-Related Variables and Risk of EPLs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Morgan, E.; Soerjomataram, I.; Rumgay, H.; Coleman, H.G.; Thrift, A.P.; Vignat, J.; Laversanne, M.; Ferlay, J.; Arnold, M. The Global Landscape of Esophageal Squamous Cell Carcinoma and Esophageal Adenocarcinoma Incidence and Mortality in 2020 and Projections to 2040: New Estimates From GLOBOCAN 2020. Gastroenterology 2022, 163, 649–658.e2. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.W.; Zhao, Z.H.; Wang, L.; Li, P.; Chen, K.S.; Zhang, J.Y.; Li, W.C.; Jiang, G.Z.; Li, X.N. MicroRNA-134 prevents the progression of esophageal squamous cell carcinoma via the PLXNA1-mediated MAPK signalling pathway. eBioMedicine 2019, 46, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.Y.A.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.S.; Chen, R.; Han, B.F.; Wang, S.M.; Li, L.; Sun, K.X.; Zeng, H.M.; Wei, W.W.; He, J. Cancer incidence and mortality in China, 2022. Zhonghua Zhong Liu Za Zhi [Chin. J. Oncol.] 2024, 46, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Arnal, M.J.D.; Arenas, A.F.; Arbeloa, A.L. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J. Gastroenterol. 2015, 21, 7933–7943. [Google Scholar] [CrossRef] [PubMed]
- Mehdi, Y.; Hornick, J.L.; Istasse, L.; Dufrasne, I. Selenium in the environment, metabolism and involvement in body functions. Molecules 2013, 18, 3292–3311. [Google Scholar] [CrossRef] [PubMed]
- Mendieta, J.M.; Fields, C.L.; Ossorio, M.A.; Roy, T.M. Respiratory failure associated with the post-polio syndrome. J. Ky. Med. Assoc. 1989, 87, 63–65. [Google Scholar] [PubMed]
- Kershaw, J.L.; Hall, A.J. Mercury in cetaceans: Exposure, bioaccumulation and toxicity. Sci. Total Environ. 2019, 694, 133683. [Google Scholar] [CrossRef]
- Ahsan, A.; Liu, Z.; Su, R.; Liu, C.; Liao, X.; Su, M. Potential Chemotherapeutic Effect of Selenium for Improved Canceration of Esophageal Cancer. Int. J. Mol. Sci. 2022, 23, 5509. [Google Scholar] [CrossRef]
- Younesian, O.; Sheikh Arabi, M.; Jafari, S.M.; Joshaghani, H. Long-Term Excessive Selenium Supplementation Affects Gene Expression in Esophageal Tissue of Rats. Biol. Trace Elem. Res. 2023, 201, 3387–3394. [Google Scholar] [CrossRef]
- Nozadi, F.; Azadi, N.; Mansouri, B.; Tavakoli, T.; Mehrpour, O. Association between trace element concentrations in cancerous and non-cancerous tissues with the risk of gastrointestinal cancers in Eastern Iran. Environ. Sci. Pollut. Res. Int. 2021, 28, 62530–62540. [Google Scholar] [CrossRef]
- Hashemi, S.M.; Mashhadi, M.; Moghaddam, A.A.; Yousefi, J.; Mofrad, A.D.; Sadeghi, M.; Allahyari, A. The Relationship between Serum Selenium and Zinc with Gastroesophageal Cancers in the Southeast of Iran. Indian J. Med. Paediatr. Oncol. 2017, 38, 169–172. [Google Scholar]
- Nouarie, M.; Pourshams, A.; Kamangar, F.; Sotoudeh, M.; Derakhshan, M.H.; Akbari, M.R.; Fakheri, H.; Zahedi, M.J.; Caldwell, K.; Abnet, C.C.; et al. Ecologic study of serum selenium and upper gastrointestinal cancers in Iran. World J. Gastroenterol. 2004, 10, 2544–2546. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Wang, C.; Yu, W.; Fan, W.; Wang, S.; Shen, N.; Wu, P.; Li, X.; Wang, F. Selenium Exposure and Cancer Risk: An Updated Meta-analysis and Meta-regression. Sci. Rep. 2016, 6, 19213. [Google Scholar] [CrossRef]
- Kang, D.; Lee, J.; Wu, C.; Guo, X.; Lee, B.J.; Chun, J.S.; Kim, J.H. The role of selenium metabolism and selenoproteins in cartilage homeostasis and arthropathies. Exp. Mol. Med. 2020, 52, 1198–1208. [Google Scholar] [CrossRef]
- Tan, L.C.; Nancharaiah, Y.V.; van Hullebusch, E.D.; Lens, P.N.L. Selenium: Environmental significance, pollution, and biological treatment technologies. Biotechnol. Adv. 2016, 34, 886–907. [Google Scholar] [CrossRef]
- Touat-Hamici, Z.; Legrain, Y.; Bulteau, A.L.; Chavatte, L. Selective up-regulation of human selenoproteins in response to oxidative stress. J. Biol. Chem. 2014, 289, 14750–14761. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Pan, X.; Wei, G.; Hua, Y. Research progress of glutathione peroxidase family (GPX) in redoxidation. Front. Pharmacol. 2023, 14, 1147414. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Flohé, L. Regulatory Phenomena in the Glutathione Peroxidase Superfamily. Antioxid. Redox Signal. 2020, 33, 498–516. [Google Scholar] [CrossRef]
- Kipp, A.P. Selenium-Dependent Glutathione Peroxidases During Tumor Development. Adv. Cancer Res. 2017, 136, 109–138. [Google Scholar] [CrossRef]
- Sun, Q.; Oltra, E.; Dijck-Brouwer, D.A.J.; Chillon, T.S.; Seemann, P.; Asaad, S.; Demircan, K.; Espejo-Oltra, J.A.; Sánchez-Fito, T.; Martín-Martínez, E.; et al. Autoantibodies to selenoprotein P in chronic fatigue syndrome suggest selenium transport impairment and acquired resistance to thyroid hormone. Redox Biol. 2023, 65, 102796. [Google Scholar] [CrossRef] [PubMed]
- Ashton, K.; Hooper, L.; Harvey, L.J.; Hurst, R.; Casgrain, A.; Fairweather-Tait, S.J. Methods of assessment of selenium status in humans: A systematic review. Am. J. Clin. Nutr. 2009, 89, 2025S–2039S. [Google Scholar] [CrossRef]
- Psathakis, D.; Wedemeyer, N.; Oevermann, E.; Krug, F.; Siegers, C.P.; Bruch, H.P. Blood selenium and glutathione peroxidase status in patients with colorectal cancer. Dis. Colon Rectum 1998, 41, 328–335. [Google Scholar] [CrossRef]
- Oh, I.J.; Kim, H.E.; Song, S.Y.; Na, K.J.; Kim, K.S.; Kim, Y.C.; Lee, S.W. Diagnostic value of serum glutathione peroxidase 3 levels in patients with lung cancer. Thorac. Cancer 2014, 5, 425–430. [Google Scholar] [CrossRef]
- Barrett, C.W.; Ning, W.; Chen, X.; Smith, J.J.; Washington, M.K.; Hill, K.E.; Coburn, L.A.; Peek, R.M.; Chaturvedi, R.; Wilson, K.T.; et al. Tumor suppressor function of the plasma glutathione peroxidase gpx3 in colitis-associated carcinoma. Cancer Res. 2013, 73, 1245–1255. [Google Scholar] [CrossRef]
- Chang, S.N.; Lee, J.M.; Oh, H.; Park, J.H. Glutathione Peroxidase 3 Inhibits Prostate Tumorigenesis in TRAMP Mice. Prostate 2016, 76, 1387–1398. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhang, Y.; Chen, Y.; Liu, Z. Promoter methylation and clinical significance of GPX3 in esophageal squamous cell carcinoma. Pathol. Res. Pract. 2019, 215, 152676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, N.; Yang, X.; Xin, X.; Jia, C.H.; Li, S.; Lu, Q.; Jiang, T.; Wang, T. Machine Learning and Novel Biomarkers Associated with Immune Infiltration for the Diagnosis of Esophageal Squamous Cell Carcinoma. J. Oncol. 2022, 2022, 6732780. [Google Scholar] [CrossRef]
- Wang, N.; Pan, D.; Wang, X.; Su, M.; Wang, X.; Yan, Q.; Sun, G.; Wang, S. NAPRT, but Not NAMPT, Provides Additional Support for NAD Synthesis in Esophageal Precancerous Lesions. Nutrients 2022, 14, 4916. [Google Scholar] [CrossRef]
- Qin, J.; Zhu, Y.; Ding, Y.; Niu, T.; Zhang, Y.; Wu, H.; Zhu, L.; Yuan, B.; Qiao, Y.; Lu, J.; et al. DNA polymerase beta deficiency promotes the occurrence of esophageal precancerous lesions in mice. Neoplasia 2021, 23, 663–675. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, M.; Ying, S.M.; Zhang, C.; Lin, R.H.; Zheng, J.X.; Zhang, G.H.; Tian, D.P.; Guo, Y.; Du, C.W.; et al. Genetic Alterations in Esophageal Tissues From Squamous Dysplasia to Carcinoma. Gastroenterology 2017, 153, 166–177. [Google Scholar] [CrossRef]
- Abnet, C.C.; Arnold, M.; Wei, W.Q. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology 2018, 154, 360–373. [Google Scholar] [CrossRef]
- Wang, S.K.; Pan, D.; Chen, Z.T.; Song, G.; Han, R.Q.; Sun, G.J.; Su, M. Trends in Incidence and Mortality of Esophageal Cancer in Huai’an District, a High-Risk Area in Northern Jiangsu Province, China. Cancer Control 2022, 29. [Google Scholar] [CrossRef]
- Pan, D.; Su, M.; Zhang, T.; Miao, C.; Fu, L.; Yang, L.; Song, G.; Raine, P.J.; Wang, S.; Sun, G. A Distinct Epidemiologic Pattern of Precancerous Lesions of Esophageal Squamous Cell Carcinoma in a High-risk Area of Huai’an, Jiangsu Province, China. Cancer Prev. Res. 2019, 12, 449–462. [Google Scholar] [CrossRef]
- Pan, D.; Wang, S.; Su, M.; Sun, G.; Zhu, X.; Ghahvechi Chaeipeima, M.; Guo, Z.; Wang, N.; Zhang, Z.; Cui, M. Vitamin B(12) may play a preventive role in esophageal precancerous lesions: A case-control study based on markers in blood and 3-day duplicate diet samples. Eur. J. Nutr. 2021, 60, 3375–3386. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board (Ed.) WHO Classification of Tumours, 5th Edition: Digestive System Tumours; International Agency for Research on Cancer: Lyon, France, 2019; Volume 1, p. 37. [Google Scholar]
- Nirgude, S.; Choudhary, B. Insights into the role of GPX3, a highly efficient plasma antioxidant, in cancer. Biochem. Pharmacol. 2021, 184, 114365. [Google Scholar] [CrossRef]
- He, Y.; Wang, Y.; Li, P.; Zhu, S.; Wang, J.; Zhang, S. Identification of GPX3 epigenetically silenced by CpG methylation in human esophageal squamous cell carcinoma. Dig. Dis. Sci. 2011, 56, 681–688. [Google Scholar] [CrossRef]
- Salta, S.; Macedo-Silva, C.; Miranda-Goncalves, V.; Lopes, N.; Gigliano, D.; Guimaraes, R.; Farinha, M.; Sousa, O.; Henrique, R.; Jeronimo, C. A DNA methylation-based test for esophageal cancer detection. Biomark. Res. 2020, 8, 68. [Google Scholar] [CrossRef]
- An, B.C.; Choi, Y.D.; Oh, I.J.; Kim, J.H.; Park, J.I.; Lee, S.W. GPx3-mediated redox signaling arrests the cell cycle and acts as a tumor suppressor in lung cancer cell lines. PLoS ONE 2018, 13, e0204170. [Google Scholar] [CrossRef]
- Chang, C.; Worley, B.L.; Phaeton, R.; Hempel, N. Extracellular Glutathione Peroxidase GPx3 and Its Role in Cancer. Cancers 2020, 12, 2197. [Google Scholar] [CrossRef]
- Guven, M.; Ozturk, B.; Sayal, A.; Ozeturk, A.; Ulutin, T. Lipid peroxidation and antioxidant system in the blood of cancerous patients with metastasis. Cancer Biochem. Biophys. 1999, 17, 155–162. [Google Scholar]
- He, S.X.; Wu, B.; Chang, X.M.; Li, H.X.; Qiao, W. Effects of selenium on peripheral blood mononuclear cell membrane fluidity, interleukin-2 production and interleukin-2 receptor expression in patients with chronic hepatitis. World J. Gastroenterol. 2004, 10, 3531–3533. [Google Scholar] [CrossRef]
- Gaschler, M.M.; Stockwell, B.R. Lipid peroxidation in cell death. Biochem. Biophys. Res. Commun. 2017, 482, 419–425. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Bernlohr, D.A. Metabolic functions of FABPs—Mechanisms and therapeutic implications. Nat. Rev. Endocrinol. 2015, 11, 592–605. [Google Scholar] [CrossRef]
- Wu, Y.L.; Peng, X.E.; Zhu, Y.B.; Yan, X.L.; Chen, W.N.; Lin, X. Hepatitis B Virus X Protein Induces Hepatic Steatosis by Enhancing the Expression of Liver Fatty Acid Binding Protein. J. Virol. 2016, 90, 1729–1740. [Google Scholar] [CrossRef]
- Yamamoto, T.; Noiri, E.; Ono, Y.; Doi, K.; Negishi, K.; Kamijo, A.; Kimura, K.; Fujita, T.; Kinukawa, T.; Taniguchi, H.; et al. Renal L-type fatty acid–binding protein in acute ischemic injury. J. Am. Soc. Nephrol. 2007, 18, 2894–2902. [Google Scholar] [CrossRef]
- Jablonska, E.; Gromadzinska, J.; Peplonska, B.; Fendler, W.; Reszka, E.; Krol, M.B.; Wieczorek, E.; Bukowska, A.; Gresner, P.; Galicki, M.; et al. Lipid peroxidation and glutathione peroxidase activity relationship in breast cancer depends on functional polymorphism of GPX1. BMC Cancer 2015, 15, 657. [Google Scholar] [CrossRef]
- Mukai, T.; Egawa, M.; Takeuchi, T.; Yamashita, H.; Kusudo, T. Silencing of FABP1 ameliorates hepatic steatosis, inflammation, and oxidative stress in mice with nonalcoholic fatty liver disease. FEBS Open Bio 2017, 7, 1009–1016. [Google Scholar] [CrossRef]
- Vinceti, M.; Filippini, T.; Del Giovane, C.; Dennert, G.; Zwahlen, M.; Brinkman, M.; Zeegers, M.P.; Horneber, M.; D’Amico, R.; Crespi, C.M. Selenium for preventing cancer. Cochrane Database Syst. Rev. 2018, 1, CD005195. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Z.; Gong, P.; Yao, W.; Ba, Q.; Wang, H. Review on the health-promoting effect of adequate selenium status. Front. Nutr. 2023, 10, 1136458. [Google Scholar] [CrossRef]
- Dinh, Q.T.; Cui, Z.; Huang, J.; Tran, T.A.T.; Wang, D.; Yang, W.; Zhou, F.; Wang, M.; Yu, D.; Liang, D. Selenium distribution in the Chinese environment and its relationship with human health: A review. Environ. Int. 2018, 112, 294–309. [Google Scholar] [CrossRef]
- Fordyce, F.M.; Johnson, C.C.; Navaratna, U.R.; Appleton, J.D.; Dissanayake, C.B. Selenium and iodine in soil, rice and drinking water in relation to endemic goitre in Sri Lanka. Sci. Total Environ. 2000, 263, 127–141. [Google Scholar] [CrossRef]
- Favorito, J.E.; Grossl, P.R.; Davis, T.Z.; Eick, M.J.; Hankes, N. Soil-plant-animal relationships and geochemistry of selenium in the Western Phosphate Resource Area (United States): A review. Chemosphere 2021, 266, 128959. [Google Scholar] [CrossRef]
- Soderlund, M.; Virkanen, J.; Holgersson, S.; Lehto, J. Sorption and speciation of selenium in boreal forest soil. J. Environ. Radioact. 2016, 164, 220–231. [Google Scholar] [CrossRef]
- Spears, J.W.; Weiss, W.P. Role of antioxidants and trace elements in health and immunity of transition dairy cows. Vet. J. 2008, 176, 70–76. [Google Scholar] [CrossRef]
- Thomson, C.D.; Robinson, M.F. Urinary and Fecal Excretions and Absorption of a Large Supplement of SELENIUM—Superiority of Selenate over Selenite. Am. J. Clin. Nutr. 1986, 44, 659–663. [Google Scholar] [CrossRef]
- Pearce, N. The globalization of epidemiology: Introductory remarks. Int. J. Epidemiol. 2004, 33, 1127–1131. [Google Scholar] [CrossRef]
- Cook, M.B.; Chow, W.H.; Devesa, S.S. Oesophageal cancer incidence in the United States by race, sex, and histologic type, 1977–2005. Br. J. Cancer 2009, 101, 855–859. [Google Scholar] [CrossRef]
- Matejcic, M.; Iqbal Parker, M. Gene-environment interactions in esophageal cancer. Crit. Rev. Clin. Lab. Sci. 2015, 52, 211–231. [Google Scholar] [CrossRef]
- van Blankenstein, M.; Looman, C.W.; Hop, W.C.; Bytzer, P. The incidence of adenocarcinoma and squamous cell carcinoma of the esophagus: Barrett’s esophagus makes a difference. Am. J. Gastroenterol. 2005, 100, 766–774. [Google Scholar] [CrossRef]
- Gilbert, S.; Jobe, B.A. Surgical therapy for Barrett’s esophagus with high-grade dysplasia and early esophageal carcinoma. Surg. Oncol. Clin. N. Am. 2009, 18, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, J.B.; Reed, C.E. Epidemiology of esophageal cancer. Surg. Clin. N. Am. 2012, 92, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Kern, F.; Sharma, N.; McKeon, F.; Xian, W.; Yeoh, K.G.; Ho, K.Y.; Teh, M. FABP1 and Hepar expression levels in Barrett’s esophagus and associated neoplasia in an Asian population. Dig. Liver Dis. 2017, 49, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Dum, D.; Ocokoljic, A.; Lennartz, M.; Hube-Magg, C.; Reiswich, V.; Hoflmayer, D.; Jacobsen, F.; Bernreuther, C.; Lebok, P.; Sauter, G.; et al. FABP1 expression in human tumors: A tissue microarray study on 17,071 tumors. Virchows Arch. 2022, 481, 945–961. [Google Scholar] [CrossRef]
- Abrams, J.A.; Del Portillo, A.; Hills, C.; Compres, G.; Friedman, R.A.; Cheng, B.; Poneros, J.; Lightdale, C.J.; De La Rue, R.; di Pietro, M.; et al. Randomized Controlled Trial of the Gastrin/CCK(2) Receptor Antagonist Netazepide in Patients with Barrett’s Esophagus. Cancer Prev. Res. 2021, 14, 675–682. [Google Scholar] [CrossRef]


| Stage | GPx3 | |||
|---|---|---|---|---|
| Low | High | p Value | ||
| n | 81 | 81 | ||
| Age, ± s | 62.49 ± 11.79 | 61.99 ± 12.31 | 0.790 | |
| T, n (%) | T1 | 11 (7.6) | 16 (11) | 0.478 |
| T2 | 19 (13.1) | 18 (12.4) | ||
| T3 | 42 (29) | 35 (24.1) | ||
| T4 | 1 (0.7) | 3 (2.1) | ||
| N, n (%) | N0 | 35 (24.3) | 31 (21.5) | 0.272 |
| N1 | 30 (20.8) | 33 (22.9) | ||
| N2 | 3 (2.1) | 6 (4.2) | ||
| N3 | 5 (3.5) | 1 (0.7) | ||
| M, n (%) | M0 | 58 (45) | 63 (48.8) | 0.486 |
| M1 | 5 (3.9) | 3 (2.3) | ||
| Median (25th–75th) | EPLs | Controls | p Value |
|---|---|---|---|
| Dietary samples (n) | 100 | 100 | |
| Dietary Selenium (μg/d) | 136.72 (94.77–180.06) | 147.20 (99.20–177.65) | 0.498 |
| Blood samples (n) | 100 | 100 | |
| Plasma Selenium (μg/L) | 49.46 (41.75–57.71) | 55.97 (43.07–68.23) | <0.001 ** |
| GPx3 (pmol/mL) | 42.78 (35.68–51.18) | 44.82 (34.48–56.56) | 0.561 |
| FABP1 (ng/L) | 1433.61 (1337.08–1512.72) | 1388.40 (1297.61–1481.82) | 0.011 * |
| Plasma Selenium | Dietary Selenium | GPx3 | FABP1 | |
|---|---|---|---|---|
| EPLs | 1.00 | 0.931 ** | 0.141 * | −0.76 |
| Controls | 1.00 | 0.924 ** | 0.239 * | −0.82 |
| Q1 | Q2 | Q3 | p for Trend | |
|---|---|---|---|---|
| OR (95% CI) | OR (95% CI) | OR (95% CI) | ||
| Dietary samples | ||||
| Dietary Selenium (μg/d) | 11.44–114.27 | 114.28–165.68 | 165.69–379.39 | |
| EPLs cases (%) | 48.48 | 52.24 | 49.25 | |
| crude model | 1.00 | 1.22 (0.56–2.67) | 1.05 (0.49–2.24) | 0.846 |
| adjusted model # | 1.00 | 1.91 (0.69–5.29) | 1.45 (0.52–4.01) | 0.545 |
| Blood samples | ||||
| Plasma Selenium (μg/L) | 5.03–45.53 | 45.54–59.26 | 59.27–115.92 | |
| EPLs cases (%) | 56.06 | 58.82 | 34.85 | |
| crude model | 1.00 | 1.29 (0.63–2.63) | 0.39 (0.19–0.82) * | 0.021 * |
| adjusted model | 1.00 | 2.04 (0.79–5.25) | 0.34 (0.13–0.93) * | 0.103 |
| GPx3 (pmol/mL) | 17.00–37.79 | 37.80–47.99 | 48.00–125.96 | |
| EPLs cases (%) | 52.24 | 61.19 | 36.36 | |
| crude model | 1.00 | 1.64 (0.78–3.47) | 0.56 (0.27–1.13) | 0.291 |
| adjusted model | 1.00 | 1.56 (0.60–4.04) | 0.27 (0.10–0.70) * | 0.035 * |
| FABP1 (ng/L) | 1192.84–1346.47 | 1346.48–1465.23 | 1465.24–1649.36 | |
| EPLs cases (%) | 73.68 | 52.24 | 55.22 | |
| crude model | 1.00 | 1.41 (0.73–2.71) | 1.54 (0.82–2.90) | 0.180 |
| adjusted model | 1.00 | 1.77 (0.77–4.09) | 2.30 (0.99–5.31) | 0.046 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Pan, D.; Zhu, X.; Ren, X.; Jin, X.; Chen, X.; Wang, Y.; Su, M.; Sun, G.; Wang, S. Selenium May Be Involved in Esophageal Squamous Cancer Prevention by Affecting GPx3 and FABP1 Expression: A Case-Control Study Based on Bioinformatic Analysis. Nutrients 2024, 16, 1322. https://doi.org/10.3390/nu16091322
Wang N, Pan D, Zhu X, Ren X, Jin X, Chen X, Wang Y, Su M, Sun G, Wang S. Selenium May Be Involved in Esophageal Squamous Cancer Prevention by Affecting GPx3 and FABP1 Expression: A Case-Control Study Based on Bioinformatic Analysis. Nutrients. 2024; 16(9):1322. https://doi.org/10.3390/nu16091322
Chicago/Turabian StyleWang, Niannian, Da Pan, Xiaopan Zhu, Xingyuan Ren, Xingyi Jin, Xiangjun Chen, Yuanyuan Wang, Ming Su, Guiju Sun, and Shaokang Wang. 2024. "Selenium May Be Involved in Esophageal Squamous Cancer Prevention by Affecting GPx3 and FABP1 Expression: A Case-Control Study Based on Bioinformatic Analysis" Nutrients 16, no. 9: 1322. https://doi.org/10.3390/nu16091322
APA StyleWang, N., Pan, D., Zhu, X., Ren, X., Jin, X., Chen, X., Wang, Y., Su, M., Sun, G., & Wang, S. (2024). Selenium May Be Involved in Esophageal Squamous Cancer Prevention by Affecting GPx3 and FABP1 Expression: A Case-Control Study Based on Bioinformatic Analysis. Nutrients, 16(9), 1322. https://doi.org/10.3390/nu16091322










