Metabolite Profiling, Biological and Molecular Analyses Validate the Nutraceutical Potential of Green Seaweed Acrosiphonia orientalis for Human Health
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Non-Targeted Metabolite Profiling: Extraction and Identification of Metabolites
2.3. Targeted Metabolite Profiling: Amino Acid Profiling
2.4. Targeted Metabolomics: Phenolic and Flavonoid Compounds
2.5. Estimation of Total Phenolic and Total Flavonoid Contents
2.6. Biological Activities
2.7. Measurement of Intracellular ROS
2.8. Nuclear Staining with Hoechst Dye 33342 and DNA Fragmentation Study
2.9. Transcript Expression Analysis by Quantitative Real-Time PCR
2.10. Microarray Based Differential Gene Expression Analysis
2.11. Data Mining and Statistical Analysis
3. Results
3.1. Metabolite Profiling (Non-Targeted)
3.2. Targeted Metabolite Profiling
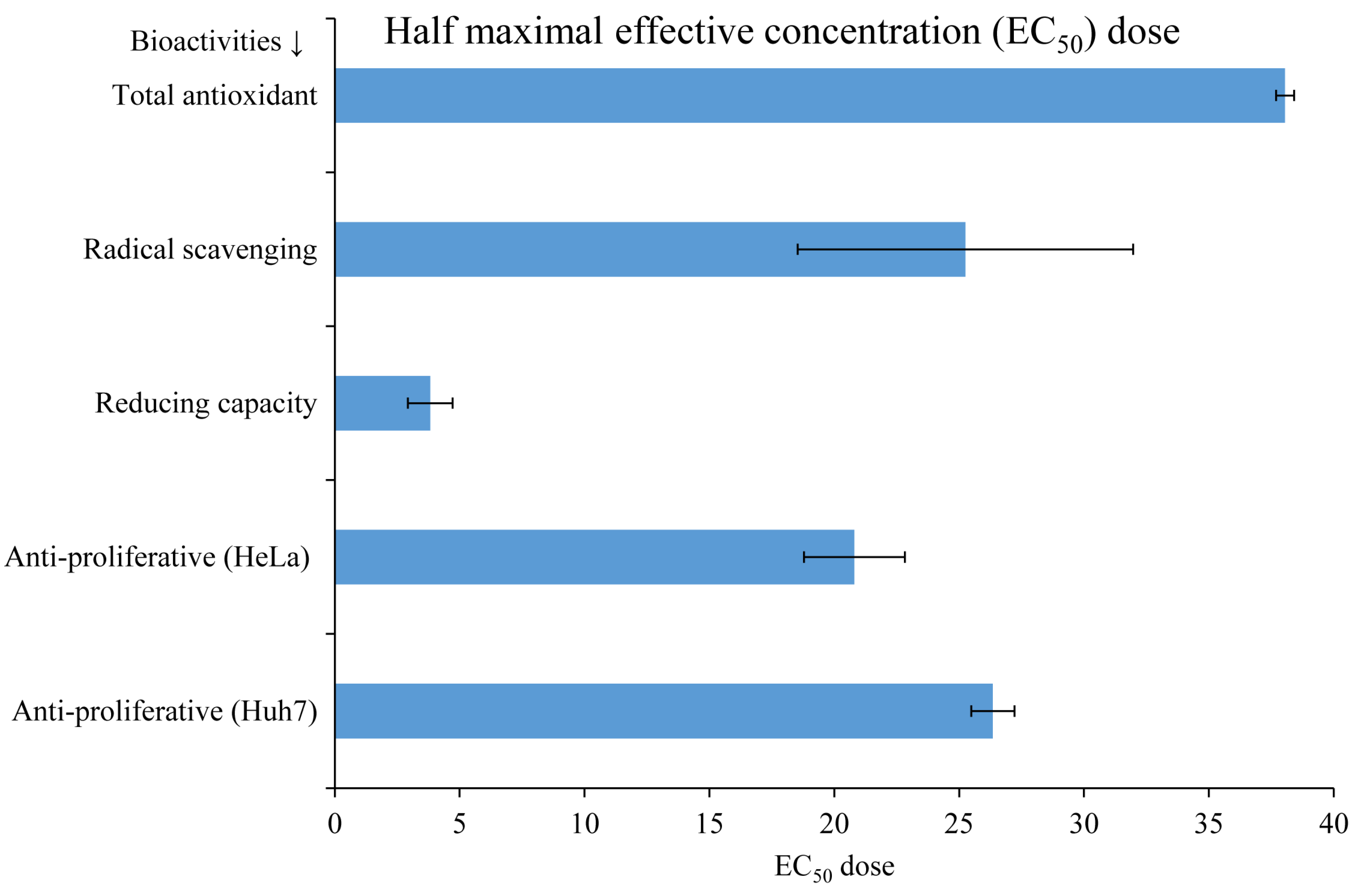
3.3. Biological Activities
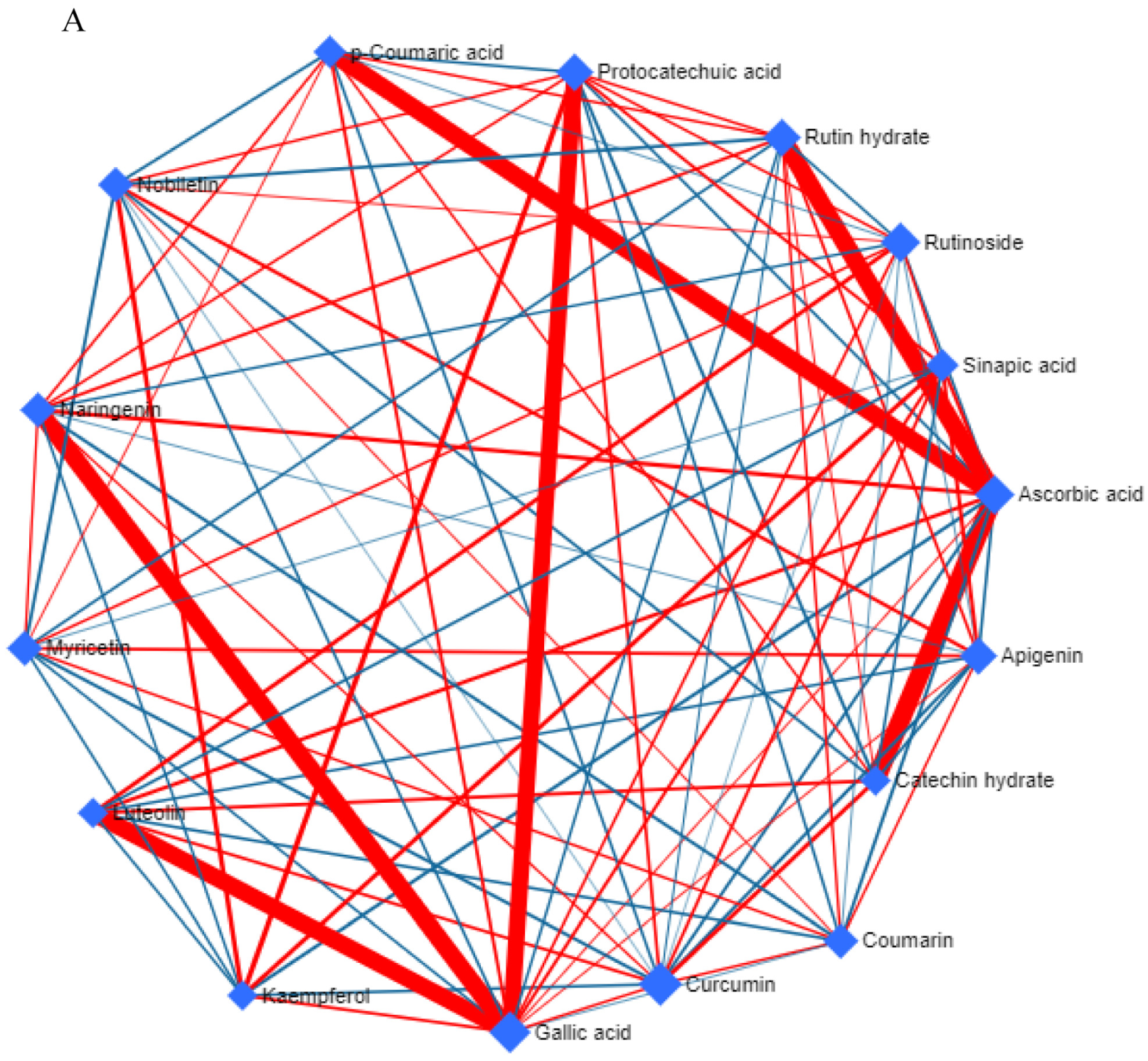
3.4. Metabolic Network and Pathway Enrichment Analysis to Elucidate the Interactions between Metabolite Changes and Metabolic Pathways
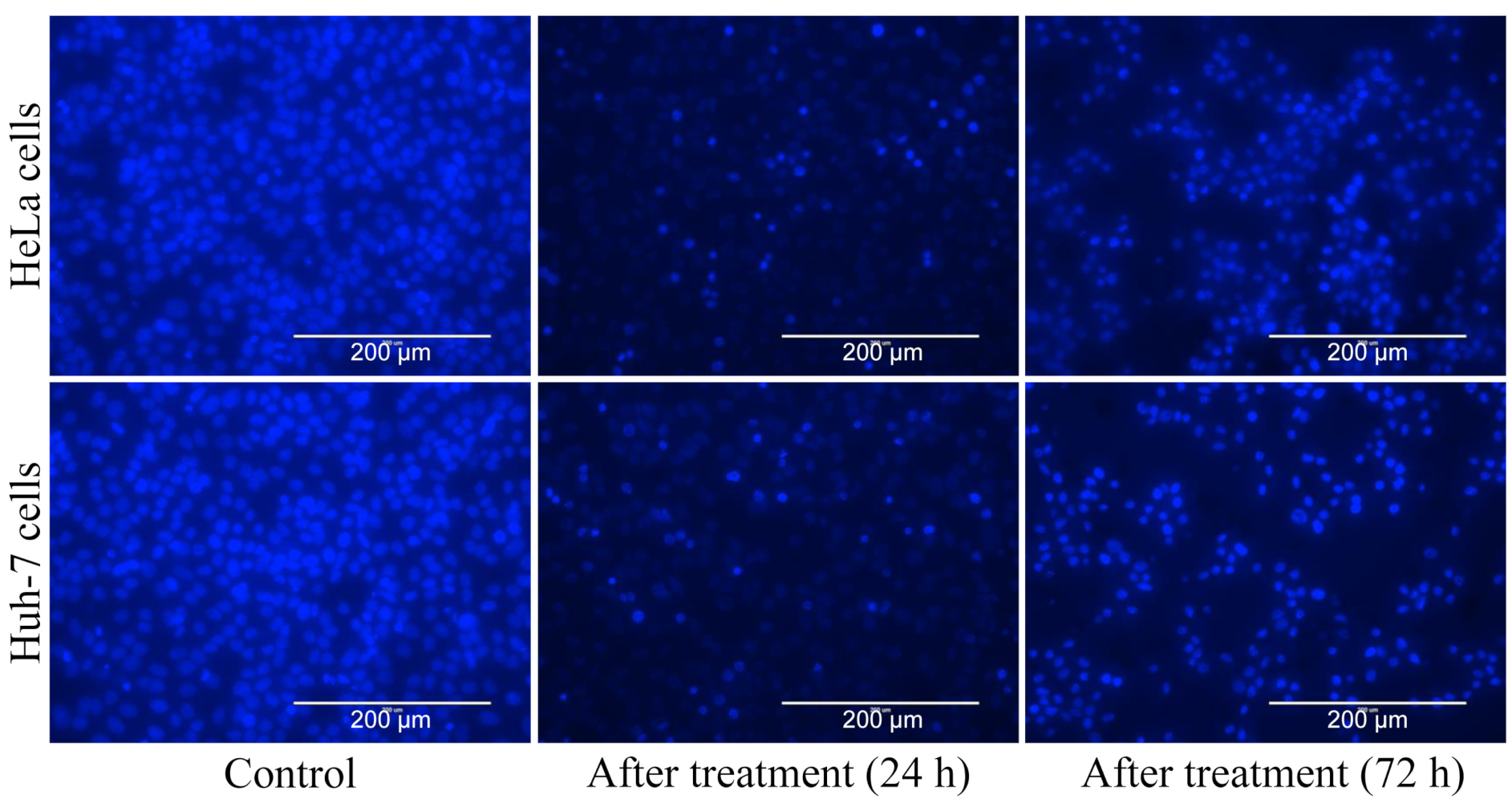
3.5. Apoptosis Analysis and ROS Inhibitory Activity
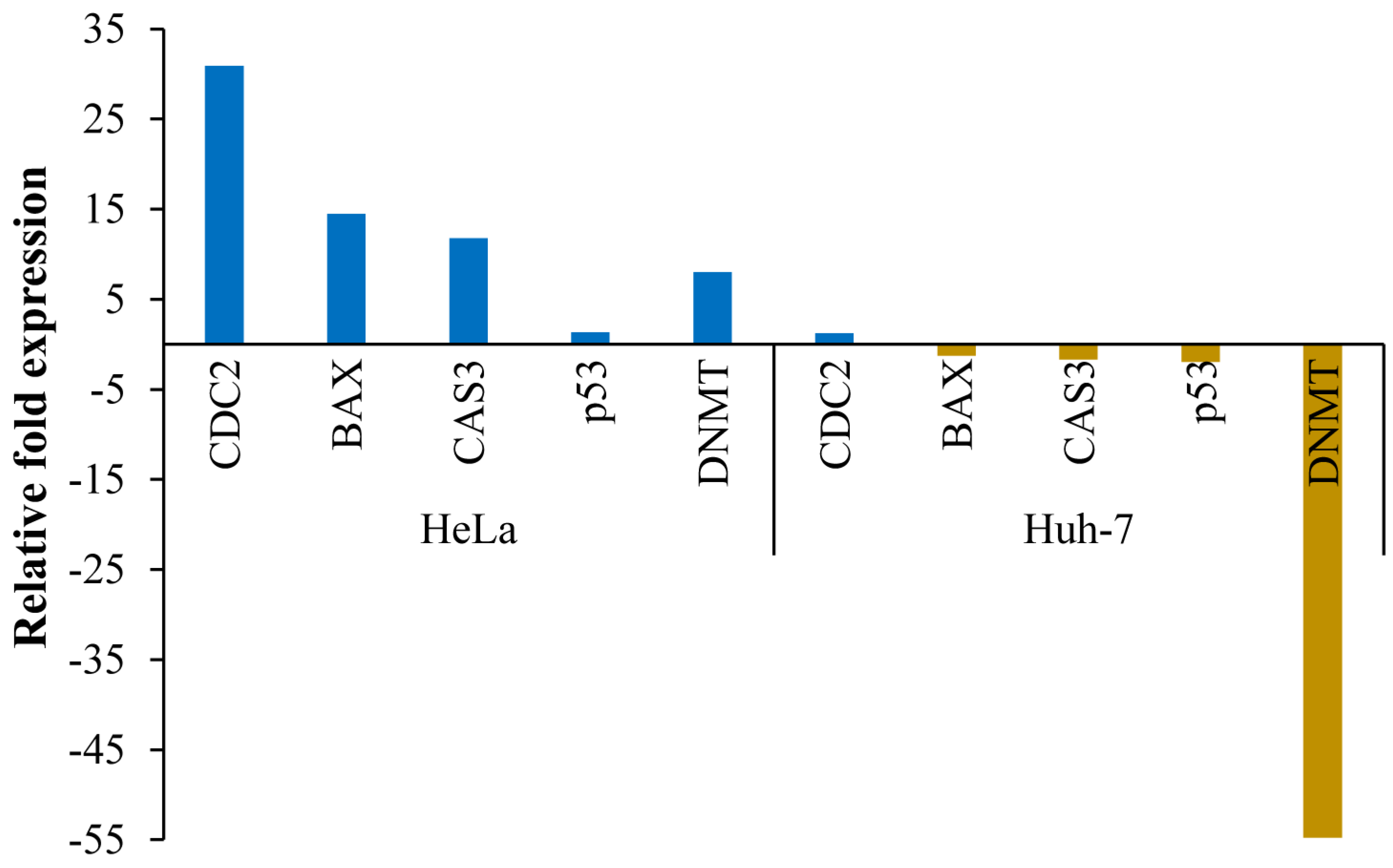
3.6. Transcript Expression Analysis by Quantitative Real-Time PCR
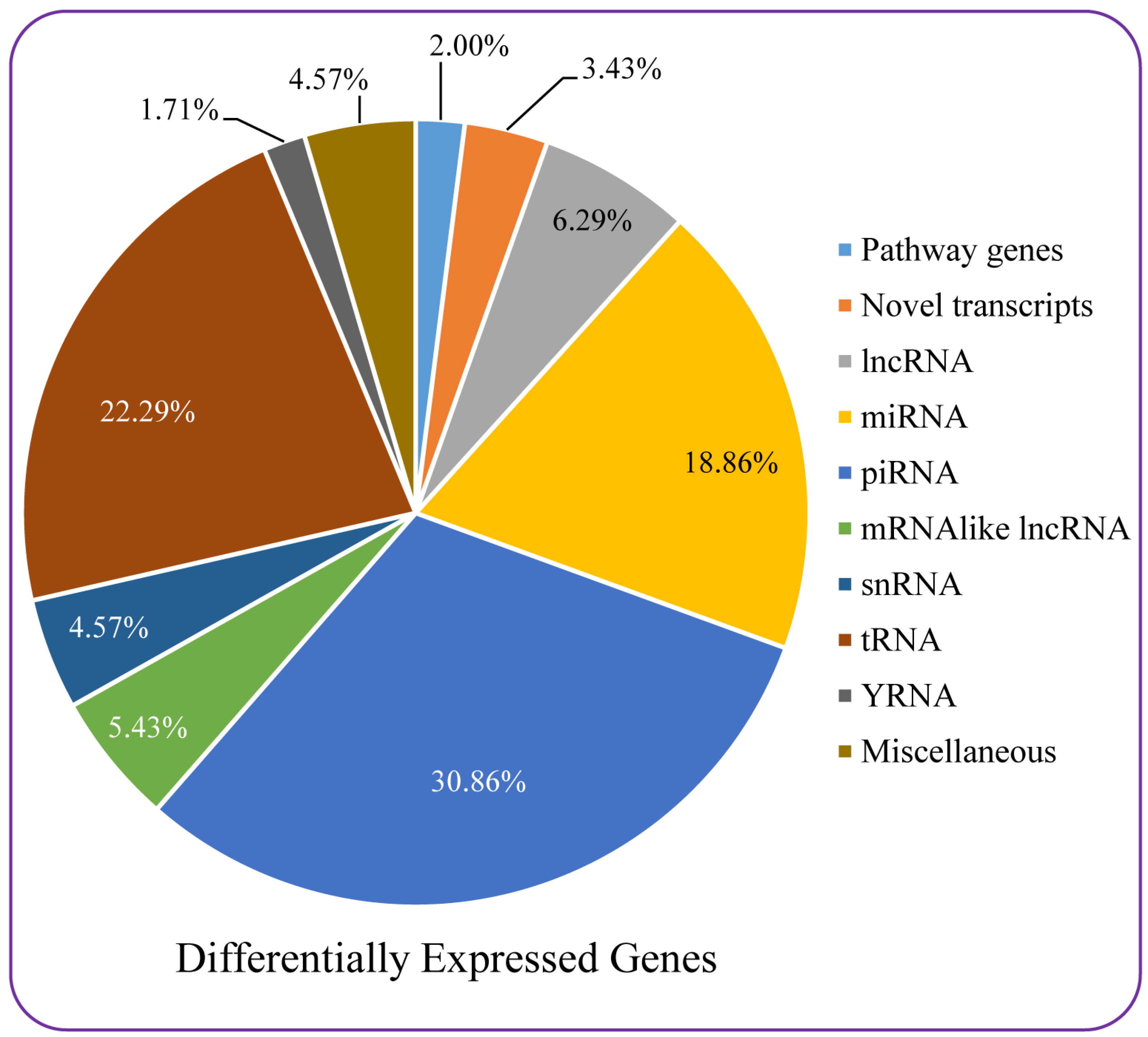
3.7. Microarray-Based Differential Gene Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smit, A.J. Medicinal and pharmaceutical uses of seaweed natural products: A review. J. Appl. Phycol. 2004, 16, 245–262. [Google Scholar] [CrossRef]
- Namvar, F.; Mohamad, R.; Baharara, J.; Zafar-Balanejad, S.; Fargahi, F.; Rahman, H.S. Antioxidant, antiproliferative, and antiangiogenesis effects of polyphenol-rich seaweed (Sargassum muticum). BioMed Res. Int. 2013, 2013, 604787. [Google Scholar] [CrossRef]
- Mishra, A. Algal Functional Foods and Nutraceuticals: Benefits, Opportunities, and Challenges; Bentham Science Publishers: Singapore, 2022. [Google Scholar] [CrossRef]
- Baghel, R.S.; Choudhary, B.; Pandey, S.; Pathak, P.K.; Patel, M.K.; Mishra, A. Rehashing Our Insight of Seaweeds as a Potential Source of Foods, Nutraceuticals, and Pharmaceuticals. Foods 2023, 12, 3642. [Google Scholar] [CrossRef]
- Tanna, B.; Brahmbhatt, H.R.; Mishra, A. Phenolic, flavonoid and amino acid compositions reveal that selected tropical seaweeds have the potential to be functional food ingredients. J. Food Process. Preserv. 2019, 43, e14266. [Google Scholar] [CrossRef]
- Sussmann, A.V.; DeWreede, R.E. Life history of Acrosiphonia (Codiolales, Chlorophyta) in southwestern British Columbia, Canada. Am. J. Bot. 2001, 88, 1535–1544. [Google Scholar] [CrossRef]
- Shanmughapriya, S.; Manilal, A.; Sujith, S.; Selvin, J.; Kiran, G.S.; Natarajaseenivasan, K. Antimicrobial activity of seaweeds extracts against multiresistant pathogens. Ann. Microbiol. 2008, 58, 535–541. [Google Scholar] [CrossRef]
- Manilal, A.; Sujith, S.; Selvin, J.; Kiran, G.S.; Shakir, C. In vivo antiviral activity of polysaccharide from the Indian green alga, Acrosiphonia orientalis (J. Agardh): Potential implication in shrimp disease management. World J. Fish Mar. Sci. 2009, 1, 278–282. Available online: https://www.idosi.org/wjfms/wjfms1(4)09/5.pdf (accessed on 16 November 2023).
- Rangaiah, G.S.; Lakshmi, P.; Sruthikeerthi, K. Antimicrobial activity of the crude extracts of Chlorophyceae seaweed Ulva, Caulerpa and Spongomorpha sps. against clinical and phytopathogens. Drug Invent. Today 2010, 2, 311–314. [Google Scholar]
- Sujatha, L.; Govardhan, T.L.; Rangaiah, G.S. Antibacterial activity of green seaweeds on oral bacteria. Indian J. Nat. Prod. Resour. 2012, 3, 328–333. [Google Scholar]
- Sarojini, Y.; Lakshminarayana, K.; Rao, P.S. Variations in distribution of flavonoids in some seaweed of Visakhapatnam coast of India. Der Pharma Chem. 2012, 4, 1481–1484. [Google Scholar]
- Manilal, A.; Selvin, J.; Sujith, S.; Panikkar, M.V. Evaluation of therapeutic efficacy of Indian green alga, Acrosiphonia orientalis (J. Agardh) in the treatment of vibriosis in Penaeus monodon. Thalassas 2012, 28, 33–46. [Google Scholar]
- Lee, H.H.; Lin, C.T.; Yang, L.L. Neuroprotection and free radical scavenging effects of Osmanthus fragrans. J. Biomed. Sci. 2007, 14, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Tanna, B.; Choudhary, B.; Mishra, A.; Chauhan, O.P.; Patel, M.K.; Shokralla, S.; El-Abedin, T.K.Z.; Elansary, H.O.; Mahmoud, E.A. Antioxidant, scavenging, reducing, and anti-proliferative activities of selected tropical brown seaweeds confirm the nutraceutical potential of Spatoglossum asperum. Foods 2021, 10, 2482. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, B.; Khandwal, D.; Gupta, N.K.; Patel, J.; Mishra, A. Nutrient composition, physicobiochemical analyses, oxidative stability and antinutritional assessment of abundant tropical seaweeds from the Arabian Sea. Plants 2023, 12, 2302. [Google Scholar] [CrossRef]
- Lourenço, S.C.; Moldão-Martins, M.; Alves, V.D. Antioxidants of natural plant origins: From sources to food industry applications. Molecules 2019, 24, 4132. [Google Scholar] [CrossRef]
- Farasat, M.; Khavari-Nejad, R.A.; Nabavi, S.M.; Namjooyan, F. Antioxidant activity, total phenolics and flavonoid contents of some edible green seaweeds from northern coasts of the Persian Gulf. Iran. J. Pharm. Res. 2014, 13, 163–170. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3985267/ (accessed on 20 December 2023).
- Wijesinghe, W.A.J.P.; Jeon, Y.J. Biological activities and potential cosmeceutical applications of bioactive components from brown seaweeds: A review. Phytochem. Rev. 2011, 10, 431–443. [Google Scholar] [CrossRef]
- Tanna, B.; Mishra, A. Metabolomics of Seaweeds: Tools and Techniques. In Plant Metabolites and Regulation under Environmental Stress; Ahmad, P., Ahanger, M.A., Singh, V.P., Tripathi, D.K., Alam, P., Alyemeni, M.N., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2018; pp. 37–52. [Google Scholar] [CrossRef]
- Kumari, P.; Bijo, A.J.; Mantri, V.A.; Reddy, C.R.K.; Jha, B. Fatty acid profiling of tropical marine macroalgae: An analysis from chemotaxonomic and nutritional perspectives. Phytochemistry 2013, 86, 44–56. [Google Scholar] [CrossRef]
- Jha, B.; Reddy, C.R.K.; Thakur, M.C.; Rao, M.U. Seaweeds of India: The Diversity and Distribution of Seaweeds of Gujarat Coast, 3rd ed.; Springer Science & Business Media: Dordrecht, The Netherlands, 2009; p. 19. [Google Scholar] [CrossRef]
- Tanna, B.; Choudhary, B.; Mishra, A. Metabolite profiling, antioxidant, scavenging and anti-proliferative activities of selected tropical green seaweeds reveal the nutraceutical potential of Caulerpa spp. Algal Res. 2018, 36, 96–105. [Google Scholar] [CrossRef]
- Saeed, N.; Khan, M.R.; Shabbir, M. Antioxidant activity, total phenolic and total flavonoid contents of whole plant extracts Torilis leptophylla L. BMC Complement. Altern. Med. 2012, 12, 221. [Google Scholar] [CrossRef]
- Tanna, B.; Choudhary, B.; Mishra, A.; Yadav, S.; Chauhan, O.P.; Elansary, H.O.; Shokralla, S.; El-Abedin, T.K.Z.; Mahmoud, E.A. Biochemical and anti-proliferative activities of seven abundant tropical red seaweeds confirm nutraceutical potential of Grateloupia indica. Arab. J. Chem. 2022, 15, 103868. [Google Scholar] [CrossRef]
- Sim Choi, H.; Woo Kim, J.; Cha, Y.N.; Kim, C. A quantitative nitroblue tetrazolium assay for determining intracellular superoxide anion production in phagocytic cells. J. Immunoass. Immunochem. 2006, 27, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Tanna, B.; Yadav, S.; Mishra, A. Anti-proliferative and ROS-inhibitory activities reveal the anticancer potential of Caulerpa species. Mol. Biol. Rep. 2020, 47, 7403–7411. [Google Scholar] [CrossRef]
- Amatori, S.; Persico, G.; Fanelli, M. Real-time quantitative PCR array to study drug-induced changes of gene expression in tumor cell lines. J. Cancer Metastatis Treat. 2017, 3, 90–99. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Alexander, A.; Singh, V.K.; Mishra, A. Introgression of a novel gene AhBINR differentially expressed during PGPR Brachybacterium saurashtrense-Arachis hypogaea interaction enhances plant performance under nitrogen starvation and salt stress in tobacco. Plant Sci. 2022, 324, 111429. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Rajubabu, D.C.H.; Srinivasulu, A.; Aruna, B.; Satyanarayana, N.; Narayana, P.V.L.; Rao, A.D.P. Nutritional and toxicological importance of selected seaweed from Visakhapatnam coast. Int. J. Sci. Res. Sci. Eng. Technol. 2017, 3, 840–850. Available online: https://ijsrset.com/IJSRSET1736208 (accessed on 21 December 2023).
- Song, J.; He, Y.; Luo, C.; Feng, B.; Ran, F.; Xu, H.; Ci, Z.; Xu, R.; Han, L.; Zhang, D. New progress in the pharmacology of protocatechuic acid: A compound ingested in daily foods and herbs frequently and heavily. Pharmacol. Res. 2020, 161, 105109. [Google Scholar] [CrossRef]
- Wolfe, R.R. Branched-chain amino acids and muscle protein synthesis in humans: Myth or reality? J. Int. Soc. Sports Nutr. 2017, 14, 30. [Google Scholar] [CrossRef]
- Roy, S.; Chikkerur, J.; Roy, S.C.; Dhali, A.; Kolte, A.P.; Sridhar, M.; Samanta, A.K. Tagatose as a potential nutraceutical: Production, properties, biological roles, and applications. J. Food Sci. 2018, 83, 2699–2709. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, H.; Li, J.; Qin, S.; Yang, W.; Ma, X.; Qiao, X.; Yang, B. Effects of genetic background and altitude on sugars, malic acid and ascorbic acid in fruits of wild and cultivated apples (Malus sp.). Foods 2021, 10, 2950. [Google Scholar] [CrossRef]
- Niklander, S.; Fuentes, F.; Sanchez, D.; Araya, V.; Chiappini, G.; Martinez, R.; Marshall, M. Impact of 1% malic acid spray on the oral health-related quality of life of patients with xerostomia. J. Oral Sci. 2018, 60, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Ocak, N.; Erener, G.; Altop, A.; Kop, C. The effect of malic acid on performance and some digestive tract traits of Japanese quails. J. Poult. Sci. 2009, 46, 25–29. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Hitomi, M.; Van Der Wee, K.; Rothenberg, F.; Fisher, S.A.; Zucker, R.; Svoboda, K.K.; Goldsmith, E.C.; Heiskanen, K.M.; Nieminen, A.L. The pros and cons of apoptosis assays for use in the study of cells, tissues, and organs. Microsc. Microanal. 2002, 8, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Wojtunik-Kulesza, K.; Oniszczuk, A.; Oniszczuk, T.; Combrzyński, M.; Nowakowska, D.; Matwijczuk, A. Influence of in vitro digestion on composition, bioaccessibility and antioxidant activity of food polyphenols—A non-systematic review. Nutrients 2020, 12, 1401. [Google Scholar] [CrossRef]
- Olsson, M.; Zhivotovsky, B. Caspases and cancer. Cell Death Differ. 2011, 18, 1441–1449. [Google Scholar] [CrossRef]
- Yi, Y.; Zhang, W.; Yi, J.; Xiao, Z.X. Role of p53 family proteins in metformin anti-cancer activities. J. Cancer 2019, 10, 2434–2442. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, J. DNA methyltransferases and their roles in tumorigenesis. Biomark. Res. 2017, 5, 1. [Google Scholar] [CrossRef]
- Chen, J.; Song, B.; Kong, G. MicroRNA-663b downregulation inhibits proliferation and induces apoptosis in bladder cancer cells by targeting TUSC2. Mol. Med. Rep. 2019, 19, 3896–3902. [Google Scholar] [CrossRef] [PubMed]
- de Almeida Chuffa, L.G.; de Souza, M.C.; Cruz, E.M.S.; Ferreira, F.B.; de Morais, J.M.B.; Seiva, F.R.F. Hepatocellular carcinoma and miRNAs: An in silico approach revealing potential therapeutic targets for polyphenols. Phytomed. Plus 2022, 2, 100259. [Google Scholar] [CrossRef]
- Wen, P.; Kong, R.; Liu, J.; Zhu, L.; Chen, X.; Li, X.; Nie, Y.; Wu, K.; Wu, J.Y. USP33, a new player in lung cancer, mediates Slit-Robo signaling. Protein Cell 2014, 5, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Kandel, S.H.; Ahmedy, I.A.; Al-Hassanin, S.A.; El-Gazzar, S.S. Evaluation of feline McDonough sarcoma-like tyrosine kinase 3 (CD135) expression in acute myeloblastic leukemia. Menoufia Med. J. 2019, 32, 784–789. [Google Scholar] [CrossRef]
- Khan, S.U.; Pushparaj, K.; Balasubramanian, B.; Meyyazhagan, A.; Alghamdi, S.; Alghamdi, A.; Kadaikunnan, S.; Alharbi, N.S.; Khaled, J.M.; Thiruvengadam, M. Marine macrolides as an efficient source of FMS-like tyrosine kinase 3 inhibitors: A comprehensive approach of in silico virtual screening. S. Afr. J. Bot. 2022, 148, 93–103. [Google Scholar] [CrossRef]
- Gupta, M.; Chandan, K.; Sarwat, M. Natural products and their derivatives as immune check point inhibitors: Targeting cytokine/chemokine signalling in cancer. Semin. Cancer Biol. 2022, 86, 214–232. [Google Scholar] [CrossRef] [PubMed]
- Méresse, S.; Fodil, M.; Fleury, F.; Chénais, B. Fucoxanthin, a marine-derived carotenoid from brown seaweeds and microalgae: A promising bioactive compound for cancer therapy. Int. J. Mol. Sci. 2020, 21, 9273. [Google Scholar] [CrossRef]
- Wu, H.L.; Fu, X.Y.; Cao, W.Q.; Xiang, W.Z.; Hou, Y.J.; Ma, J.K.; Wang, Y.; Fan, C.D. Induction of apoptosis in human glioma cells by fucoxanthin via triggering of ros-mediated oxidative damage and regulation of mapks and pi3k–akt pathways. J. Agric. Food Chem. 2019, 67, 2212–2219. [Google Scholar] [CrossRef]




| Metabolites | Name | Amount ^ |
|---|---|---|
| Amino acid | Asparagine | 12 ± 7 |
| Amino acid | Aspartic acid | 8 ± 0.5 |
| Amino acid | Glutamic acid | 50 ± 18 |
| Amino acid | Glutamine | 2 ± 0.2 |
| Amino acid | Glycine | 7 ± 2 |
| Amino acid | Leucine | 4 ± 0.05 |
| Amino acid | Lysine | 15 ± 3 |
| Amino acid | Phenylalanine | 3 ± 0.5 |
| Amino acid | Proline | 1 ± 0.2 |
| Amino acid | Serine | 3 ± 0.1 |
| Amino acid | Threonine | 3 ± 0.02 |
| Amino acid | Threonine | 4 ± 1 |
| Amino acid | Tyrosine | 3 ± 0.3 |
| Amino acid | Valine | 6 ± 1 |
| Amino acid derivative | Pyroglutamic acid | 18 ± 8 |
| Dicarboxylic acid | Succinic acid | 4 ± 0.4 |
| Fatty acid | Heptadecanoic acid | 1 ± 0.3 |
| Fatty acid | Myristic acid | 6 ± 2 |
| Fatty acid | Palmitic acid | 57 ± 8 |
| Fatty acid | Stearic acid | 50 ± 8 |
| Organic acid | 2-Propenoic acid | 13 ± 5 |
| Organic compound | Allylamine | 4 ± 1 |
| Organic compound | Citric acid | 5 ± 1 |
| Organic compound | Malic acid | 175 ± 5 |
| Organic acid | Pipecolic acid | 36 ± 3 |
| Organic compound | Pyrazine | 8 ± 4 |
| Organic compound | Succinic anhydride | 1 ± 0.1 |
| Phenolic acid | Protocatechuic acid | 630 ± 15 |
| Polyol compound | Glyceryl-glycoside | 24 ± 2 |
| Sugar | 2-Deoxy-D-ribose | 10 ± 2 |
| Sugar | Arabinopyranose | 20 ± 4 |
| Sugar | Glucose | 2 ± 0.5 |
| Sugar | Maltose | 1430 ± 30 |
| Sugar | Mannose | 11 ± 2 |
| Sugar | Melibiose | 1 ± 0.01 |
| Sugar | Myo-Inositol | 12 ± 1 |
| Sugar | Sucrose | 1830 ± 145 |
| Sugar | Tagatofuranose | 4 ± 0.4 |
| Sugar | Tagatose | 100 ± 10 |
| Sugar | Trehalose | 80 ± 8 |
| Sugar acid | Gluconolactone | 13 ± 1 |
| Sugar acid | Glyceric acid | 5 ± 1 |
| Sugar acid | Glycerol | 8 ± 0.4 |
| Sugar acid | Ribonic acid | 6 ± 2 |
| Sugar acid | Threonic acid | 24 ± 8 |
| Sugar alcohol | Mannitol | 28 ± 2 |
| Sugar alcohol | Scyllo-Inositol | 18 ± 2 |
| Amino Acids | Amount ^ |
|---|---|
| Essential amino acids (EAA) | |
| Histidine | 0.3 ± 0.1 |
| Isoleucine | 4 ± 2 |
| Leucine | 9 ± 0.5 |
| Lysine | 8 ± 2 |
| Methionine | 1 ± 0.1 |
| Phenylalanine | 50 ± 2 |
| Threonine | 2 ± 0.1 |
| Valine | 0.01 ± 0.0 |
| Conditional EAA | |
| Arginine | 2 ± 0.2 |
| Cysteine | 5 ± 1 |
| Glutamic acid | 0.3 ± 0.05 |
| Glycine | 0.03 ± 0.00 |
| Proline | 2 ± 0.1 |
| Tyrosine | 40 ± 10 |
| Others amino acids | |
| Alanine | 0.3 ± 0.1 |
| Aspartic acid | 17 ± 1 |
| Serine | 2 ± 0.1 |
| Metabolites | Amount ^ |
|---|---|
| Apigenin | 0.01 |
| Ascorbic acid | 10 |
| Catechin hydrate | 20 |
| Coumarin | 0.03 |
| Curcumin | 0.1 |
| Gallic acid | 7 |
| Kaempferol | 0.01 |
| Luteolin | 0.05 |
| Myricetin | 0.1 |
| Naringenin | 0.01 |
| Nobiletin | 0.01 |
| p-Coumaric acid | 0.002 |
| Protocatechuic acid | 6 |
| Rutin hydrate | 0.2 |
| Rutinoside | 0.01 |
| Sinapic acid | 0.01 |
| Microarray Gene Prob Id | Fold Change Log 2 | Chromosome Number | Strand | Group | Gene Description/Role |
|---|---|---|---|---|---|
| TC10001334.hg.1 | 2.4 | chr10 | - | Coding | Annexin II |
| TC22000851.hg.1 | 2.07 | chr22 | - | Coding | ATP synthase, H+ transportation |
| TC0X001435.hg.1 | −2.25 | chrX | - | Coding | CDR1 gene encoding for cerebellar degeneration-related protein 1 |
| TC04001578.hg.1 | 2.08 | chr4 | - | Coding | Protein phosphatase 1, regulatory (inhibitor) subunit |
| TC07002266.hg.1 | −2.79 | chr7 | + | Non-coding | scaRNA (small Cajal body-specific RNAs) |
| TC05001641.hg.1 | −2.12 | chr5 | - | Coding | Solute carrier organic anion transporter (SLCO4C1) |
| TC11000565.hg.1 | −2.15 | chr11 | + | Coding | Transmembrane protein 179B (TMEM179B) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanna, B.; Yadav, S.; Patel, M.K.; Mishra, A. Metabolite Profiling, Biological and Molecular Analyses Validate the Nutraceutical Potential of Green Seaweed Acrosiphonia orientalis for Human Health. Nutrients 2024, 16, 1222. https://doi.org/10.3390/nu16081222
Tanna B, Yadav S, Patel MK, Mishra A. Metabolite Profiling, Biological and Molecular Analyses Validate the Nutraceutical Potential of Green Seaweed Acrosiphonia orientalis for Human Health. Nutrients. 2024; 16(8):1222. https://doi.org/10.3390/nu16081222
Chicago/Turabian StyleTanna, Bhakti, Sonam Yadav, Manish Kumar Patel, and Avinash Mishra. 2024. "Metabolite Profiling, Biological and Molecular Analyses Validate the Nutraceutical Potential of Green Seaweed Acrosiphonia orientalis for Human Health" Nutrients 16, no. 8: 1222. https://doi.org/10.3390/nu16081222
APA StyleTanna, B., Yadav, S., Patel, M. K., & Mishra, A. (2024). Metabolite Profiling, Biological and Molecular Analyses Validate the Nutraceutical Potential of Green Seaweed Acrosiphonia orientalis for Human Health. Nutrients, 16(8), 1222. https://doi.org/10.3390/nu16081222