Effects of a High-Fat Diet on Insulin-Related miRNAs in Plasma and Brain Tissue in APPSwe/PS1dE9 and Wild-Type C57BL/6J Mice
Abstract
1. Introduction
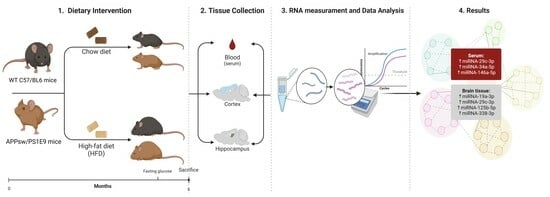
2. Materials and Methods
2.1. Animals and Dietary Treatment
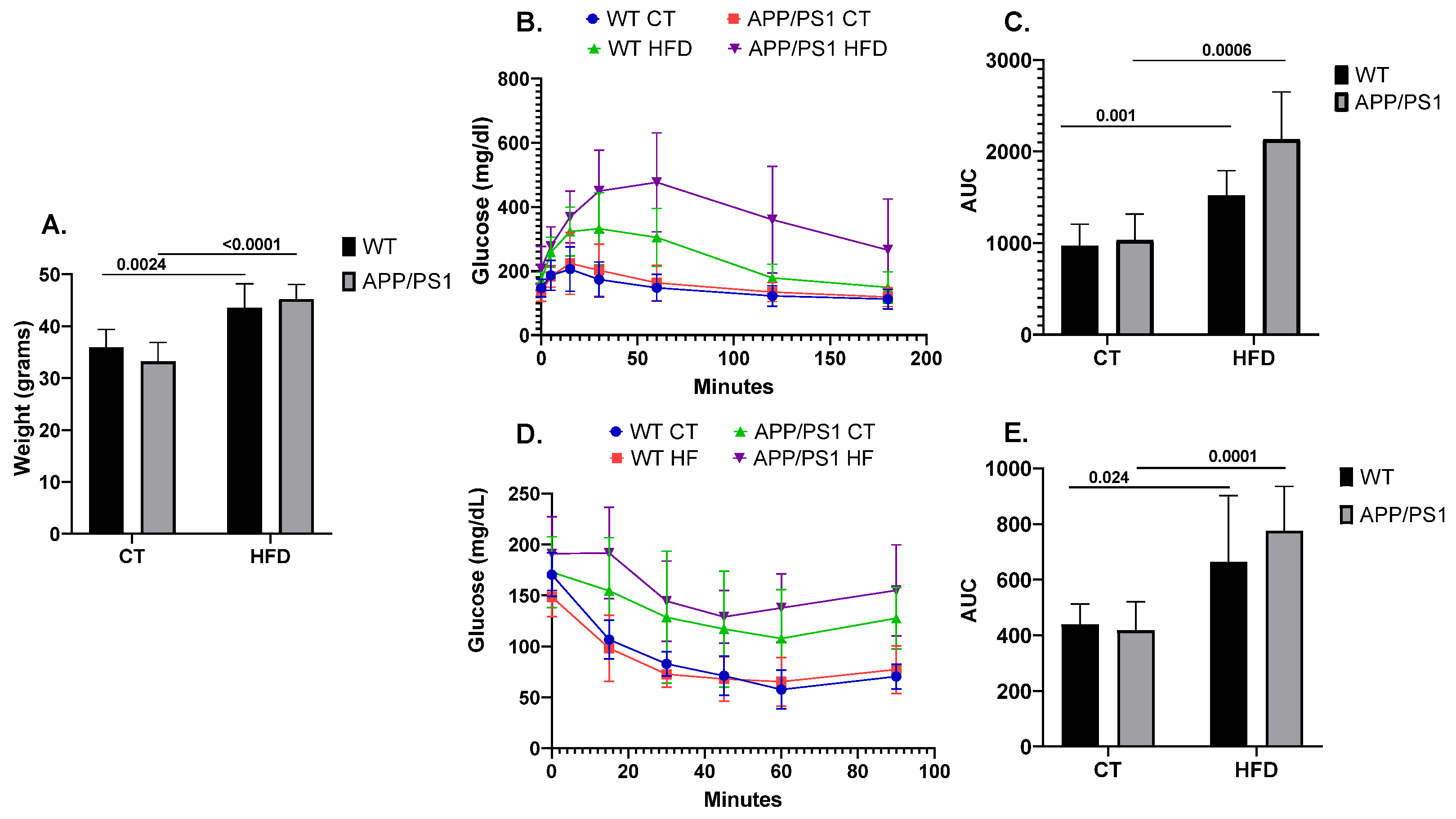
2.2. Glucose and Insulin Tolerance Tests
2.3. RNA Extraction and cDNA Synthesis
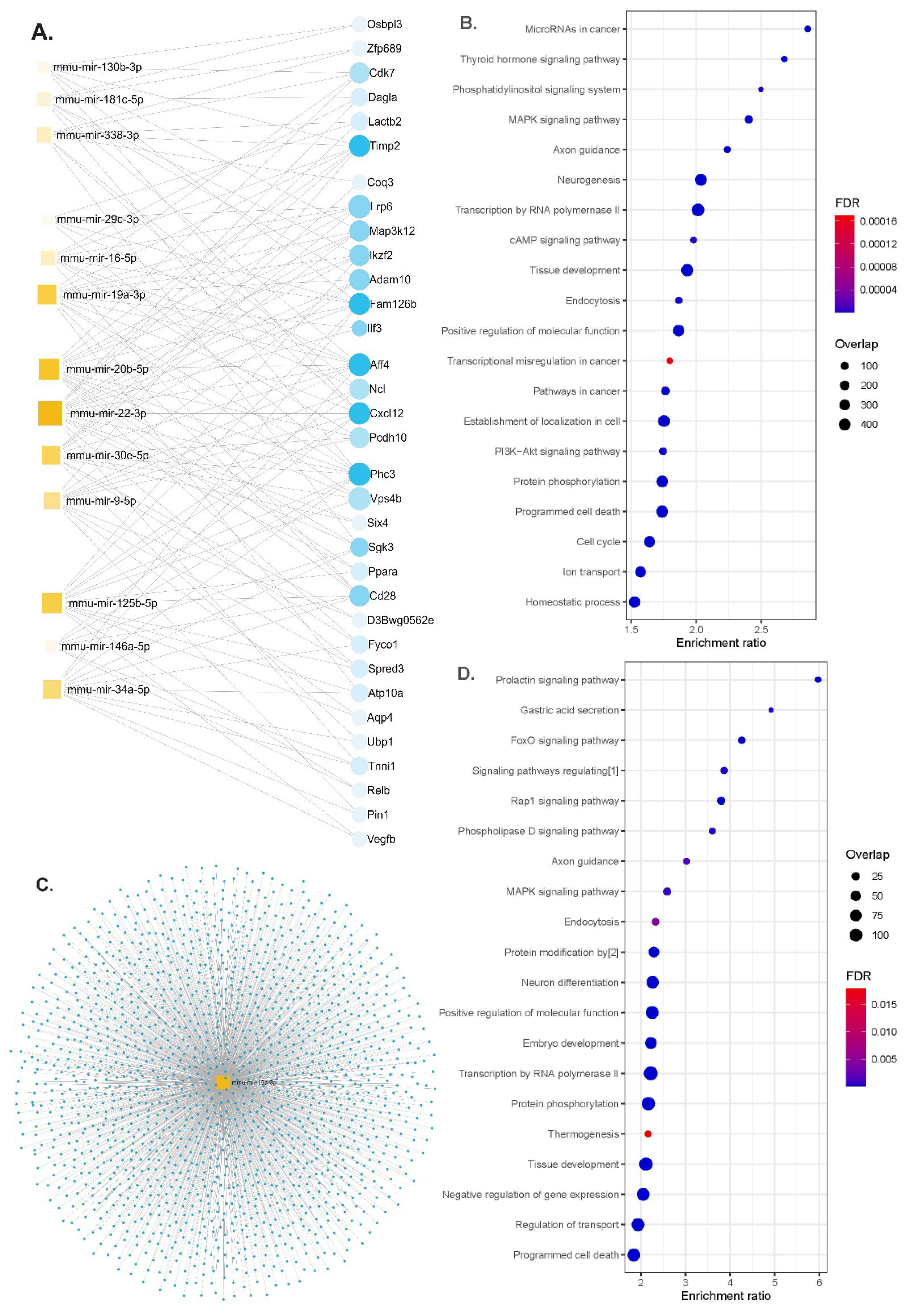
2.4. Functional Enrichment Analysis
2.5. Statistical Analysis
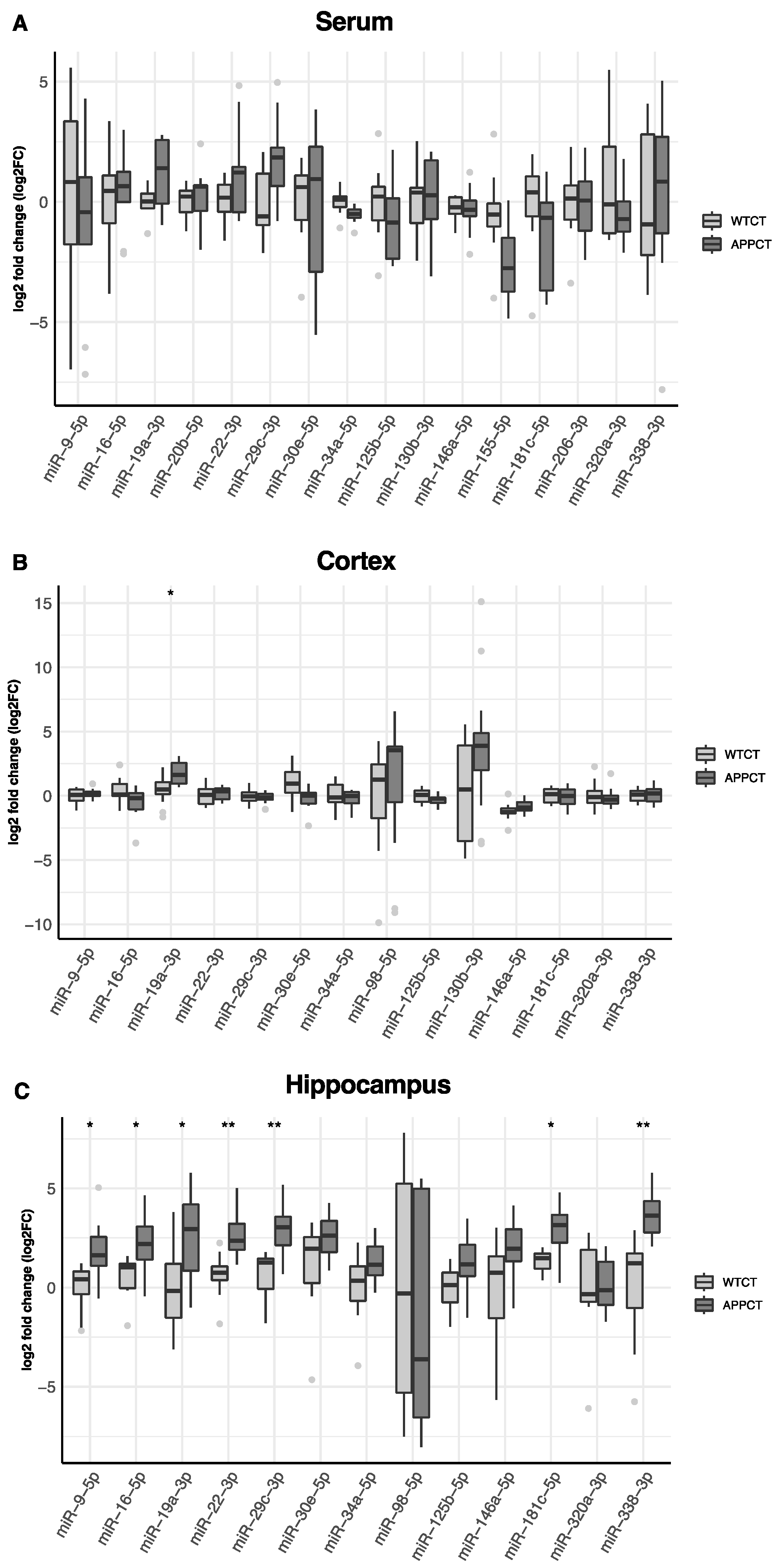
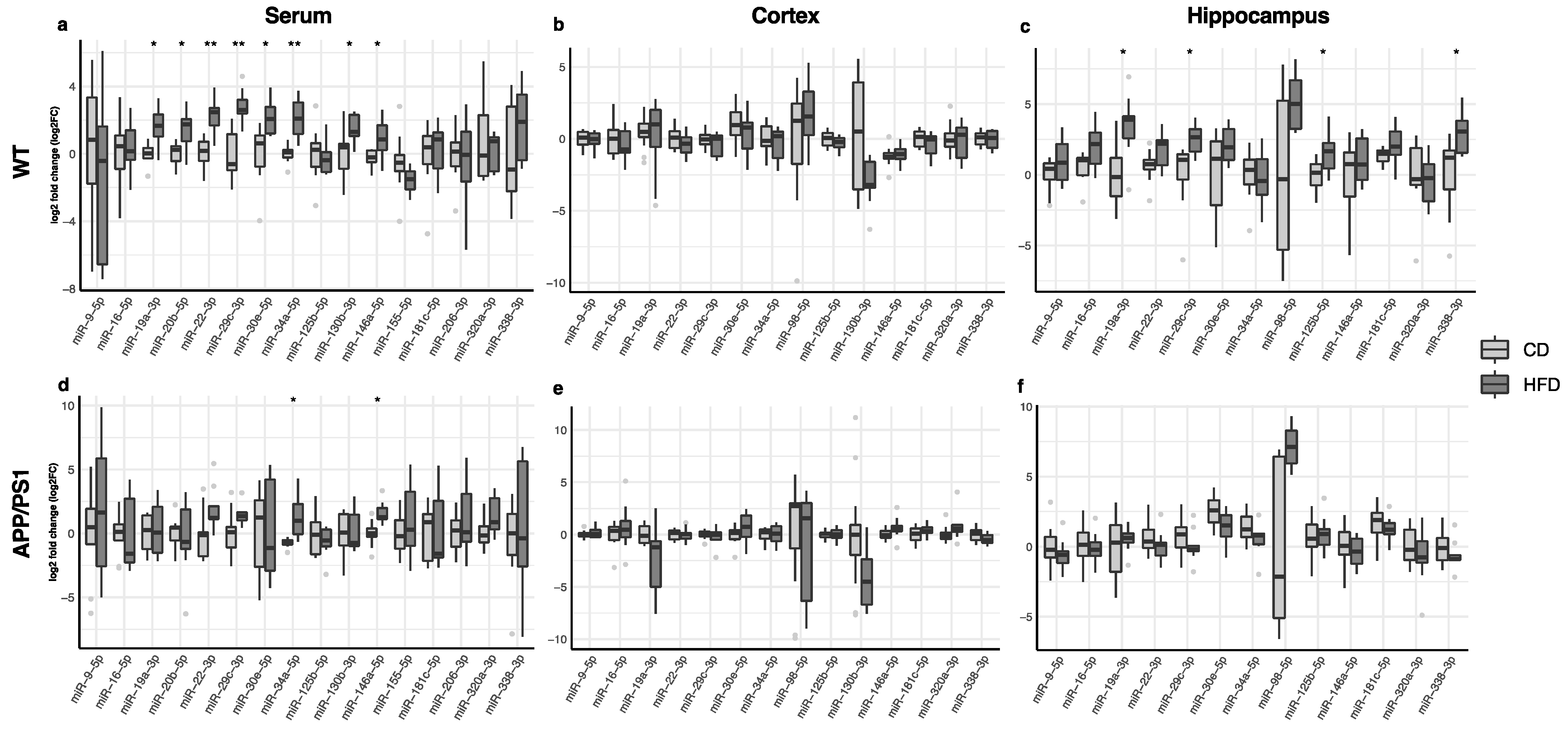
3. Results
3.1. Differential Expression of miRNAs in the Serum, Cortex and Hippocampus of APP/PS1 vs. WT Mice
3.2. Effect of the HFD on Serum, Cortex and Hippocampus miRNA Expression
3.3. Functional Enrichment Analysis of miRNAs Differentially Modulated by the HFD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pedditizi, E.; Peters, R.; Beckett, N. The risk of overweight/obesity in mid-life and late life for the development of dementia: A systematic review and meta-analysis of longitudinal studies. Age Ageing 2016, 45, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, K.; Lovestone, S. The dementias. Lancet 2002, 360, 1759–1766. [Google Scholar] [CrossRef] [PubMed]
- Wiesmann, M.; Kiliaan, A.J.; Claassen, J.A. Vascular aspects of cognitive impairment and dementia. J. Cereb. Blood Flow Metab. 2013, 33, 1696–1706. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-L.; Pan, C.-Y.; Chen, F.-C.; Huang, T.-H.; Tsai, M.-C.; Chuang, C.-Y. Differences in neurocognitive performance and metabolic and inflammatory indices in male adults with obesity as a function of regular exercise. Exp. Physiol. 2019, 104, 1650–1660. [Google Scholar] [CrossRef] [PubMed]
- Papaetis, G.S.; Papakyriakou, P.; Panagiotou, T.N. State of the art paper Central obesity, type 2 diabetes and insulin: Exploring a pathway full of thorns. Arch. Med. Sci. 2015, 3, 463–482. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.Y.; Zhao, W.; Lu, M.; Zhang, X.; Zhang, P.; Xin, Z.; Sun, R.; Tian, W.; Cardoso, M.A.; Yang, J.K.; et al. Relationship between Central Obesity and the incidence of Cognitive Impairment and Dementia from Cohort Studies Involving 5,060,687 Participants. Neurosci. Biobehav. Rev. 2021, 130, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Rhea, E.M.; Banks, W.A.; Raber, J. Insulin Resistance in Peripheral Tissues and the Brain: A Tale of Two Sites. Biomedicines 2022, 10, 1582. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, M.; Fusco, S.; Mainardi, M.; Scala, F.; Natale, F.; Lapenta, R.; Mattera, A.; Rinaudo, M.; Li Puma, D.D.; Ripoli, C.; et al. Brain insulin resistance impairs hippocampal synaptic plasticity and memory by increasing GluA1 palmitoylation through FoxO3a. Nat. Commun. 2017, 8, 2009. [Google Scholar] [CrossRef] [PubMed]
- Nelson, T.J.; Sun, M.-K.; Hongpaisan, J.; Alkon, D.L. Insulin, PKC signaling pathways and synaptic remodeling during memory storage and neuronal repair. Eur. J. Pharmacol. 2008, 585, 76–87. [Google Scholar] [CrossRef]
- Gonçalves, R.A.; Wijesekara, N.; Fraser, P.E.; De Felice, F.G. The Link between Tau and Insulin Signaling: Implications for Alzheimer’s Disease and Other Tauopathies. Front. Cell. Neurosci. 2019, 13, 17. [Google Scholar] [CrossRef]
- Yamamoto, N.; Matsubara, T.; Sobue, K.; Tanida, M.; Kasahara, R.; Naruse, K.; Taniura, H.; Sato, T.; Suzuki, K. Brain insulin resistance accelerates Aβ fibrillogenesis by inducing GM1 ganglioside clustering in the presynaptic membranes. J. Neurochem. 2012, 121, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Kanach, C.; Blusztajn, J.K.; Fischer, A.; Delalle, I. MicroRNAs as Candidate Biomarkers for Alzheimer’s Disease. Non-Coding RNA 2021, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Epple, R.; Krüger, D.; Berulava, T.; Brehm, G.; Ninov, M.; Islam, R.; Köster, S.; Fischer, A. The Coding and Small Non-coding Hippocampal Synaptic RNAome. Mol. Neurobiol. 2021, 58, 2940–2953. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Dong, Q.; Wu, T.; Guo, J. MicroRNA-20b-5p aggravates neuronal apoptosis induced by β-Amyloid via down-regulation of Ras homolog family member C in Alzheimer’s disease. Neurosci. Lett. 2021, 742, 135542. [Google Scholar] [CrossRef] [PubMed]
- Zhan, F.; Yang, J.; Lin, S.; Chen, L. miRNA-Based Signature to Predict the Development of Alzheimer’s Disease. Comb. Chem. High Throughput Screen 2022, 25, 2103–2111. [Google Scholar]
- Gutierrez-Tordera, L.; Papandreou, C.; Novau-Ferré, N.; García-González, P.; Rojas, M.; Marquié, M.; Chapado, L.A.; Papagiannopoulos, C.; Fernàndez-Castillo, N.; Valero, S.; et al. Exploring small non-coding RNAs as blood-based biomarkers to predict Alzheimer’s disease. Cell Biosci. 2024, 14, 8. [Google Scholar] [CrossRef]
- Kaur, P.; Kotru, S.; Singh, S.; Behera, B.S.; Munshi, A. Role of miRNAs in the pathogenesis of T2DM, insulin secretion, insulin resistance, and β cell dysfunction: The story so far. J. Physiol. Biochem. 2020, 76, 485–502. [Google Scholar] [CrossRef]
- Kim, M.; Zhang, X. The Profiling and Role of miRNAs in Diabetes Mellitus. J. Diabetes Clin. Res. 2019, 1, 5–23. [Google Scholar] [CrossRef]
- Chen, K.; Yu, T.; Wang, X. Inhibition of Circulating Exosomal miRNA-20b-5p Accelerates Diabetic Wound Repair. Int. J. Nanomed. 2021, 16, 371–381. [Google Scholar] [CrossRef]
- Yu, C.-Y.; Yang, C.-Y.; Rui, Z.-L. MicroRNA-125b-5p improves pancreatic β-cell function through inhibiting JNK signaling pathway by targeting DACT1 in mice with type 2 diabetes mellitus. Life Sci. 2019, 224, 67–75. [Google Scholar] [CrossRef]
- Sierksma, A.; Lu, A.; Salta, E.; Vanden Eynden, E.; Callaerts-Vegh, Z.; D’Hooge, R.; Blum, D.; Buée, L.; Fiers, M.; De Strooper, B. Deregulation of neuronal miRNAs induced by amyloid-β or TAU pathology. Mol. Neurodegener. 2018, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Yu, J.-T.; Tan, M.-S.; Liu, Q.-Y.; Wang, H.-F.; Zhang, W.; Jiang, T.; Tan, L. Genome-Wide Serum microRNA Expression Profiling Identifies Serum Biomarkers for Alzheimer’s Disease. J. Alzheimers Dis. 2014, 40, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Corral-Fernández, N.E.; Salgado-Bustamante, M.; Martínez-Leija, M.E.; Cortez-Espinosa, N.; García-Hernández, M.H.; Reynaga-Hernández, E.; Quezada-Calvillo, R.; Portales-Pérez, D.P. Dysregulated miR-155 expression in peripheral blood mononuclear cells from patients with type 2 diabetes. Exp. Clin. Endocrinol. Diabetes 2013, 121, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Espejo, S.; Santos-Zorrozua, B.; Álvarez-González, P.; Lopez-Lopez, E.; Garcia-Orad, Á. A Systematic Review of MicroRNA Expression as Biomarker of Late-Onset Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 8376–8391. [Google Scholar] [CrossRef]
- Li, Y.; Luo, T.; Wang, L.; Wu, J.; Guo, S. MicroRNA-19a-3p enhances the proliferation and insulin secretion, while it inhibits the apoptosis of pancreatic β cells via the inhibition of SOCS3. Int. J. Mol. Med. 2016, 38, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, X.; Chen, L.; Zhang, M.; Lei, L.; Gao, W.; Shi, Z.; Dong, Y.; Wang, Z.; Li, X.; et al. Hepatic miR-125b inhibits insulin signaling pathway by targeting PIK3CD. J. Cell. Physiol. 2018, 233, 6052–6066. [Google Scholar] [CrossRef]
- Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Boutati, E.; Sideris, D.C.; Fragoulis, E.G.; Christodoulou, M.-I. Decreased expression of microRNAs targeting type-2 diabetes susceptibility genes in peripheral blood of patients and predisposed individuals. Endocrine 2019, 66, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-T.; Chu, K.; Jung, K.-H.; Kim, J.H.; Huh, J.-Y.; Yoon, H.; Park, D.-K.; Lim, J.-Y.; Kim, J.-M.; Jeon, D.; et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 2012, 72, 269–277. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Kim, S.H.; Park, Y.; Park, J.; Lee, J.H.; Kim, B.C.; Song, W.K. miR-16-5p is upregulated by amyloid β deposition in Alzheimer’s disease models and induces neuronal cell apoptosis through direct targeting and suppression of BCL-2. Exp. Gerontol. 2020, 136, 110954. [Google Scholar] [CrossRef]
- Lei, X.; Lei, L.; Zhang, Z.; Zhang, Z.; Cheng, Y. Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer’s disease. Int. J. Clin. Exp. Pathol. 2015, 8, 1565–1574. [Google Scholar] [PubMed]
- Li, Q.; Li, X.; Wang, L.; Zhang, Y.; Chen, L. miR-98-5p Acts as a Target for Alzheimer’s Disease by Regulating Aβ Production Through Modulating SNX6 Expression. J. Mol. Neurosci. 2016, 60, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; He, H.; Shen, X.; Zhao, J.; Cao, X.; Han, S.; Cui, C.; Chen, Y.; Wei, Y.; Xia, L.; et al. miR-9-5p Inhibits Skeletal Muscle Satellite Cell Proliferation and Differentiation by Targeting IGF2BP3 through the IGF2-PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2020, 21, 1655. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.-L.; Jiang, H.; Li, C.-Y.; Gao, T.; Liu, M.-R.; Li, H.-W. MicroRNA-338 in MSCs-derived exosomes inhibits cardiomyocyte apoptosis in myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 10107–10117. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Wang, Z.-Y.; Ma, L.-N.; Zhang, T.-T.; Cao, Y.; Li, H. MicroRNAs in Alzheimer’s Disease: Function and Potential Applications as Diagnostic Biomarkers. Front. Mol. Neurosci. 2020, 13, 160. [Google Scholar] [CrossRef]
- Swarbrick, S.; Wragg, N.; Ghosh, S.; Stolzing, A. Systematic Review of miRNA as Biomarkers in Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 6156–6167. [Google Scholar] [CrossRef] [PubMed]
- Reiserer, R.S.; Harrison, F.E.; Syverud, D.C.; McDonald, M.P. Impaired spatial learning in the APP Swe + PSEN1ΔE9 bigenic mouse model of Alzheimer’s disease. Genes Brain Behav. 2007, 6, 54–65. [Google Scholar] [CrossRef]
- Ayala, J.E.; Samuel, V.T.; Morton, G.J.; Obici, S.; Croniger, C.M.; Shulman, G.I.; Wasserman, D.H.; McGuinness, O.P. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis. Model. Mech. 2010, 3, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-D.; Lin, F.-M.; Wu, W.-Y.; Liang, C.; Huang, W.-C.; Chan, W.-L.; Tsai, W.-T.; Chen, G.-Z.; Lee, C.-J.; Chiu, C.-M.; et al. miRTarBase: A database curates experimentally validated microRNA–target interactions. Nucleic Acids Res. 2011, 39, D163–D169. [Google Scholar] [CrossRef]
- Sheinerman, K.S.; Tsivinsky, V.G.; Abdullah, L.; Crawford, F.; Umansky, S.R. Plasma microRNA biomarkers for detection of mild cognitive impairment: Biomarker Validation Study. Aging 2013, 5, 925–938. [Google Scholar] [CrossRef]
- Chang, L.; Xia, J. MicroRNA Regulatory Network Analysis Using miRNet 2.0. In Transcription Factor Regulatory Networks; Song, Q., Tao, Z., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2023; pp. 185–204. ISBN 978-1-07-162815-7. [Google Scholar]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Quackenbush, J. Microarray data normalization and transformation. Nat. Genet. 2002, 32, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.N.; Therneau, T.M.; Zhang, Y.; Poland, G.A.; Kocher, J.-P. Calculating Sample Size Estimates for RNA Sequencing Data. J. Comput. Biol. 2013, 20, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Ettcheto, M.; Cano, A.; Manzine, P.R.; Busquets, O.; Verdaguer, E.; Castro-Torres, R.D.; García, M.L.; Beas-Zarate, C.; Olloquequi, J.; Auladell, C.; et al. Epigallocatechin-3-Gallate (EGCG) Improves Cognitive Deficits Aggravated by an Obesogenic Diet Through Modulation of Unfolded Protein Response in APPswe/PS1dE9 Mice. Mol. Neurobiol. 2020, 57, 1814–1827. [Google Scholar] [CrossRef]
- Ettcheto, M.; Sánchez-López, E.; Gómez-Mínguez, Y.; Cabrera, H.; Busquets, O.; Beas-Zarate, C.; García, M.L.; Carro, E.; Casadesus, G.; Auladell, C.; et al. Peripheral and Central Effects of Memantine in a Mixed Preclinical Mice Model of Obesity and Familial Alzheimer’s Disease. Mol. Neurobiol. 2018, 55, 7327–7339. [Google Scholar] [CrossRef] [PubMed]
- Petrov, D.; Pedrós, I.; Artiach, G.; Sureda, F.X.; Barroso, E.; Pallàs, M.; Casadesús, G.; Beas-Zarate, C.; Carro, E.; Ferrer, I.; et al. High-fat diet-induced deregulation of hippocampal insulin signaling and mitochondrial homeostasis deficiences contribute to Alzheimer disease pathology in rodents. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2015, 1852, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Więckowska-Gacek, A.; Mietelska-Porowska, A.; Wydrych, M.; Wojda, U. Western diet as a trigger of Alzheimer’s disease: From metabolic syndrome and systemic inflammation to neuroinflammation and neurodegeneration. Ageing Res. Rev. 2021, 70, 101397. [Google Scholar] [CrossRef] [PubMed]
- Busquets, O.; Ettcheto, M.; Pallàs, M.; Beas-Zarate, C.; Verdaguer, E.; Auladell, C.; Folch, J.; Camins, A. Long-term exposition to a high fat diet favors the appearance of β-amyloid depositions in the brain of C57BL/6J mice. A potential model of sporadic Alzheimer’s disease. Mech. Ageing Dev. 2017, 162, 38–45. [Google Scholar] [CrossRef]
- Guo, Y.; Ma, X.; Li, P.; Dong, S.; Huang, X.; Ren, X.; Yuan, L. High-fat diet induced discrepant peripheral and central nervous systems insulin resistance in APPswe/PS1dE9 and wild-type C57BL/6J mice. Aging 2020, 13, 1236–1250. [Google Scholar] [CrossRef]
- Burgos, K.; Malenica, I.; Metpally, R.; Courtright, A.; Rakela, B.; Beach, T.; Shill, H.; Adler, C.; Sabbagh, M.; Villa, S.; et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS ONE 2014, 9, e94839. [Google Scholar] [CrossRef]
- Wu, A.; Ying, Z.; Gomez-Pinilla, F. Dietary omega-3 fatty acids normalize BDNF levels, reduce oxidative damage, and counteract learning disability after traumatic brain injury in rats. J. Neurotrauma 2004, 21, 1457–1467. [Google Scholar] [CrossRef]
- Zhang, N.; Li, W.-W.; Lv, C.-M.; Gao, Y.-W.; Liu, X.-L.; Zhao, L. miR-16-5p and miR-19b-3p prevent amyloid β-induced injury by targeting BACE1 in SH-SY5Y cells. NeuroReport 2020, 31, 205. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Maguire, B.; Brodaty, H.; O’Leary, F. Dietary Patterns and Cognitive Health in Older Adults: A Systematic Review. J. Alzheimers Dis. 2019, 67, 583–619. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Muddashetty, R.S. Emerging Role of microRNAs in Dementia. J. Mol. Biol. 2019, 431, 1743–1762. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, H.; Dong, W.; Quan, X.; Zhu, H.; Xu, Y.; Huang, L.; Ma, C.; Qin, C. miR-29c regulates BACE1 protein expression. Brain Res. 2011, 1395, 108–115. [Google Scholar] [CrossRef]
- Hébert, S.S.; Horré, K.; Nicolaï, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/β-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef]
- Wu, L.; Feng, X.; Li, T.; Sun, B.; Khan, M.Z.; He, L. Risperidone ameliorated Aβ1-42-induced cognitive and hippocampal synaptic impairments in mice. Behav. Brain Res. 2017, 322, 145–156. [Google Scholar] [CrossRef]
- Sun, W.; Wang, Q.; Guo, Y.; Zhao, Y.; Wang, X.; Zhang, Z.; Deng, G.; Guo, M. Selenium suppresses inflammation by inducing microRNA-146a in Staphylococcus aureus-infected mouse mastitis model. Oncotarget 2017, 8, 110949–110964. [Google Scholar] [CrossRef] [PubMed]
- Demirsoy, İ.H.; Ertural, D.Y.; Balci, Ş.; Çınkır, Ü.; Sezer, K.; Tamer, L.; Aras, N. Profiles of Circulating MiRNAs Following Metformin Treatment in Patients with Type 2 Diabetes. J. Med. Biochem. 2018, 37, 499–506. [Google Scholar] [CrossRef]
- Alicka, M.; Major, P.; Wysocki, M.; Marycz, K. Adipose-Derived Mesenchymal Stem Cells Isolated from Patients with Type 2 Diabetes Show Reduced “Stemness” through an Altered Secretome Profile, Impaired Anti-Oxidative Protection, and Mitochondrial Dynamics Deterioration. J. Clin. Med. 2019, 8, 765. [Google Scholar] [CrossRef]
- Deng, Z.; Zhang, M.; Zhu, T.; Zhili, N.; Liu, Z.; Xiang, R.; Zhang, W.; Xu, Y. Dynamic changes in peripheral blood lymphocyte subsets in adult patients with COVID-19. Int. J. Infect. Dis. 2020, 98, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Engler-Chiurazzi, E.B.; Cavendis, J.Z.; Povroznik, J.M.; Russell, A.E.; Quintana, D.D.; Mathers, P.H.; Simpkins, J.W. Over-expression of miR-34a induces rapid cognitive impairment and Alzheimer’s disease-like pathology. Brain Res. 2019, 1721, 146327. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Doss, C.G.P.; Bandyopadhyay, S.; Agoramoorthy, G. Influence of miRNA in insulin signaling pathway and insulin resistance: Micro-molecules with a major role in type-2 diabetes. WIREs RNA 2014, 5, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhu, J.; Han, W.; Jiang, X.; Xu, M.; Zhao, Y.; Dong, Q.; Pang, Z.; Guan, Q.; Gao, L.; et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011, 48, 61–69. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojas-Criollo, M.; Novau-Ferré, N.; Gutierrez-Tordera, L.; Ettcheto, M.; Folch, J.; Papandreou, C.; Panisello, L.; Cano, A.; Mostafa, H.; Mateu-Fabregat, J.; et al. Effects of a High-Fat Diet on Insulin-Related miRNAs in Plasma and Brain Tissue in APPSwe/PS1dE9 and Wild-Type C57BL/6J Mice. Nutrients 2024, 16, 955. https://doi.org/10.3390/nu16070955
Rojas-Criollo M, Novau-Ferré N, Gutierrez-Tordera L, Ettcheto M, Folch J, Papandreou C, Panisello L, Cano A, Mostafa H, Mateu-Fabregat J, et al. Effects of a High-Fat Diet on Insulin-Related miRNAs in Plasma and Brain Tissue in APPSwe/PS1dE9 and Wild-Type C57BL/6J Mice. Nutrients. 2024; 16(7):955. https://doi.org/10.3390/nu16070955
Chicago/Turabian StyleRojas-Criollo, Melina, Nil Novau-Ferré, Laia Gutierrez-Tordera, Miren Ettcheto, Jaume Folch, Christopher Papandreou, Laura Panisello, Amanda Cano, Hamza Mostafa, Javier Mateu-Fabregat, and et al. 2024. "Effects of a High-Fat Diet on Insulin-Related miRNAs in Plasma and Brain Tissue in APPSwe/PS1dE9 and Wild-Type C57BL/6J Mice" Nutrients 16, no. 7: 955. https://doi.org/10.3390/nu16070955
APA StyleRojas-Criollo, M., Novau-Ferré, N., Gutierrez-Tordera, L., Ettcheto, M., Folch, J., Papandreou, C., Panisello, L., Cano, A., Mostafa, H., Mateu-Fabregat, J., Carrasco, M., Camins, A., & Bulló, M. (2024). Effects of a High-Fat Diet on Insulin-Related miRNAs in Plasma and Brain Tissue in APPSwe/PS1dE9 and Wild-Type C57BL/6J Mice. Nutrients, 16(7), 955. https://doi.org/10.3390/nu16070955