A Free Amino Acid Diet Alleviates Colorectal Tumorigenesis through Modulating Gut Microbiota and Metabolites
Abstract
1. Introduction
2. Materials and Methods
2.1. Colorectal Cancer Mouse Models and Treatments
2.2. Disease Activity Index (DAI)
2.3. Histopathological Analysis
2.4. Cell Culture and Drug Treatment
2.5. Quantitative Real-Time Polymerase Chain Reaction Analysis
2.6. Multiplex Immunohistochemical (mIHC)
2.7. 16S rRNA Gene Sequencing
2.8. Colon Transcriptome Profiling
2.9. Metabolic Profiling of Colon Content
3. Results
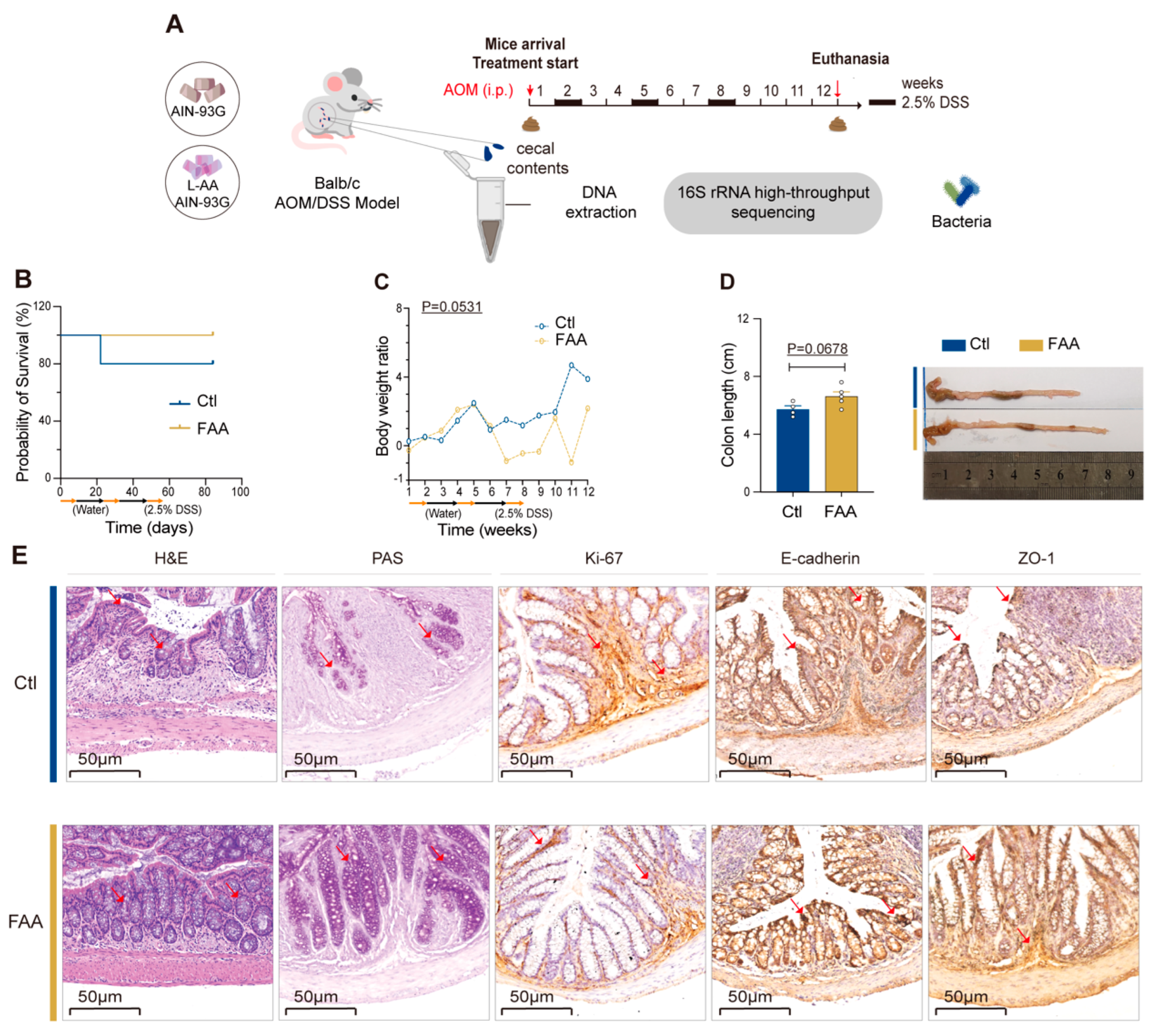
3.1. Free Amino Acid-Based Diet Attenuates the Tumor Progression in AOM/DSS-Induced Colorectal Cancer Mouse Model
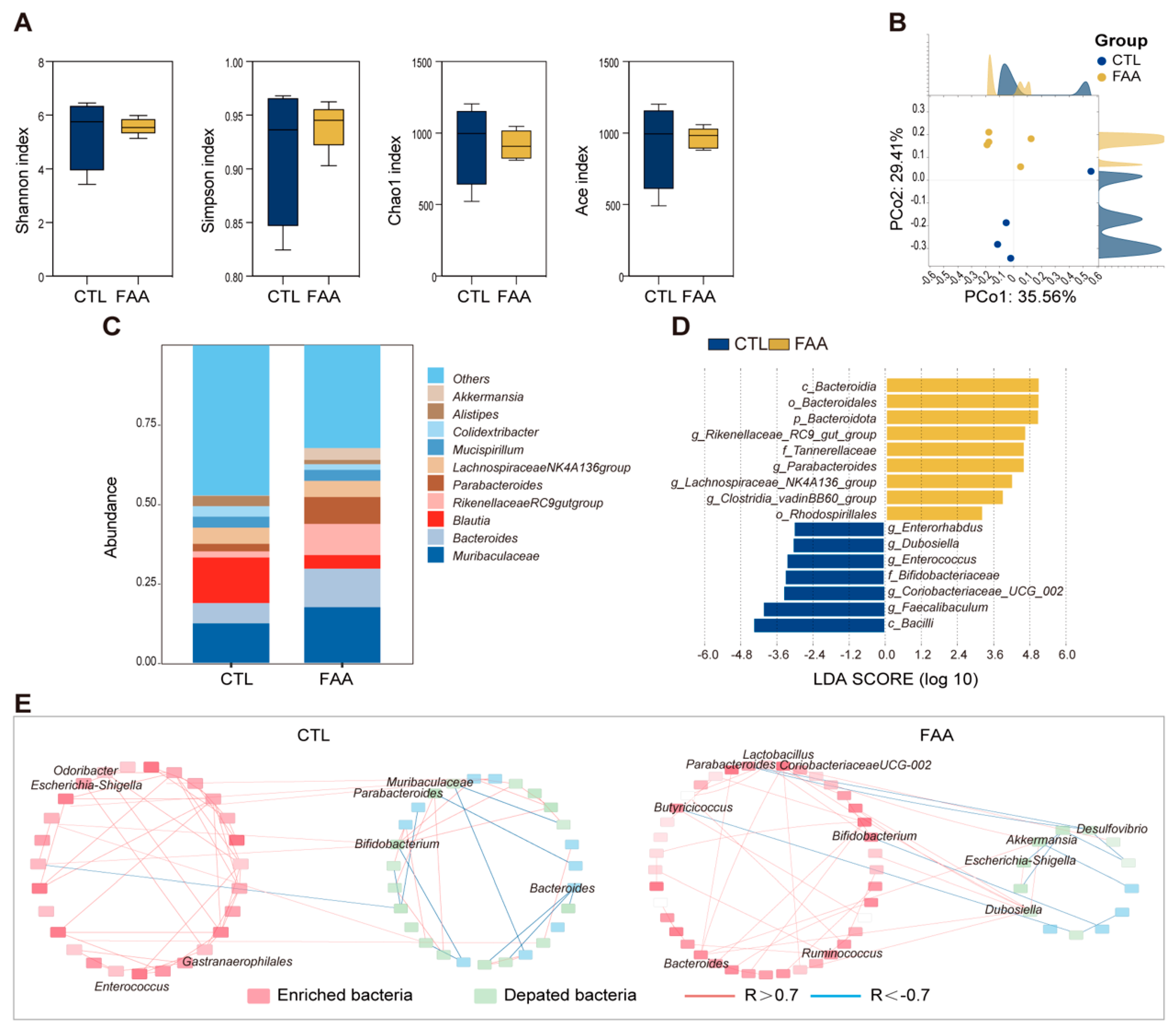
3.2. Free Amino Acid-Based Diet Modulates Gut Microbiota in AOM/DSS-Induced Colorectal Cancer Mouse Model
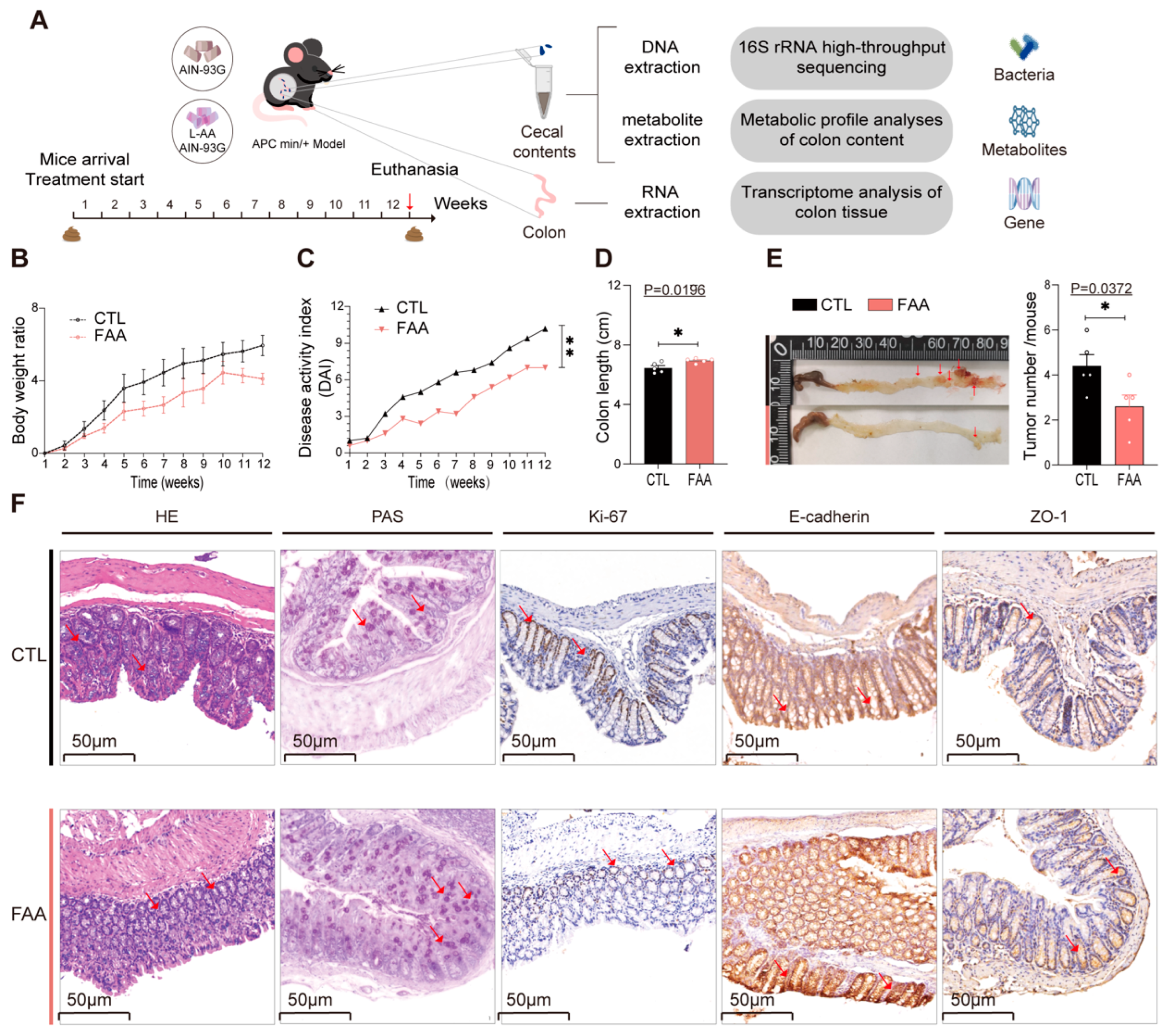
3.3. Free Amino Acid-Based Dietary Intervention Attenuates Tumor Progression in Spontaneous Genetically Induced Colorectal Cancer Mouse Model
3.4. Free Amino Acid-Based Dietary Intervention Revert Gut Microbiota Dysbiosis in Spontaneous Genetically Induced Colorectal Cancer Mouse Model
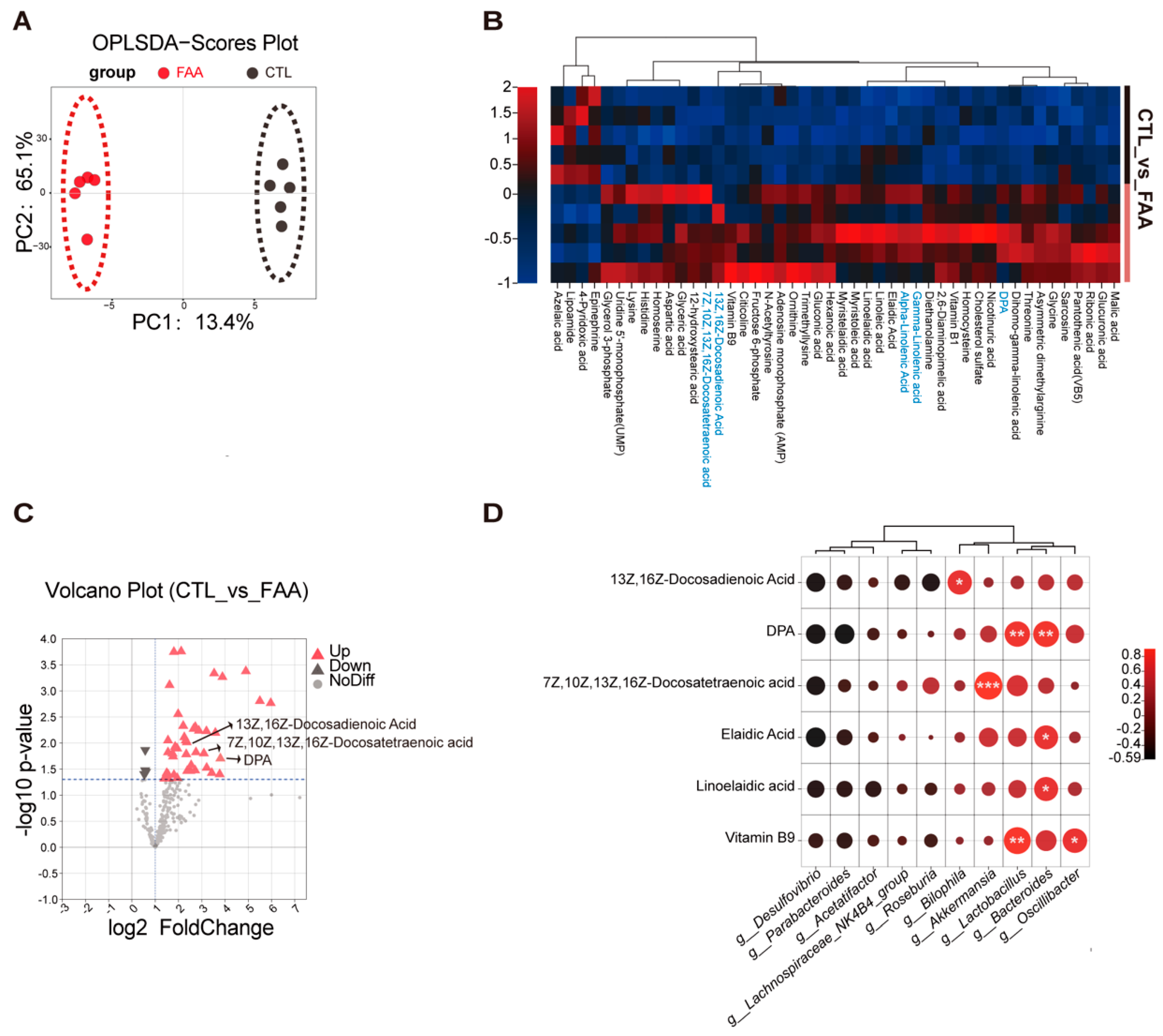
3.5. Free Amino Acid-Based Dietary Intervention Modulate Intestinal Metabolites in Spontaneous Genetically Induced Colorectal Cancer Mouse Model
3.6. Free Amino Acid-Based Dietary Intervention Suppresses Carcinogenic Signaling in Spontaneous Genetically Induced Colorectal Cancer Mouse Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends—An Update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Shafie, N.H.; Mohd Esa, N.; Ithnin, H.; Md Akim, A.; Saad, N.; Pandurangan, A.K. Preventive Inositol Hexaphosphate Extracted from Rice Bran Inhibits Colorectal Cancer through Involvement of Wnt/β-Catenin and COX-2 Pathways. BioMed Res. Int. 2013, 2013, 681027. [Google Scholar] [CrossRef] [PubMed]
- Center, M.M.; Jemal, A.; Ward, E. International Trends in Colorectal Cancer Incidence Rates. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1688–1694. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Torre, L.A.; Soerjomataram, I.; Hayes, R.B.; Bray, F.; Weber, T.K.; Jemal, A. Global patterns and trends in colorectal cancer incidence in young adults. Gut 2019, 68, 2179–2185. [Google Scholar] [CrossRef]
- Wang, Q.; Lu, W.; Yin, T.; Lu, L. Calycosin suppresses TGF-β-induced epithelial-to-mesenchymal transition and migration by upregulating BATF2 to target PAI-1 via the Wnt and PI3K/Akt signaling pathways in colorectal cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 240. [Google Scholar] [CrossRef]
- Wong, M.C.; Huang, J.; Lok, V.; Wang, J.; Fung, F.; Ding, H.; Zheng, Z.-J. Differences in Incidence and Mortality Trends of Colorectal Cancer Worldwide Based on Sex, Age, and Anatomic Location. Clin. Gastroenterol. Hepatol. 2021, 19, 955–966.e61. [Google Scholar] [CrossRef]
- Silva, V.R.; Santos LD, S.; Dias, R.B.; Quadros, C.A.; Bezerra, D.P. Emerging agents that target signaling pathways to eradicate colorectal cancer stem cells. Cancer Commun. 2021, 41, 1275–1313. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Dai, X.; Gao, J.; Li, Y.; Hou, Z.; Chang, Z.; Wang, Y. Curcumin suppresses colorectal tumorigenesis via the Wnt/β-catenin signaling pathway by downregulating Axin2. Oncol. Lett. 2021, 21, 186. [Google Scholar] [CrossRef]
- Wang, K.; Wu, W.; Wang, Q.; Yang, L.; Bian, X.; Jiang, X.; Lv, L.; Yan, R.; Xia, J.; Han, S.; et al. The negative effect of Akkermansia muciniphila-mediated post-antibiotic reconstitution of the gut microbiota on the development of colitis-associated colorectal cancer in mice. Front. Microbiol. 2022, 13, 932047. [Google Scholar] [CrossRef]
- Lau, H.C.H.; Yu, J. Gut microbiome alters functions of mutant p53 to promote tumorigenesis. Signal Transduct. Target. Ther. 2020, 5, 232. [Google Scholar] [CrossRef]
- Xia, J.; Lv, L.; Liu, B.; Wang, S.; Zhang, S.; Wu, Z.; Yang, L.; Bian, X.; Wang, Q.; Wang, K.; et al. Akkermansia muciniphila Ameliorates Acetaminophen-Induced Liver Injury by Regulating Gut Microbial Composition and Metabolism. Microbiol. Spectr. 2022, 10, e0159621. [Google Scholar] [CrossRef]
- Rodriguez, J.; Hiel, S.; Delzenne, N.M. Metformin: Old friend, new ways of action–implication of the gut microbiome? Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 294–301. [Google Scholar] [CrossRef]
- Liu, T.; Song, X.; An, Y.; Wu, X.; Zhang, W.; Li, J.; Sun, Y.; Jin, G.; Liu, X.; Guo, Z.; et al. Lactobacillus rhamnosus GG Colonization in Early Life Ameliorates Inflammaging of Offspring by Activating SIRT1/AMPK/PGC-1α Pathway. Oxidative Med. Cell. Longev. 2021, 2021, 3328505. [Google Scholar] [CrossRef]
- Ogata, Y.; Ishibashi, N.; Yamaguchi, K.; Uchida, S.; Kamei, H.; Nakayama, G.; Hirakawa, H.; Tanigawa, M.; Akagi, Y. Preventive effects of amino-acid-rich elemental diet Elental® on chemotherapy-induced oral mucositis in patients with colorectal cancer: A prospective pilot study. Support. Care Cancer 2016, 24, 783–789. [Google Scholar] [CrossRef][Green Version]
- Alhhazmi, A.A.; Almutawif, Y.A.; Mumena, W.A.; Alhazmi, S.M.; Abujamel, T.S.; Alhusayni, R.M.; Aloufi, R.; Al-Hejaili, R.R.; Alhujaily, R.; Alrehaili, L.M.; et al. Identification of Gut Microbiota Profile Associated with Colorectal Cancer in Saudi Population. Cancers 2023, 15, 5019. [Google Scholar] [CrossRef]
- Fang, Y.; Yan, C.; Zhao, Q.; Xu, J.; Liu, Z.; Gao, J.; Zhu, H.; Dai, Z.; Wang, D.; Tang, D. The roles of microbial products in the development of colorectal cancer: A review. Bioengineered 2021, 12, 720–735. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xu, C.; Yang, W.; Chen, J.; Ou, Y.; Guan, Y.; Guan, J.; Liu, Y. E3 ligase RNF167 and deubiquitinase STAMBPL1 modulate mTOR and cancer progression. Mol. Cell 2022, 82, 770–784.e9. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.T.; Rochell, S.J. Precision intestinal nutrition: Knowledge and gaps regarding the role of amino acids during an enteric challenge. Poult. Sci. 2022, 101, 101674. [Google Scholar] [CrossRef]
- Gao, R.; Wu, C.; Zhu, Y.; Kong, C.; Zhu, Y.; Gao, Y.; Zhang, X.; Yang, R.; Zhong, H.; Xiong, X.; et al. Integrated Analysis of Colorectal Cancer Reveals Cross-Cohort Gut Microbial Signatures and Associated Serum Metabolites. Gastroenterology 2022, 163, 1024–1037.e9. [Google Scholar] [CrossRef]
- Zeng, X.; Xing, X.; Gupta, M.; Keber, F.C.; Lopez, J.G.; Lee, Y.-C.J.; Roichman, A.; Wang, L.; Neinast, M.D.; Donia, M.S.; et al. Gut bacterial nutrient preferences quantified in vivo. Cell 2022, 185, 3441–3456.e19. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Nie, Y.; Yu, J.; Wong, C.C. Microbial Metabolites in Colorectal Cancer: Basic and Clinical Implications. Metabolites 2021, 11, 159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhao, C.; Zhang, X.; Li, X.; Zhang, Y.; Liu, X.; Yin, J.; Li, X.; Wang, J.; Wang, S. An Elemental Diet Enriched in Amino Acids Alters the Gut Microbial Community and Prevents Colonic Mucus Degradation in Mice with Colitis. mSystems 2022, 7, e0088322. [Google Scholar] [CrossRef] [PubMed]
- Lannagan, T.R.M.; Lee, Y.K.; Wang, T.; Roper, J.; Bettington, M.L.; Fennell, L.; Vrbanac, L.; Jonavicius, L.; Somashekar, R.; Gieniec, K.; et al. Genetic editing of colonic organoids provides a molecularly distinct and orthotopic preclinical model of serrated carcinogenesis. Gut 2019, 68, 684–692. [Google Scholar] [CrossRef]
- Diether, N.; Willing, B. Microbial Fermentation of Dietary Protein: An Important Factor in Diet–Microbe–Host Interaction. Microorganisms 2019, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Coker, O.O.; Liu, C.; Wu, W.K.K.; Wong, S.H.; Jia, W.; Sung, J.J.Y.; Yu, J. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome 2022, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Aglago, E.K.; Huybrechts, I.; Murphy, N.; Casagrande, C.; Nicolas, G.; Pischon, T.; Fedirko, V.; Severi, G.; Boutron-Ruault, M.-C.; Fournier, A.; et al. Consumption of Fish and Long-chain n-3 Polyunsaturated Fatty Acids Is Associated With Reduced Risk of Colorectal Cancer in a Large European Cohort. Clin. Gastroenterol. Hepatol. 2020, 18, 654–666.e6. [Google Scholar] [CrossRef] [PubMed]
- Kantor, E.D.; Lampe, J.W.; Peters, U.; Vaughan, T.L.; White, E. Long-Chain Omega-3 Polyunsaturated Fatty Acid Intake and Risk of Colorectal Cancer. Nutr. Cancer 2014, 66, 716–727. [Google Scholar] [CrossRef]
- Newell, M.; Mazurak, V.; Postovit, L.M.; Field, C.J. N-3 Long-Chain Polyunsaturated Fatty Acids, Eicosapentaenoic and Docosahexaenoic Acid, and the Role of Supplementation during Cancer Treatment: A Scoping Review of Current Clinical Evidence. Cancers 2021, 13, 1206. [Google Scholar] [CrossRef]
- Da Cruz, R.S.; Carney, E.J.; Clarke, J.; Cao, H.; Cruz, M.I.; Benitez, C.; Jin, L.; Fu, Y.; Cheng, Z.; Wang, Y.; et al. Paternal malnutrition programs breast cancer risk and tumor metabolism in offspring. Breast Cancer Res. 2018, 20, 99. [Google Scholar] [CrossRef]
- Greene, E.; Cauble, R.; Dhamad, A.E.; Kidd, M.T.; Kong, B.; Howard, S.M.; Castro, H.F.; Campagna, S.R.; Bedford, M.; Dridi, S. Muscle Metabolome Profiles in Woody Breast-(un)Affected Broilers: Effects of Quantum Blue Phytase-Enriched Diet. Front. Vet. Sci. 2020, 7, 458. [Google Scholar] [CrossRef] [PubMed]
- Warburton, D. YAP and TAZ in Lung Development: The Timing Is Important. Am. J. Respir. Cell Mol. Biol. 2020, 62, 141–142. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.-M.; Li, G.-F.; Ren, Y.-L.; Xu, X.-Y.; Xu, Z.-H.; Geng, Y.; Mao, Y. A Free Amino Acid Diet Alleviates Colorectal Tumorigenesis through Modulating Gut Microbiota and Metabolites. Nutrients 2024, 16, 1040. https://doi.org/10.3390/nu16071040
Yu Y-M, Li G-F, Ren Y-L, Xu X-Y, Xu Z-H, Geng Y, Mao Y. A Free Amino Acid Diet Alleviates Colorectal Tumorigenesis through Modulating Gut Microbiota and Metabolites. Nutrients. 2024; 16(7):1040. https://doi.org/10.3390/nu16071040
Chicago/Turabian StyleYu, Yang-Meng, Gui-Fang Li, Yi-Lin Ren, Xin-Yi Xu, Zheng-Hong Xu, Yan Geng, and Yong Mao. 2024. "A Free Amino Acid Diet Alleviates Colorectal Tumorigenesis through Modulating Gut Microbiota and Metabolites" Nutrients 16, no. 7: 1040. https://doi.org/10.3390/nu16071040
APA StyleYu, Y.-M., Li, G.-F., Ren, Y.-L., Xu, X.-Y., Xu, Z.-H., Geng, Y., & Mao, Y. (2024). A Free Amino Acid Diet Alleviates Colorectal Tumorigenesis through Modulating Gut Microbiota and Metabolites. Nutrients, 16(7), 1040. https://doi.org/10.3390/nu16071040






