microRNA Expression Profile in Obesity-Induced Kidney Disease Driven by High-Fat Diet in Mice
Abstract
1. Introduction
2. Results
2.1. Metabolic and Physiological Parameters Induced by High-Fat Diet
2.2. Kidney Injury and Dysfunction in OIKD Model
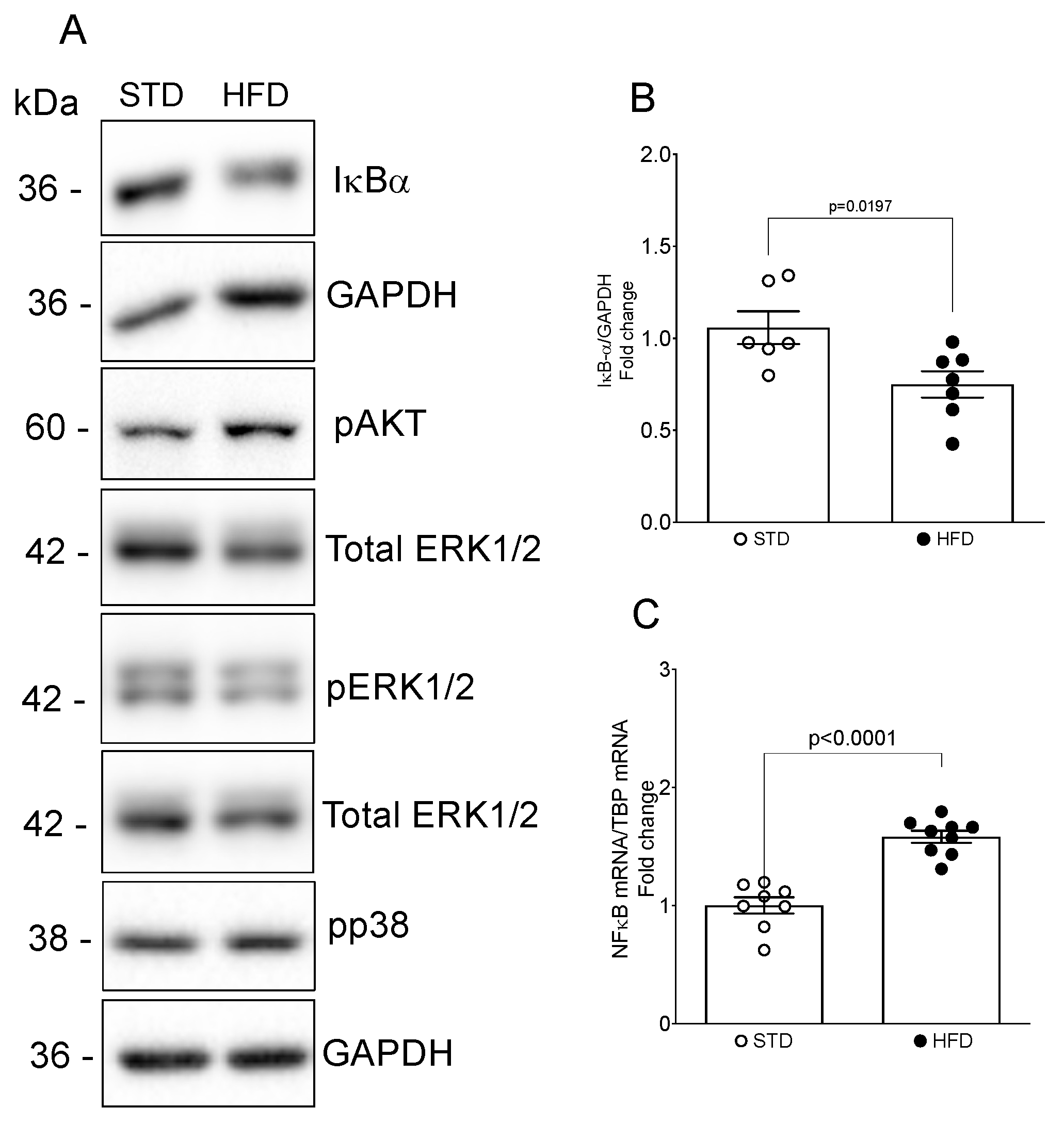
2.3. Differential microRNA Expression Profile in Kidneys of OIKD Model
2.4. Correlation between miRNA Expression, Total Serum Cholesterol and Renal Lipid Content
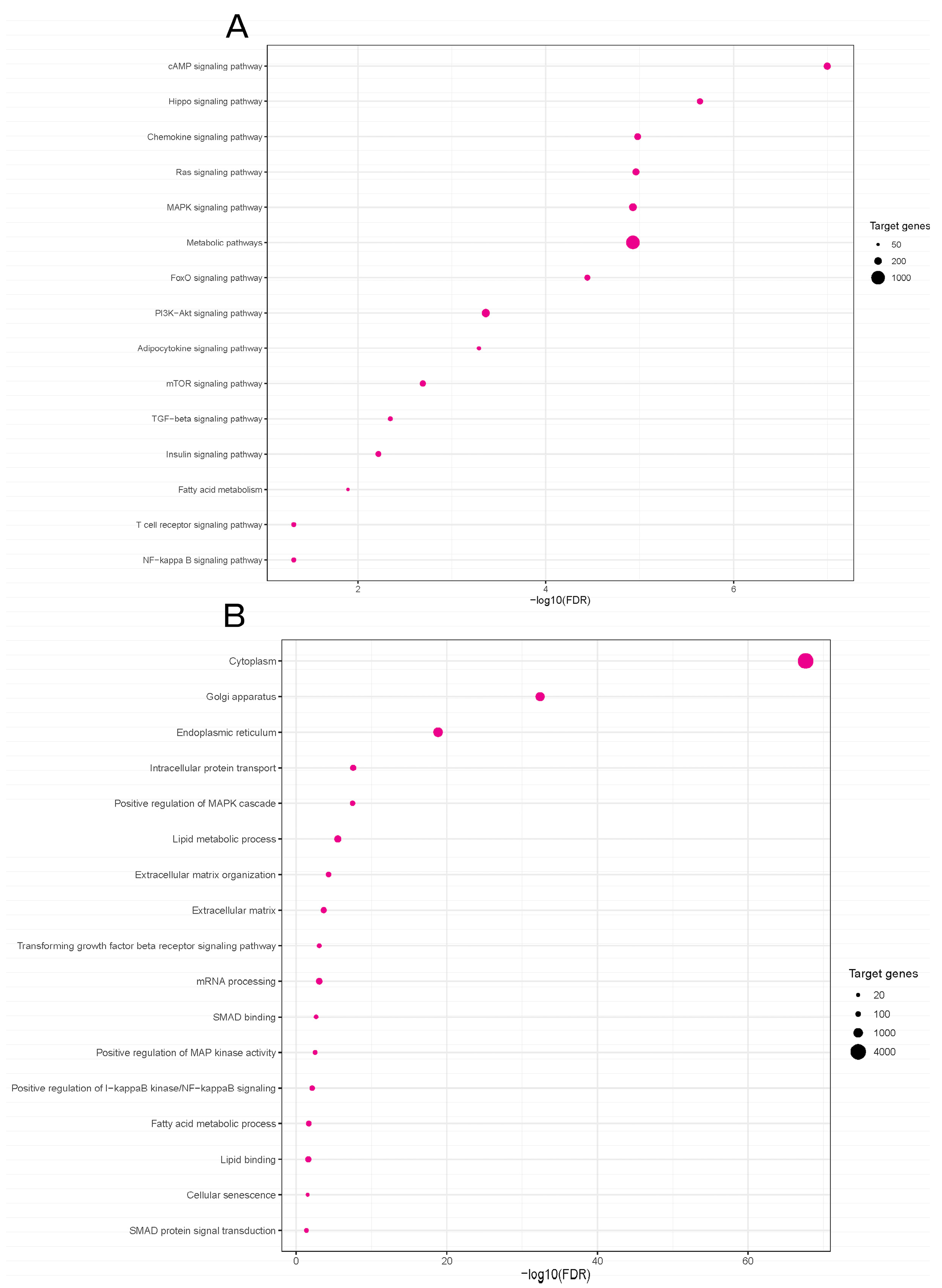
2.5. Functional Enrichment Analyses
3. Discussion
4. Materials and Methods
4.1. Animals and Experimental Protocol
4.2. Biochemical Analysis
4.3. Histopathological Analysis
4.4. Quantitative Real-Time PCR for mRNA Quantitation
4.5. microRNA Sequencing and Data Analysis
4.5.1. Sample Preparation
4.5.2. Library Preparation and Sequencing
4.5.3. Read Mapping and Quantification of Gene Expression
4.6. Quantitative Real-Time PCR for miRNA Validation
4.7. Western Blot Analysis
4.8. Statistical Analysis and Bioinformatics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, J.C.; Carson, T.L.; Thompson, H.J.; Agurs-Collins, T. The Triple Health Threat of Diabetes, Obesity, and Cancer—Epidemiology, Disparities, Mechanisms, and Interventions. Obesity 2021, 29, 954–959. [Google Scholar] [CrossRef] [PubMed]
- Di Angelantonio, E.; Bhupathiraju, S.N.; Wormser, D.; Gao, P.; Kaptoge, S.; de Gonzalez, A.B.; Cairns, B.J.; Huxley, R.; Jackson, C.L.; Joshy, G.; et al. Body-mass index and all-cause mortality: Individual-participant-data meta-analysis of 239 prospective studies in four continents. Lancet 2016, 388, 776–786. [Google Scholar] [CrossRef] [PubMed]
- Burki, T. European Commission classifies obesity as a chronic disease. Lancet Diabetes Endocrinol. 2021, 9, 418. [Google Scholar] [CrossRef] [PubMed]
- Ejerblad, E.; Fored, C.M.; Lindblad, P.; Fryzek, J.; McLaughlin, J.K.; Nyrén, O. Obesity and Risk for Chronic Renal Failure. J. Am. Soc. Nephrol. 2006, 17, 1695–1702. [Google Scholar] [CrossRef]
- Hsu, C.-Y.; McCulloch, C.E.; Iribarren, C.; Darbinian, J.; Go, A.S. Body Mass Index and Risk for End-Stage Renal Disease. Ann. Intern. Med. 2006, 144, 21–28. [Google Scholar] [CrossRef]
- Sharma, I.; Liao, Y.; Zheng, X.; Kanwar, Y.S. New Pandemic: Obesity and Associated Nephropathy. Front. Med. 2021, 8, 673556. [Google Scholar] [CrossRef]
- Li, Z.; Li, J.; Miao, X.; Cui, W.; Miao, L.; Cai, L. A minireview: Role of AMP-activated protein kinase (AMPK) signaling in obesity-related renal injury. Life Sci. 2021, 265, 118828. [Google Scholar] [CrossRef]
- Caus, M.; Eritja, À.; Bozic, M. Role of microRNAs in Obesity-Related Kidney Disease. Int. J. Mol. Sci. 2021, 22, 11416. [Google Scholar] [CrossRef]
- Izquierdo-Lahuerta, A.; Martínez-García, C.; Medina-Gómez, G. Lipotoxicity as a trigger factor of renal disease. J. Nephrol. 2016, 29, 603–610. [Google Scholar] [CrossRef]
- Weinberg, J. Lipotoxicity. Kidney Int. 2006, 70, 1560–1566. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, J.E. Lipotoxicity: When tissues overeat. Curr. Opin. Infect. Dis. 2003, 14, 281–287. [Google Scholar] [CrossRef]
- Bobulescu, I.A. Renal lipid metabolism and lipotoxicity. Curr. Opin. Nephrol. Hypertens. 2010, 19, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Eom, D.-W.; Jung, Y.; Yamabe, N.; Lee, S.; Jeon, Y.; Hwang, Y.R.; Lee, J.H.; Kim, Y.K.; Kang, K.S.; et al. Dendrobium moniliforme Attenuates High-Fat Diet-Induced Renal Damage in Mice through the Regulation of Lipid-Induced Oxidative Stress. Am. J. Chin. Med. 2012, 40, 1217–1228. [Google Scholar] [CrossRef]
- Lee, E.S.; Kwon, M.H.; Kim, H.M.; Kim, N.; Kim, Y.M.; Kim, H.S.; Lee, E.Y.; Chung, C.H. Dibenzoylmethane ameliorates lipid-induced inflammation and oxidative injury in diabetic nephropathy. J. Endocrinol. 2019, 240, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2018, 76, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.-C.; Chen, C.-C.; Chen, Y.-C.; Chang, Y.-S.; Chu, P.-H. MicroRNAs in acute kidney injury. Hum. Genom. 2016, 10, 29. [Google Scholar] [CrossRef]
- Chandrasekaran, K.; Karolina, D.S.; Sepramaniam, S.; Armugam, A.; Wintour, E.M.; Bertram, J.F.; Jeyaseelan, K. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012, 81, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yao, D.; Yan, H.; Chen, X.; Wang, L.; Zhan, H. The Role of MicroRNAs in the Pathogenesis of Diabetic Nephropathy. Int. J. Endocrinol. 2019, 2019, 8719060. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Mukhadi, S.; Hull, R.; Mbita, Z.; Dlamini, Z. The Role of MicroRNAs in Kidney Disease. Non-Coding RNA 2015, 1, 192–221. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.-W.; Lee, Y.S.; Kim, J.B. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARγ expression. Biochem. Biophys. Res. Commun. 2010, 392, 323–328. [Google Scholar] [CrossRef]
- Sun, L.; Xie, H.; Mori, M.A.; Alexander, R.; Yuan, B.; Hattangadi, S.M.; Liu, Q.; Kahn, C.R.; Lodish, H.F. Mir193b–365 is essential for brown fat differentiation. Nature 2011, 13, 958–965. [Google Scholar] [CrossRef]
- Fang, S.; Feng, J.; Zhang, H.; Li, P.; Zhang, Y.; Zeng, Y.; Cai, Y.; Lin, X.; Xue, Y.; Guan, M. MiR-455 targeting SOCS3 improve liver lipid disorders in diabetic mice. Adipocyte 2020, 9, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Peng, R.; Peng, H.; Liu, H.; Wen, L.; Wu, T.; Yi, H.; Li, A.; Zhang, Z. miR-451 suppresses the NF-kappaB-mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. Mol. Cell. Endocrinol. 2016, 433, 75–86. [Google Scholar] [CrossRef]
- Sun, D.; Chen, J.; Wu, W.; Tang, J.; Luo, L.; Zhang, K.; Jin, L.; Lin, S.; Gao, Y.; Yan, X.; et al. MiR-802 causes nephropathy by suppressing NF-κB-repressing factor in obese mice and human. J. Cell. Mol. Med. 2019, 23, 2863–2871. [Google Scholar] [CrossRef]
- Jaswani, P.; Prakash, S.; Dhar, A.; Sharma, R.K.; Prasad, N.; Agrawal, S. MicroRNAs involvement in renal pathophysiology: A bird’s eye view. Indian J. Nephrol. 2017, 27, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, K.; Padmanabhan, G. MiRNAs as potential biomarker of kidney diseases: A review. Cell Biochem. Funct. 2020, 38, 990–1005. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Komers, R.; Carew, R.; Winbanks, C.E.; Xu, B.; Herman-Edelstein, M.; Koh, P.; Thomas, M.; Jandeleit-Dahm, K.; Gregorevic, P.; et al. Suppression of microRNA-29 Expression by TGF-β1 Promotes Collagen Expression and Renal Fibrosis. J. Am. Soc. Nephrol. 2012, 23, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Van der Hauwaert, C.; Glowacki, F.; Pottier, N.; Cauffiez, C. Non-Coding RNAs as New Therapeutic Targets in the Context of Renal Fibrosis. Int. J. Mol. Sci. 2019, 20, 1977. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Shu, S.; Cai, J.; Tang, C.; Dong, Z. Non-coding RNAs in kidney injury and repair. Am. J. Physiol. Physiol. 2019, 317, C177–C188. [Google Scholar] [CrossRef] [PubMed]
- Chau, B.N.; Xin, C.; Hartner, J.; Ren, S.; Castano, A.P.; Linn, G.; Li, J.; Tran, P.T.; Kaimal, V.; Huang, X.; et al. MicroRNA-21 Promotes Fibrosis of the Kidney by Silencing Metabolic Pathways. Sci. Transl. Med. 2012, 4, 121ra18. [Google Scholar] [CrossRef]
- Chung, A.C.; Lan, H.Y. MicroRNAs in renal fibrosis. Front. Physiol. 2015, 6, 50. [Google Scholar] [CrossRef]
- Gai, Z.; Wang, T.; Visentin, M.; Kullak-Ublick, G.A.; Fu, X.; Wang, Z. Lipid Accumulation and Chronic Kidney Disease. Nutrients 2019, 11, 722. [Google Scholar] [CrossRef]
- Dorotea, D.; Koya, D.; Ha, H. Recent Insights into SREBP as a Direct Mediator of Kidney Fibrosis via Lipid-Independent Pathways. Front. Pharmacol. 2020, 11, 265. [Google Scholar] [CrossRef]
- Qiu, P.; Wang, H.; Zhang, M.; Peng, R.; Zhao, Q.; Liu, J. FATP2-targeted therapies—A role beyond fatty liver disease. Pharmacol. Res. 2020, 161, 105228. [Google Scholar] [CrossRef]
- Tastsoglou, S.; Skoufos, G.; Miliotis, M.; Karagkouni, D.; Koutsoukos, I.; Karavangeli, A.; Kardaras, F.S.; Hatzigeorgiou, A.G. DIANA-miRPath v4.0: Expanding target-based miRNA functional analysis in cell-type and tissue contexts. Nucleic Acids Res. 2023, 51, W154–W159. [Google Scholar] [CrossRef]
- Bozic, M.; Caus, M.; Rodrigues-Diez, R.R.; Pedraza, N.; Ruiz-Ortega, M.; Garí, E.; Gallel, P.; Panadés, M.J.; Martinez, A.; Fernández, E.; et al. Protective role of renal proximal tubular alpha-synuclein in the pathogenesis of kidney fibrosis. Nat. Commun. 2020, 11, 1943. [Google Scholar] [CrossRef]
- Bozic, M.; Álvarez, Á.; de Pablo, C.; Sanchez-Niño, M.-D.; Ortiz, A.; Dolcet, X.; Encinas, M.; Fernandez, E.; Valdivielso, J.M. Impaired Vitamin D Signaling in Endothelial Cell Leads to an Enhanced Leukocyte-Endothelium Interplay: Implications for Atherosclerosis Development. PLoS ONE 2015, 10, e0136863. [Google Scholar] [CrossRef] [PubMed]
- Wicks, S.E.; Nguyen, T.-T.; Breaux, C.; Kruger, C.; Stadler, K. Diet-induced obesity and kidney disease—In search of a susceptible mouse model. Biochimie 2015, 124, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Wang, Z.; Proctor, G.; Moskowitz, S.; Liebman, S.E.; Rogers, T.; Lucia, M.S.; Li, J.; Levi, M. Diet-induced Obesity in C57BL/6J Mice Causes Increased Renal Lipid Accumulation and Glomerulosclerosis via a Sterol Regulatory Element-binding Protein-1c-dependent Pathway. J. Biol. Chem. 2005, 280, 32317–32325. [Google Scholar] [CrossRef]
- Declèves, A.E.; Zolkipli, Z.; Satriano, J.; Wang, L.; Nakayama, T.; Rogac, M.; Le, T.P.; Nortier, J.L.; Farquhar, M.G.; Naviaux, R.K.; et al. Regulation of lipid accumulation by AMK-activated kinase in high fat diet–induced kidney injury. Kidney Int. 2014, 85, 611–623. [Google Scholar] [CrossRef]
- Deji, N.; Kume, S.; Araki, S.-I.; Soumura, M.; Sugimoto, T.; Isshiki, K.; Chin-Kanasaki, M.; Sakaguchi, M.; Koya, D.; Haneda, M.; et al. Structural and functional changes in the kidneys of high-fat diet-induced obese mice. Am. J. Physiol. Physiol. 2009, 296, F118–F126. [Google Scholar] [CrossRef]
- Stemmer, K.; Perez-Tilve, D.; Ananthakrishnan, G.; Bort, A.; Seeley, R.J.; Tschöp, M.H.; Dietrich, D.R.; Pfluger, P.T. High-fat-diet-induced obesity causes an inflammatory and tumor-promoting microenvironment in the rat kidney. Dis. Model. Mech. 2012, 5, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. BioTechniques 2004, 37, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, C.I.G.D.; Anderson, J.R.; Fang, Y.; Milner, P.I.; Goljanek-Whysall, K.; House, L.; Clegg, P.D.; Peffers, M.J. Mouse microRNA signatures in joint ageing and post-traumatic osteoarthritis. Osteoarthr. Cartil. Open 2021, 3, 100186. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, Y.; Ke, X.; Guo, Y.; Yao, C.; Tang, N.; Pang, P.; Xie, G.; Fang, L.; Zhang, Z.; et al. Transcriptome Sequencing Unravels Potential Biomarkers at Different Stages of Cerebral Ischemic Stroke. Front. Genet. 2019, 10, 814. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Yang, F.; Huang, Z.; Jin, Y.; Sheng, Y.; Ji, L. Serum microRNA-122-3p, microRNA-194-5p and microRNA-5099 are potential toxicological biomarkers for the hepatotoxicity induced by Airpotato yam. Toxicol. Lett. 2017, 280, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lin, H.-L.; Wang, N.; Sun, Y.-L.; Kan, Y.; Guo, H.; Chen, J.-L.; Fang, M. Inhibition of microRNA-30a prevents puromycin aminonucleoside-induced podocytic apoptosis by upregulating the glucocorticoid receptor α. Mol. Med. Rep. 2015, 12, 6043–6052. [Google Scholar] [CrossRef]
- Bai, S.-Y.; Ji, R.; Wei, H.; Guo, Q.-H.; Yuan, H.; Chen, Z.-F.; Wang, Y.-P.; Liu, Z.; Yang, X.-Y.; Zhou, Y.-N.; et al. Serum miR-551b-3p is a potential diagnostic biomarker for gastric cancer. Turk. J. Gastroenterol. 2019, 30, 415–419. [Google Scholar] [CrossRef]
- Rogucki, M.; Sidorkiewicz, I.; Niemira, M.; Dzięcioł, J.B.; Buczyńska, A.; Adamska, A.; Siewko, K.; Kościuszko, M.; Maliszewska, K.; Wójcicka, A.; et al. Expression Profile and Diagnostic Significance of MicroRNAs in Papillary Thyroid Cancer. Cancers 2022, 14, 2679. [Google Scholar] [CrossRef] [PubMed]
- Qiao, D.; He, X.; Deng, X.; Ji, Y.; Yang, H.; Cheng, L.; Zhou, X. Aberrant expression of five miRNAs in papillary thyroid carcinomas. J. Clin. Lab. Anal. 2021, 35, e23907. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-J.; Qiu, B.-Q.; Yuan, M.-M.; Zou, H.-X.; Gong, C.-W.; Huang, H.; Lai, S.-Q.; Liu, J.-C. Identification and Validation of Dilated Cardiomyopathy-Related Genes via Bioinformatics Analysis. Int. J. Gen. Med. 2022, 15, 3663–3676. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, Ş.G.; Bozkurt, H.; Ndadza, A.; Thomford, N.E.; Karaoğlan, M.; Keskin, M.; Benlier, N.; Dandara, C. Childhood Obesity Risk in Relationship to Perilipin 1 (PLIN1) Gene Regulation by Circulating microRNAs. OMICS 2020, 24, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Kuryłowicz, A.; Wicik, Z.; Owczarz, M.; Jonas, M.I.; Kotlarek, M.; Świerniak, M.; Lisik, W.; Jonas, M.; Noszczyk, B.; Puzianowska-Kuźnicka, M. NGS Reveals Molecular Pathways Affected by Obesity and Weight Loss-Related Changes in miRNA Levels in Adipose Tissue. Int. J. Mol. Sci. 2017, 19, 66. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Zhou, J.; Gu, X.; Zhou, Y.; Wang, L.; Si, N.; Fan, X.; Bian, B.; Wang, H.; Zhao, H. A multi-network comparative analysis of whole-transcriptome and translatome reveals the effect of high-fat diet on APP/PS1 mice and the intervention with Chinese medicine. Front. Nutr. 2022, 9, 974333. [Google Scholar] [CrossRef]
- Lee, H.W.; Khan, S.Q.; Khaliqdina, S.; Altintas, M.M.; Grahammer, F.; Zhao, J.L.; Koh, K.H.; Tardi, N.J.; Faridi, M.H.; Geraghty, T.; et al. Absence of miR-146a in Podocytes Increases Risk of Diabetic Glomerulopathy via Up-regulation of ErbB4 and Notch-1. J. Biol. Chem. 2017, 292, 732–747. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, Y.; Li, L.; Su, B.; Yang, L.; Fan, W.; Yin, Q.; Chen, L.; Cui, T.; Zhang, J.; et al. Involvement of inflammation-related miR-155 and miR-146a in diabetic nephropathy: Implications for glomerular endothelial injury. BMC Nephrol. 2014, 15, 142. [Google Scholar] [CrossRef]
- Morrison, M.C.; Yakala, G.K.; Liang, W.; Wielinga, P.Y.; Salic, K.; van Koppen, A.; Tomar, T.; Kleemann, R.; Heeringa, P.; Kooistra, T. Protective effect of rosiglitazone on kidney function in high-fat challenged human-CRP transgenic mice: A possible role for adiponectin and miR-21? Sci. Rep. 2017, 7, 2915. [Google Scholar] [CrossRef]
- Gholaminejad, A.; Tehrani, H.A.; Fesharaki, M.G. Identification of candidate microRNA biomarkers in renal fibrosis: A meta-analysis of profiling studies. Biomarkers 2018, 23, 713–724. [Google Scholar] [CrossRef]
- Taïbi, F.; Meuth, V.M.-L.; Massy, Z.A.; Metzinger, L. miR-223: An inflammatory oncomiR enters the cardiovascular field. Biochim. Biophys. Acta 2014, 1842, 1001–1009. [Google Scholar] [CrossRef]
- Taïbi, F.; Meuth, V.M.-L.; M’Baya-Moutoula, E.; Djelouat, M.S.e.I.; Louvet, L.; Bugnicourt, J.-M.; Poirot, S.; Bengrine, A.; Chillon, J.-M.; Massy, Z.A.; et al. Possible involvement of microRNAs in vascular damage in experimental chronic kidney disease. Biochim. Biophys. Acta 2014, 1842, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Ulbing, M.; Kirsch, A.; Leber, B.; Lemesch, S.; Münzker, J.; Schweighofer, N.; Hofer, D.; Trummer, O.; Rosenkranz, A.; Müller, H.; et al. MicroRNAs 223-3p and 93-5p in patients with chronic kidney disease before and after renal transplantation. Bone 2017, 95, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Anglicheau, D.; Sharma, V.K.; Ding, R.; Hummel, A.; Snopkowski, C.; Dadhania, D.; Seshan, S.V.; Suthanthiran, M. MicroRNA expression profiles predictive of human renal allograft status. Proc. Natl. Acad. Sci. USA 2009, 106, 5330–5335. [Google Scholar] [CrossRef]
- Coimbra, T.M.; Janssen, U.; Gröne, H.J.; Ostendorf, T.; Kunter, U.; Schmidt, H.; Brabant, G.; Floege, J. Early events leading to renal injury in obese Zucker (fatty) rats with type II diabetes. Kidney Int. 2000, 57, 167–182. [Google Scholar] [CrossRef]
- Anders, H.-J.; Vielhauer, V.; Schlöndorff, D. Chemokines and chemokine receptors are involved in the resolution or progression of renal disease. Kidney Int. 2003, 63, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Fearn, A.; Situmorang, G.R.; Fox, C.; Oakley, F.; Howarth, R.; Wilson, C.L.; Kiosia, A.; Robson, M.G.; A Mann, D.; Moles, A.; et al. The NF-κB1 is a key regulator of acute but not chronic renal injury. Cell Death Dis. 2017, 8, e2883. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, S.-C. NF-κB in inflammation and renal diseases. Cell Biosci. 2015, 5, 63. [Google Scholar] [CrossRef]
- Bozic, M.; de Rooij, J.; Parisi, E.; Ortega, M.R.; Fernandez, E.; Valdivielso, J.M. Glutamatergic Signaling Maintains the Epithelial Phenotype of Proximal Tubular Cells. J. Am. Soc. Nephrol. 2011, 22, 1099–1111. [Google Scholar] [CrossRef]
- Phanish, M.K.; Wahab, N.A.; Colville-Nash, P.; Hendry, B.M.; Dockrell, M.E.C. The differential role of Smad2 and Smad3 in the regulation of pro-fibrotic TGFβ1 responses in human proximal-tubule epithelial cells. Biochem. J. 2005, 393, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Walton, K.L.; Johnson, K.E.; Harrison, C.A. Targeting TGF-β Mediated SMAD Signaling for the Prevention of Fibrosis. Front. Pharmacol. 2017, 8, 461. [Google Scholar] [CrossRef] [PubMed]
- Valdivielso, J.M.; Eritja, À.; Caus, M.; Bozic, M. Glutamate-Gated NMDA Receptors: Insights into the Function and Signaling in the Kidney. Biomolecules 2020, 10, 1051. [Google Scholar] [CrossRef]
- Wen, X.; Zhang, B.; Wu, B.; Xiao, H.; Li, Z.; Li, R.; Xu, X.; Li, T. Signaling pathways in obesity: Mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2022, 7, 298. [Google Scholar] [CrossRef]
- Zhang, X.; Gu, S.; Shen, S.; Luo, T.; Zhao, H.; Liu, S.; Feng, J.; Yang, M.; Yi, L.; Fan, Z.; et al. Identification of Circular RNA Profiles in the Liver of Diet-Induced Obese Mice and Construction of the ceRNA Network. Genes 2023, 14, 688. [Google Scholar] [CrossRef]
- Huang, X.; Liu, G.; Guo, J.; Su, Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci. 2018, 14, 1483–1496. [Google Scholar] [CrossRef]
- Lovis, P.; Roggli, E.; Laybutt, D.R.; Gattesco, S.; Yang, J.-Y.; Widmann, C.; Abderrahmani, A.; Regazzi, R. Alterations in MicroRNA Expression Contribute to Fatty Acid–Induced Pancreatic β-Cell Dysfunction. Diabetes 2008, 57, 2728–2736. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, H.; Wang, Y.; Peng, X.; Li, J.; Gu, W.; He, T.; Chen, M. Regulation and mechanism of miR-146 on renal ischemia reperfusion injury. Pharmazie 2018, 73, 29–34. [Google Scholar] [CrossRef]
- Yu, J.; Xue, J.; Liu, C.; Zhang, A.; Qin, L.; Liu, J.; Yang, Y. MiR-146a-5p accelerates sepsis through dendritic cell activation and glycolysis via targeting ATG7. J. Biochem. Mol. Toxicol. 2022, 36, e23151. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tan, X.-R.; Li, S.-J.; Zhang, X.-X. LncRNA NEAT1 promotes hepatic lipid accumulation via regulating miR-146a-5p/ROCK1 in nonalcoholic fatty liver disease. Life Sci. 2019, 235, 116829. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.-P.; Seemann, J.; Goldstein, J.L.; Brown, M.S. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 6519–6526. [Google Scholar] [CrossRef]
- Eid, W.; Dauner, K.; Courtney, K.C.; Gagnon, A.; Parks, R.J.; Sorisky, A.; Zha, X. mTORC1 activates SREBP-2 by suppressing cholesterol trafficking to lysosomes in mammalian cells. Proc. Natl. Acad. Sci. USA 2017, 114, 7999–8004. [Google Scholar] [CrossRef]
- Declèves, A.-E.; Mathew, A.V.; Cunard, R.; Sharma, K. AMPK Mediates the Initiation of Kidney Disease Induced by a High-Fat Diet. J. Am. Soc. Nephrol. 2011, 22, 1846–1855. [Google Scholar] [CrossRef]
- Bozic, M.; Guzmán, C.; Benet, M.; Sánchez-Campos, S.; García-Monzón, C.; Gari, E.; Gatius, S.; Valdivielso, J.M.; Jover, R. Hepatocyte vitamin D receptor regulates lipid metabolism and mediates experimental diet-induced steatosis. J. Hepatol. 2016, 65, 748–757. [Google Scholar] [CrossRef]
- Potočnjak, I.; Domitrović, R. Carvacrol attenuates acute kidney injury induced by cisplatin through suppression of ERK and PI3K/Akt activation. Food Chem. Toxicol. 2016, 98, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Smyth, G.K. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 2007, 9, 321–332. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]




| Parameters | STD | HFD | p |
|---|---|---|---|
| Body weight (g) | 32.78 ± 0.662 | 41.78 ± 1.164 | <0.0001 |
| Food intake (g/mouse/24 h) | 3.69 ± 0.193 | 2.90 ± 0.050 | <0.0001 |
| Total cholesterol (mg/dL) | 100.1 ± 4.478 | 148.4 ± 6.679 | <0.0001 |
| LDL cholesterol (mg/dL) | 10.65 ± 1.369 | 16.75 ± 2.238 | 0.0356 |
| HDL cholesterol (mg/dL) | 78.11 ± 3.728 | 114.2 ± 4.983 | <0.0001 |
| Triglycerides (mg/dL) | 64.13 ± 3.404 | 94.78 ± 10.80 | 0.0008 |
| Glycemia (mg/dL) | 148.2 ± 9.843 | 197.4 ± 15.98 | 0.0229 |
| BUN (mg/dL) | 26.33 ± 1.373 | 21.09 ± 0.7058 | 0.0037 |
| AST/GOT (IU/L) | 8.54 ± 1.268 | 17.65 ± 1.877 | 0.0014 |
| ALT/GPT (IU/L) | 1.60 ± 0.549 | 5.50 ± 1.654 | 0.0508 |
| C-peptide (pM) | 33.72 ± 3.845 | 75.04 ± 28.10 | 0.0288 |
| HOMA-IR index | 12.92 ± 1.931 | 39.48 ± 17.94 | 0.0206 |
| miRNA | Identifier | Fold Change | FDR p-Value (q-Value) |
|---|---|---|---|
| mmu-miR-5099 | URS000056F567_10090 | −2.624887175 | 6.4841 × 10−14 |
| mmu-miR-223-3p | URS00000B7E30_10090 | 1.481082774 | 4.41731 × 10−5 |
| mmu-miR-551b-3p | URS000008C563_10090 | −1.813165613 | 0.000856739 |
| mmu-miR-21a-3p | URS000015930E_10090 | 1.745000997 | 0.005360045 |
| mmu-miR-146a-3p | URS0000121576_10090 | 2.179056951 | 0.005360045 |
| mmu-miR-129-5p | URS00004E1410_10090 | 2.420949709 | 0.012797245 |
| mmu-miR-142a-5p | URS00001E0AEA_10090 | 1.285697411 | 0.02634235 |
| mmu-miR-144-3p | URS000037C5A8_10090 | −3.132081075 | 0.028191062 |
| mmu-miR-802-5p | URS00005B2C05_10090 | 1.682896286 | 0.034999635 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eritja, À.; Caus, M.; Belmonte, T.; de Gonzalo-Calvo, D.; García-Carrasco, A.; Martinez, A.; Martínez, M.; Bozic, M. microRNA Expression Profile in Obesity-Induced Kidney Disease Driven by High-Fat Diet in Mice. Nutrients 2024, 16, 691. https://doi.org/10.3390/nu16050691
Eritja À, Caus M, Belmonte T, de Gonzalo-Calvo D, García-Carrasco A, Martinez A, Martínez M, Bozic M. microRNA Expression Profile in Obesity-Induced Kidney Disease Driven by High-Fat Diet in Mice. Nutrients. 2024; 16(5):691. https://doi.org/10.3390/nu16050691
Chicago/Turabian StyleEritja, Àuria, Maite Caus, Thalia Belmonte, David de Gonzalo-Calvo, Alicia García-Carrasco, Ana Martinez, Montserrat Martínez, and Milica Bozic. 2024. "microRNA Expression Profile in Obesity-Induced Kidney Disease Driven by High-Fat Diet in Mice" Nutrients 16, no. 5: 691. https://doi.org/10.3390/nu16050691
APA StyleEritja, À., Caus, M., Belmonte, T., de Gonzalo-Calvo, D., García-Carrasco, A., Martinez, A., Martínez, M., & Bozic, M. (2024). microRNA Expression Profile in Obesity-Induced Kidney Disease Driven by High-Fat Diet in Mice. Nutrients, 16(5), 691. https://doi.org/10.3390/nu16050691







