Effects of Pterostilbene on the Cell Division Cycle of a Neuroblastoma Cell Line
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. RNA Extraction, and Cyclin Expression Analysis
2.3. Indirect Immunofluorescence Analysis
2.4. Statistical Analysis
2.5. Genomic DNA Isolation
2.6. Epigenomic Assessment of DNA Methylation
3. Results
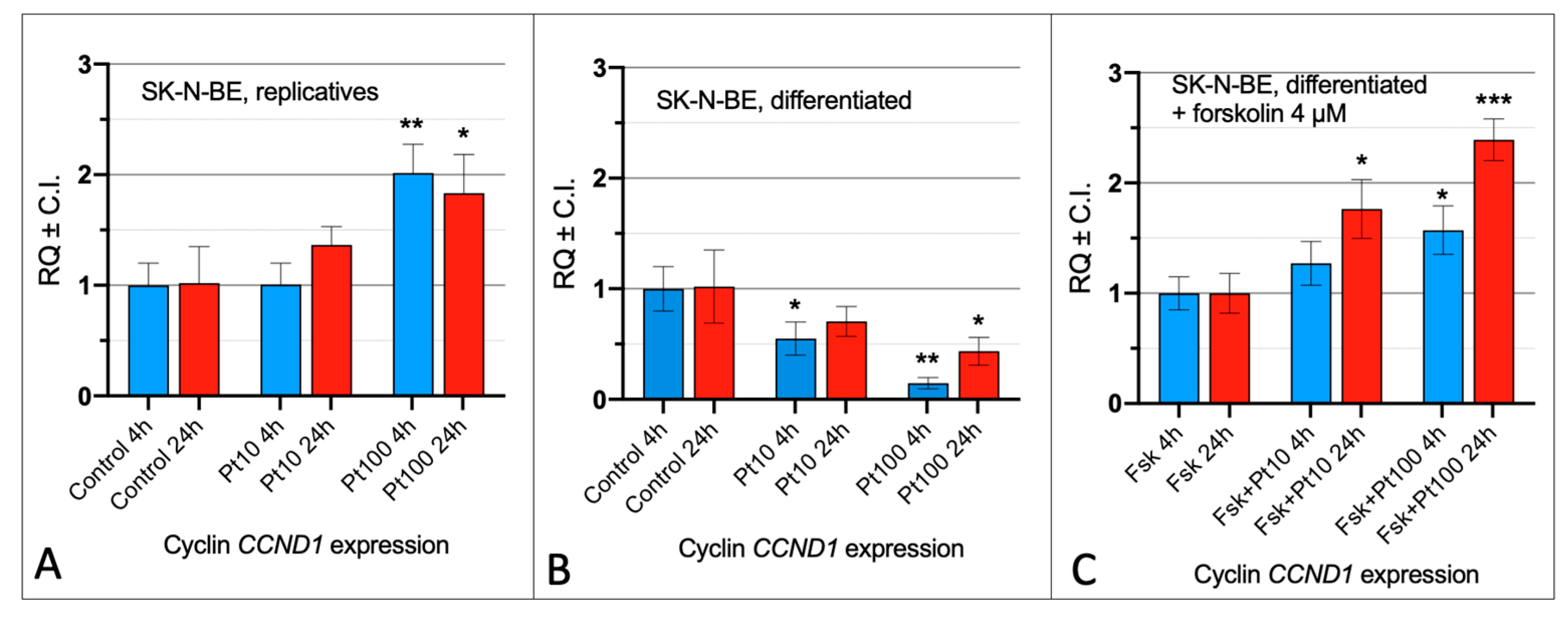
3.1. Effect of Pterostilbene on the Cell Cycle by Cyclin CCND1 Expression
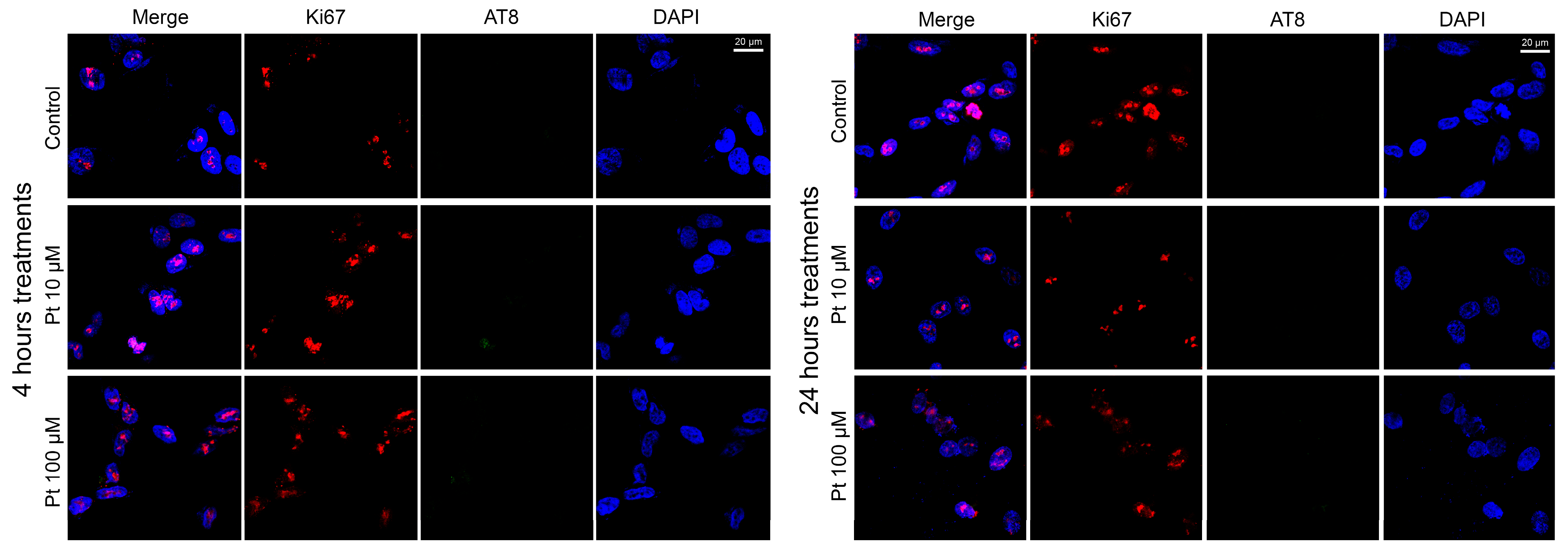
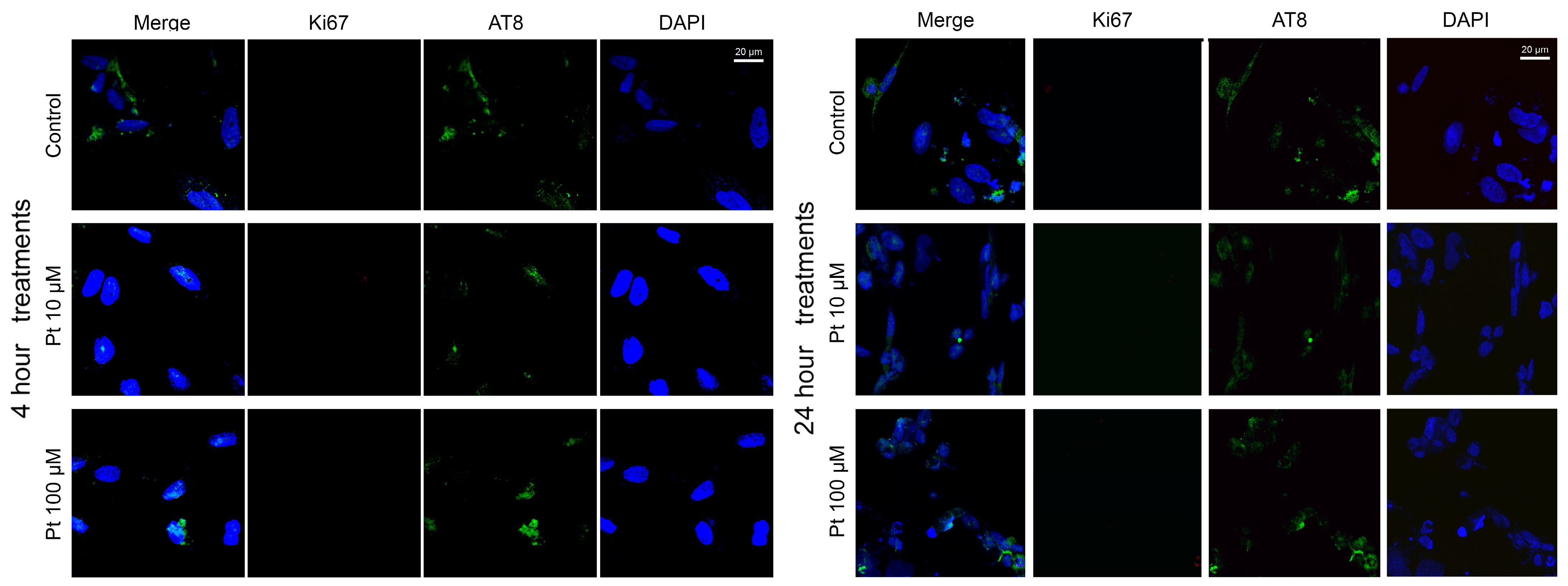
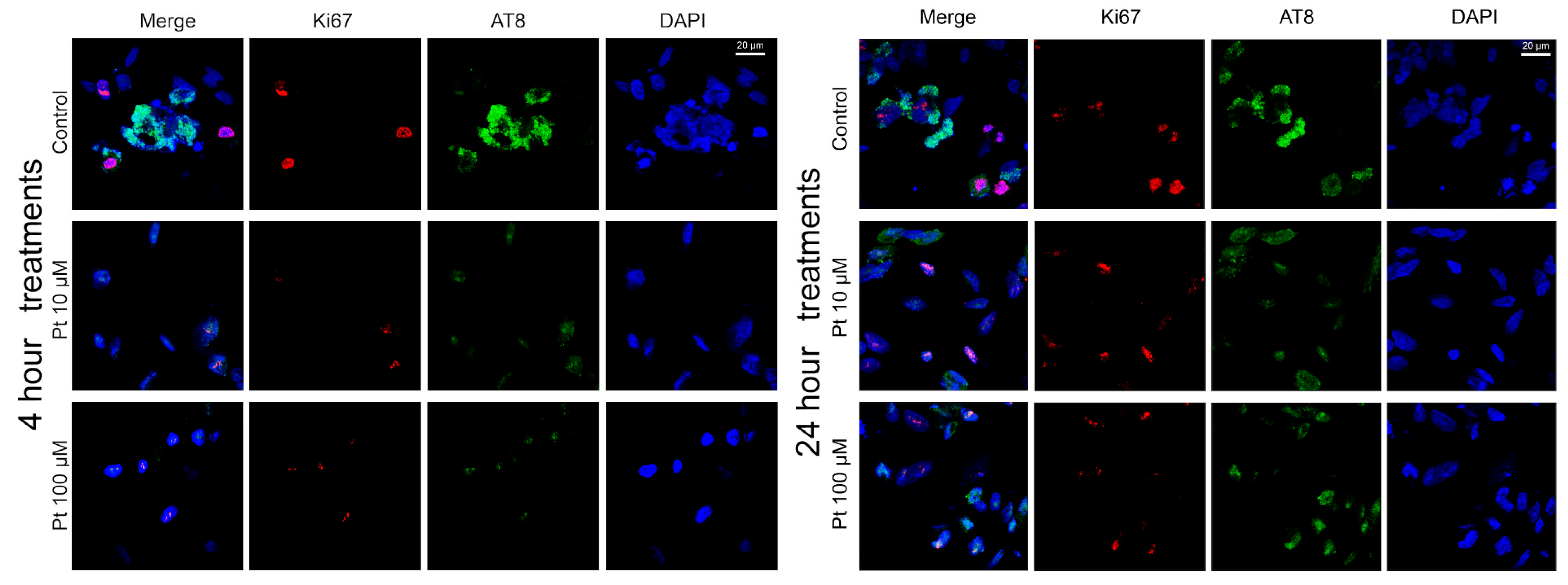
3.2. Effect of Pterostilbene on the Cell Cycle by Detection of Ki67 and AT8 Markers
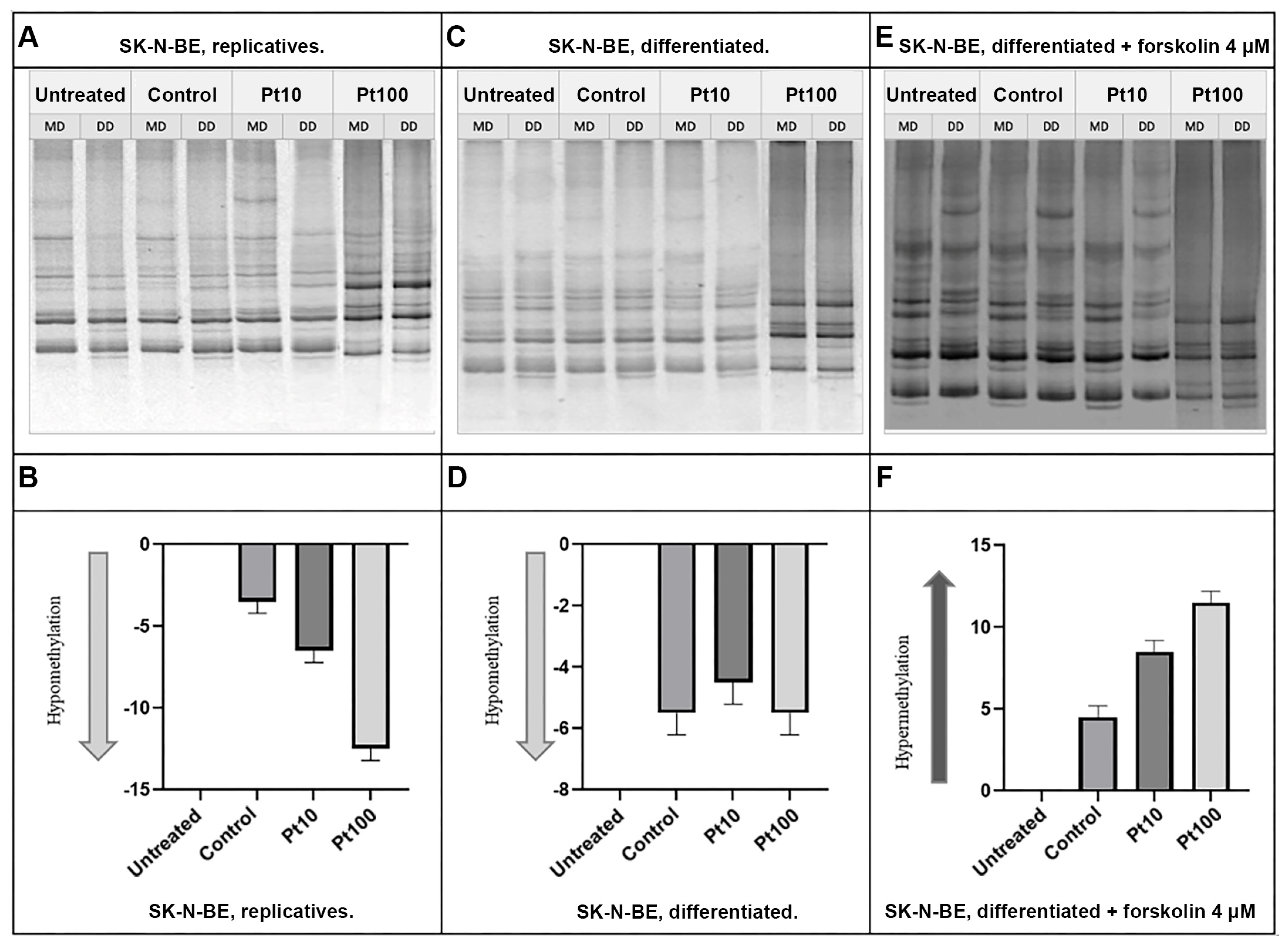
3.3. Effects of Pterostilbene on the Genome-Wide DNA Methylation Pattern
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Z.; Zhang, Z.; Ren, Y.; Wang, Y.; Fang, J.; Yue, H.; Ma, S.; Guan, F. Aging and Age-related Diseases: From Mechanisms to Therapeutic Strategies. Biogerontology 2021, 22, 165–187. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Jha, A.; Ambasta, R.K.; Kumar, P. Regulatory Mechanism of Cyclins and Cyclin-Dependent Kinases in Post-Mitotic Neuronal Cell Division. Life Sci. 2021, 285, 120006. [Google Scholar] [CrossRef] [PubMed]
- Ashrafian, H.; Zadeh, E.H.; Khan, R.H. Review on Alzheimer’s Disease: Inhibition of Amyloid Beta and Tau Tangle Formation. Int. J. Biol. Macromol. 2021, 167, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.R.; Barbosa, D.J.; Remião, F.; Silva, R. Alzheimer’s Disease: Insights and New Prospects in Disease Pathophysiology, Biomarkers and Disease-Modifying Drugs. Biochem. Pharmacol. 2023, 211, 115522. [Google Scholar] [CrossRef]
- Frost, B. Alzheimer’s Disease and Related Tauopathies: Disorders of Disrupted Neuronal Identity. Trends Neurosci. 2023, 46, 797–813. [Google Scholar] [CrossRef]
- Zhu, X.; Lee, H.; Perry, G.; Smith, M.A. Alzheimer Disease, the Two-Hit Hypothesis: An Update. Biochim. Et Biophys. Acta (BBA)—Mol. Basis Dis. 2007, 1772, 494–502. [Google Scholar] [CrossRef]
- Joseph, C.; Mangani, A.S.; Gupta, V.; Chitranshi, N.; Shen, T.; Dheer, Y.; Kb, D.; Mirzaei, M.; You, Y.; Graham, S.L.; et al. Cell Cycle Deficits in Neurodegenerative Disorders: Uncovering Molecular Mechanisms to Drive Innovative Therapeutic Development. Aging Dis. 2020, 11, 946. [Google Scholar] [CrossRef]
- Zhang, X.; Song, S.; Peng, W. Cell Cycle Deregulation in Neurodegenerative Diseases. Int. J. Neurosci. 2023, 133, 408–416. [Google Scholar] [CrossRef]
- Moh, C.; Kubiak, J.Z.; Bajic, V.P.; Zhu, X.; Smith, M.A.; Lee, H. Cell Cycle Deregulation in the Neurons of Alzheimer’s Disease. In Cell Cycle in Development; Kubiak, J.Z., Ed.; Results and Problems in Cell Differentiation; Springer: Berlin/Heidelberg, Germany, 2011; pp. 565–576. ISBN 978-3-642-19064-3. [Google Scholar]
- Kozlov, S.; Afonin, A.; Evsyukov, I.; Bondarenko, A. Alzheimer’s Disease: As It Was in the Beginning. Rev. Neurosci. 2017, 28, 825–843. [Google Scholar] [CrossRef]
- Koseoglu, M.M.; Norambuena, A.; Sharlow, E.R.; Lazo, J.S.; Bloom, G.S. Aberrant Neuronal Cell Cycle Re-Entry: The Pathological Confluence of Alzheimer’s Disease and Brain Insulin Resistance, and Its Relation to Cancer. J. Alzheimers Dis. 2019, 67, 1–11. [Google Scholar] [CrossRef]
- Van Leeuwen, L.A.G.; Hoozemans, J.J.M. Physiological and Pathophysiological Functions of Cell Cycle Proteins in Post-Mitotic Neurons: Implications for Alzheimer’s Disease. Acta Neuropathol. 2015, 129, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Gil, L.; Federico, C.; Pinedo, F.; Bruno, F.; Rebolledo, A.B.; Montoya, J.J.; Olazabal, I.M.; Ferrer, I.; Saccone, S. Aging Dependent Effect of Nuclear Tau. Brain Res. 2017, 1677, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Federico, C.; Gil, L.; Bruno, F.; D’Amico, A.G.; D’Agata, V.; Saccone, S. Phosphorylated Nucleolar Tau Protein Is Related to the Neuronal in Vitro Differentiation. Gene 2018, 664, 1–11. [Google Scholar] [CrossRef]
- Younas, N.; Saleem, T.; Younas, A.; Zerr, I. Nuclear Face of Tau: An inside Player in Neurodegeneration. Acta Neuropathol. Commun. 2023, 11, 196. [Google Scholar] [CrossRef]
- Sturiale, V.; Bruno, F.; Brancato, D.; D’Amico, A.G.; Maugeri, G.; D’Agata, V.; Saccone, S.; Federico, C. Cell Cycle Reactivation, at the Start of Neurodegeneration, Induced by Forskolin and Aniline in Differentiated Neuroblastoma Cells. Int. J. Mol. Sci. 2023, 24, 14373. [Google Scholar] [CrossRef]
- Lange, K.W.; Li, S. Resveratrol, Pterostilbene, and Dementia. BioFactors 2018, 44, 83–90. [Google Scholar] [CrossRef]
- Volpes, S.; Cruciata, I.; Ceraulo, F.; Schimmenti, C.; Naselli, F.; Pinna, C.; Mauro, M.; Picone, P.; Dallavalle, S.; Nuzzo, D.; et al. Nutritional Epigenomic and DNA-Damage Modulation Effect of Natural Stilbenoids. Sci. Rep. 2023, 13. [Google Scholar] [CrossRef]
- Mohd Sairazi, N.S.; Sirajudeen, K.N.S. Natural Products and Their Bioactive Compounds: Neuroprotective Potentials against Neurodegenerative Diseases. Evid.-Based Complement. Altern. Med. 2020, 2020, 1–30. [Google Scholar] [CrossRef]
- Noori, T.; Dehpour, A.R.; Sureda, A.; Sobarzo-Sanchez, E.; Shirooie, S. Role of Natural Products for the Treatment of Alzheimer’s Disease. Eur. J. Pharmacol. 2021, 898, 173974. [Google Scholar] [CrossRef]
- Duta-Bratu, C.-G.; Nitulescu, G.M.; Mihai, D.P.; Olaru, O.T. Resveratrol and Other Natural Oligomeric Stilbenoid Compounds and Their Therapeutic Applications. Plants 2023, 12, 2935. [Google Scholar] [CrossRef]
- Surien, O.; Masre, S.F.; Basri, D.F.; Ghazali, A.R. Potential Chemopreventive Role of Pterostilbene in Its Modulation of the Apoptosis Pathway. Int. J. Mol. Sci. 2023, 24, 9707. [Google Scholar] [CrossRef] [PubMed]
- Akinwumi, B.; Bordun, K.-A.; Anderson, H. Biological Activities of Stilbenoids. Int. J. Mol. Sci. 2018, 19, 792. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.; Lin, C. Effect of Resveratrol and Pterostilbene on Aging and Longevity. BioFactors 2018, 44, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y.-C.; Ho, C.-T.; Pan, M.-H. Recent Advances in Health Benefits of Stilbenoids. J. Agric. Food Chem. 2021, 69, 10036–10057. [Google Scholar] [CrossRef]
- Milenkovic, D.; Ruskovska, T. Mechanistic Insights into Dietary (Poly)Phenols and Vascular Dysfunction-Related Diseases Using Multi-Omics and Integrative Approaches: Machine Learning as a next Challenge in Nutrition Research. Mol. Asp. Med. 2023, 89, 101101. [Google Scholar] [CrossRef]
- Liu, Y.; You, Y.; Lu, J.; Chen, X.; Yang, Z. Recent Advances in Synthesis, Bioactivity, and Pharmacokinetics of Pterostilbene, an Important Analog of Resveratrol. Molecules 2020, 25, 5166. [Google Scholar] [CrossRef]
- Nagarajan, S.; Mohandas, S.; Ganesan, K.; Xu, B.; Ramkumar, K.M. New Insights into Dietary Pterostilbene: Sources, Metabolism, and Health Promotion Effects. Molecules 2022, 27, 6316. [Google Scholar] [CrossRef]
- Kapetanovic, I.M.; Muzzio, M.; Huang, Z.; Thompson, T.N.; McCormick, D.L. Pharmacokinetics, Oral Bioavailability, and Metabolic Profile of Resveratrol and Its Dimethylether Analog, Pterostilbene, in Rats. Cancer Chemother. Pharmacol. 2011, 68, 593–601. [Google Scholar] [CrossRef]
- Liu, H.; Wu, X.; Luo, J.; Wang, X.; Guo, H.; Feng, D.; Zhao, L.; Bai, H.; Song, M.; Liu, X.; et al. Pterostilbene Attenuates Astrocytic Inflammation and Neuronal Oxidative Injury After Ischemia-Reperfusion by Inhibiting NF-κB Phosphorylation. Front. Immunol. 2019, 10, 2408. [Google Scholar] [CrossRef]
- Ciccone, L.; Nencetti, S.; Marino, M.; Battocchio, C.; Iucci, G.; Venditti, I.; Marsotto, M.; Montalesi, E.; Socci, S.; Bargagna, B.; et al. Pterostilbene Fluorescent Probes as Potential Tools for Targeting Neurodegeneration in Biological Applications. J. Enzym. Inhib. Med. Chem. 2022, 37, 1812–1820. [Google Scholar] [CrossRef]
- Islam, F.; Nafady, M.H.; Islam, M.R.; Saha, S.; Rashid, S.; Akter, A.; Or-Rashid, M.H.; Akhtar, M.F.; Perveen, A.; Md Ashraf, G.; et al. Resveratrol and Neuroprotection: An Insight into Prospective Therapeutic Approaches against Alzheimer’s Disease from Bench to Bedside. Mol. Neurobiol. 2022, 59, 4384–4404. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Zhang, L.; Wang, L. Pterostilbene as a Therapeutic Alternative for Central Nervous System Disorders: A Review of the Current Status and Perspectives. J. Agric. Food Chem. 2023, 71, 14432–14457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Tang, Z. Therapeutic Potential of Natural Molecules against Alzheimer’s Disease via SIRT1 Modulation. Biomed. Pharmacother. 2023, 161, 114474. [Google Scholar] [CrossRef] [PubMed]
- De Jager, P.L.; Srivastava, G.; Lunnon, K.; Burgess, J.; Schalkwyk, L.C.; Yu, L.; Eaton, M.L.; Keenan, B.T.; Ernst, J.; McCabe, C.; et al. Alzheimer’s Disease: Early Alterations in Brain DNA Methylation at ANK1, BIN1, RHBDF2 and Other Loci. Nat. Neurosci. 2014, 17, 1156–1163. [Google Scholar] [CrossRef]
- Pihlstrøm, L.; Berge, V.; Rengmark, A.; Toft, M. Parkinson’s Disease Correlates with Promoter Methylation in the A-synuclein Gene. Mov. Disord. 2015, 30, 577–580. [Google Scholar] [CrossRef]
- Charlton, C.G.; Crowell, B. Striatal Dopamine Depletion, Tremors, and Hypokinesia Following the Intracranial Injection ofS-Adenosylmethionine: A Possible Role of Hypermethylation in Parkinsonism. Mol. Chem. Neuropathol. 1995, 26, 269–284. [Google Scholar] [CrossRef]
- Fernández-Santiago, R.; Merkel, A.; Castellano, G.; Heath, S.; Raya, Á.; Tolosa, E.; Martí, M.-J.; Consiglio, A.; Ezquerra, M. Whole-Genome DNA Hyper-Methylation in iPSC-Derived Dopaminergic Neurons from Parkinson’s Disease Patients. Clin. Epigenet. 2019, 11, 108. [Google Scholar] [CrossRef]
- Biedler, J.L.; Roffler-Tarlov, S.; Schachner, M.; Freedman, L.S. Multiple Neurotransmitter Synthesis by Human Neuroblastoma Cell Lines and Clones. Cancer Res. 1978, 38, 3751–3757. [Google Scholar]
- Andres, D.; Keyser, B.M.; Petrali, J.; Benton, B.; Hubbard, K.S.; McNutt, P.M.; Ray, R. Morphological and Functional Differentiation in BE (2)-M17 Human Neuroblastoma Cells by Treatment with Trans-Retinoic Acid. BMC Neurosci. 2013, 14, 49. [Google Scholar] [CrossRef]
- Leotta, C.G.; Federico, C.; Brundo, M.V.; Tosi, S.; Saccone, S. HLXB9 Gene Expression, and Nuclear Location during In Vitro Neuronal Differentiation in the SK-N-BE Neuroblastoma Cell Line. PLoS ONE 2014, 9, e105481. [Google Scholar] [CrossRef][Green Version]
- Naselli, F.; Belshaw, N.J.; Gentile, C.; Tutone, M.; Tesoriere, L.; Livrea, M.A.; Caradonna, F. Phytochemical Indicaxanthin Inhibits Colon Cancer Cell Growth and Affects the DNA Methylation Status by Influencing Epigenetically Modifying Enzyme Expression and Activity. J. Nutr. Nutr. 2015, 8, 114–127. [Google Scholar] [CrossRef]
- Caradonna, F.; Cruciata, I.; Schifano, I.; La Rosa, C.; Naselli, F.; Chiarelli, R.; Perrone, A.; Gentile, C. Methylation of cytokines gene promoters in IL-1β-treated human intestinal epithelial cells. Inflamm. Res. 2018, 67, 327–337. [Google Scholar] [CrossRef]
- Librizzi, M.; Chiarelli, R.; Bosco, L.; Sansook, S.; Gascon, J.M.; Spencer, J.; Caradonna, F.; Luparello, C. The Histone Deacetylase Inhibitor JAHA Down-Regulates pERK and Global DNA Methylation in MDA-MB231 Breast Cancer Cells. Materials 2015, 8, 7041–7047. [Google Scholar] [CrossRef]
- Tiwari, S.; Atluri, V.; Kaushik, A.; Yndart, A.; Nair, M. Alzheimer’s Disease: Pathogenesis, Diagnostics, and Therapeutics. IJN 2019, 14, 5541–5554. [Google Scholar] [CrossRef]
- Behrens, M.; Lendon, C.; Roe, C. A Common Biological Mechanism in Cancer and Alzheimers Disease? CAR 2009, 6, 196–204. [Google Scholar] [CrossRef]
- Cruickshanks, H.A.; Adams, P.D. Chromatin: A Molecular Interface between Cancer and Aging. Curr. Opin. Genet. Dev. 2011, 21, 100–106. [Google Scholar] [CrossRef]
- Driver, J.A.; Zhou, X.Z.; Lu, K.P. Regulation of Protein Conformation by Pin1 Offers Novel Disease Mechanisms and Therapeutic Approaches in Alzheimer’s Disease. Discov. Med. 2014, 17, 93–99. [Google Scholar]
- Choi, S.-W.; Friso, S. Epigenetics: A New Bridge between Nutrition and Health. Adv. Nutr. 2010, 1, 8–16. [Google Scholar] [CrossRef]
- Stallings, R.L.; Foley, N.H.; Bray, I.M.; Das, S.; Buckley, P.G. MicroRNA and DNA methylation alterations mediating retinoic acid induced neuroblastoma cell differentiation. Semin. Cancer Biol. 2011, 21, 283–290. [Google Scholar] [CrossRef]
- Orticello, M.; Cavallaro, R.A.; Antinori, D.; Raia, T.; Lucarelli, M.; Fuso, A. Amyloidogenic and Neuroinflammatory Molecular Pathways Are Contrasted Using Menaquinone 4 (MK4) and Reduced Menaquinone 7 (MK7R) in Association with Increased DNA Methylation in SK-N-BE Neuroblastoma Cell Line. Cells 2023, 13, 58. [Google Scholar] [CrossRef]
- Gerdes, J.; Lemke, H.; Baisch, H.; Wacker, H.-H.; Schwab, U.; Stein, H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 1984, 133, 1710–1715. [Google Scholar] [CrossRef] [PubMed]
- Fodder, K.; De Silva, R.; Warner, T.T.; Bettencourt, C. The Contribution of DNA Methylation to the (Dys)Function of Oligodendroglia in Neurodegeneration. Acta Neuropathol. Commun. 2023, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Stoccoro, A.; Coppedè, F. Mitochondrial DNA Methylation and Human Diseases. Int. J. Mol. Sci. 2021, 22, 4594. [Google Scholar] [CrossRef] [PubMed]
- Caradonna, F.; Barbata, G.; Sciandrello, G. Genomewide Hypomethylation and Pthrp Gene Hypermethylation as A Model for the Prediction of Cancer Risk in Rheumatoid Arthritis. In C. Luparello, Novel Aspects of PTHrP Physiopathology; Nova Science Publishers Inc.: New York, NY, USA, 2007; pp. 305–320. Available online: https://hdl.handle.net/10447/3857 (accessed on 25 October 2024).
- Fernández-Santiago, R.; Garrido, A.; Infante, J.; González-Aramburu, I.; Sierra, M.; Fernández, M.; Valldeoriola, F.; Muñoz, E.; Compta, Y.; Martí, M.; et al. A-synuclein (SNCA) but Not Dynamin 3 (DNM3) Influences Age at Onset of Leucine-rich Repeat Kinase 2 (LRRK2) Parkinson’s Disease in Spain. Mov. Disord. 2018, 33, 637–641. [Google Scholar] [CrossRef]
- Wang, P.; Sang, S. Metabolism and Pharmacokinetics of Resveratrol and Pterostilbene. BioFactors 2018, 44, 16–25. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bruno, F.; Naselli, F.; Brancato, D.; Volpes, S.; Cardinale, P.S.; Saccone, S.; Federico, C.; Caradonna, F. Effects of Pterostilbene on the Cell Division Cycle of a Neuroblastoma Cell Line. Nutrients 2024, 16, 4152. https://doi.org/10.3390/nu16234152
Bruno F, Naselli F, Brancato D, Volpes S, Cardinale PS, Saccone S, Federico C, Caradonna F. Effects of Pterostilbene on the Cell Division Cycle of a Neuroblastoma Cell Line. Nutrients. 2024; 16(23):4152. https://doi.org/10.3390/nu16234152
Chicago/Turabian StyleBruno, Francesca, Flores Naselli, Desiree Brancato, Sara Volpes, Paola Sofia Cardinale, Salvatore Saccone, Concetta Federico, and Fabio Caradonna. 2024. "Effects of Pterostilbene on the Cell Division Cycle of a Neuroblastoma Cell Line" Nutrients 16, no. 23: 4152. https://doi.org/10.3390/nu16234152
APA StyleBruno, F., Naselli, F., Brancato, D., Volpes, S., Cardinale, P. S., Saccone, S., Federico, C., & Caradonna, F. (2024). Effects of Pterostilbene on the Cell Division Cycle of a Neuroblastoma Cell Line. Nutrients, 16(23), 4152. https://doi.org/10.3390/nu16234152






