Extracellular Vesicles of the Probiotic Escherichia coli Nissle 1917 Reduce PepT1 Levels in IL-1β-Treated Caco-2 Cells via Upregulation of miR-193a-3p
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Isolation of EVs
2.2. Cell Culture and Stimulation Conditions
2.3. Cell Viability Assays
2.4. RNA Extraction and Quantitative Reverse Transcription-Polymerase Chain Reaction (RT-qPCR)
2.5. Quantification of Mature miRNA Expression
2.6. Quantification of Cytokines by ELISA
2.7. Quantification of PepT1 by ELISA
2.8. Immunofluorescence, Confocal Microscopy, and Image Analysis
2.9. Statistical Analysis
3. Results
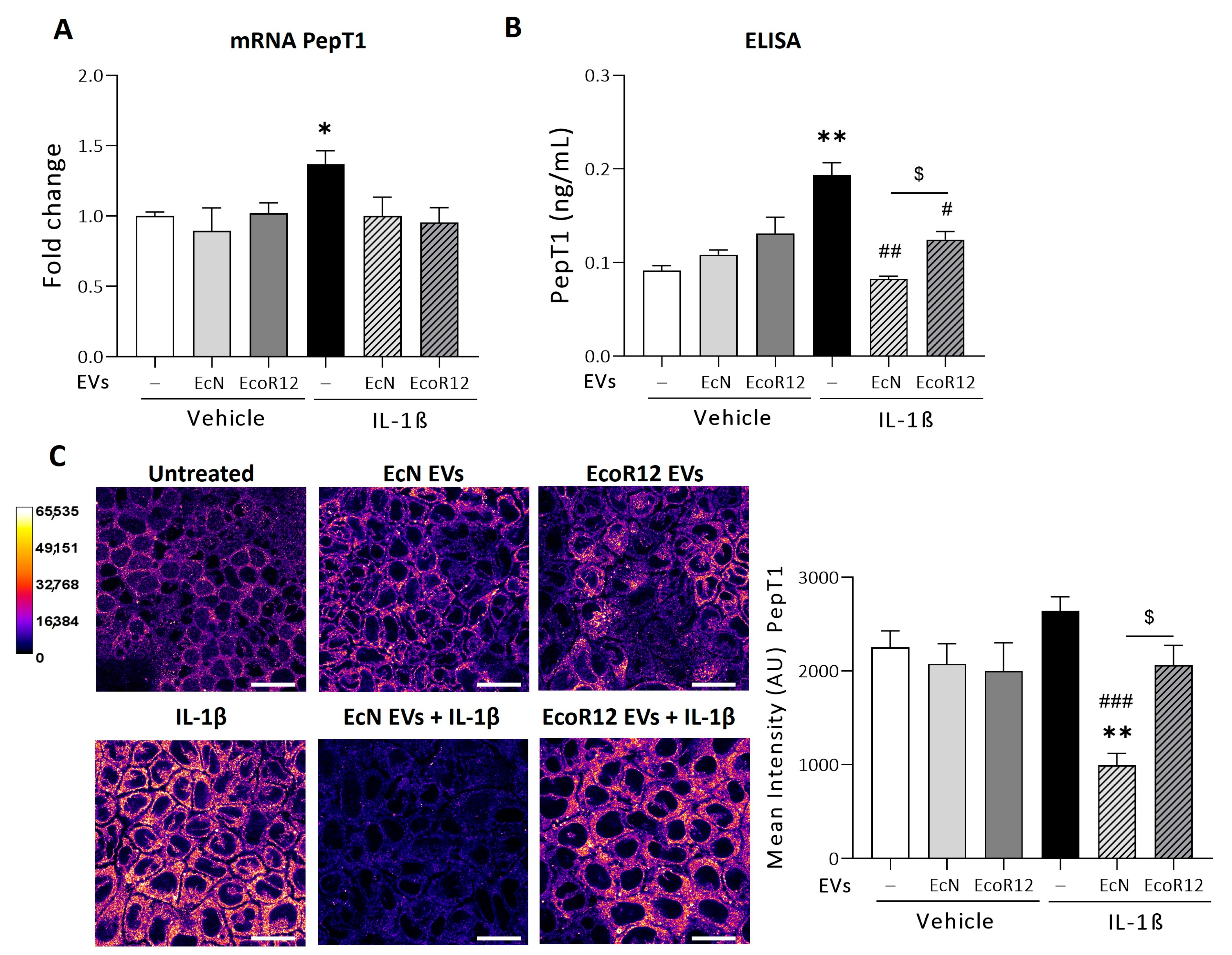
3.1. Modulation of PepT1 by EcN or EcoR12 EVs Under Conditions of IL-1β-Induced Inflammation
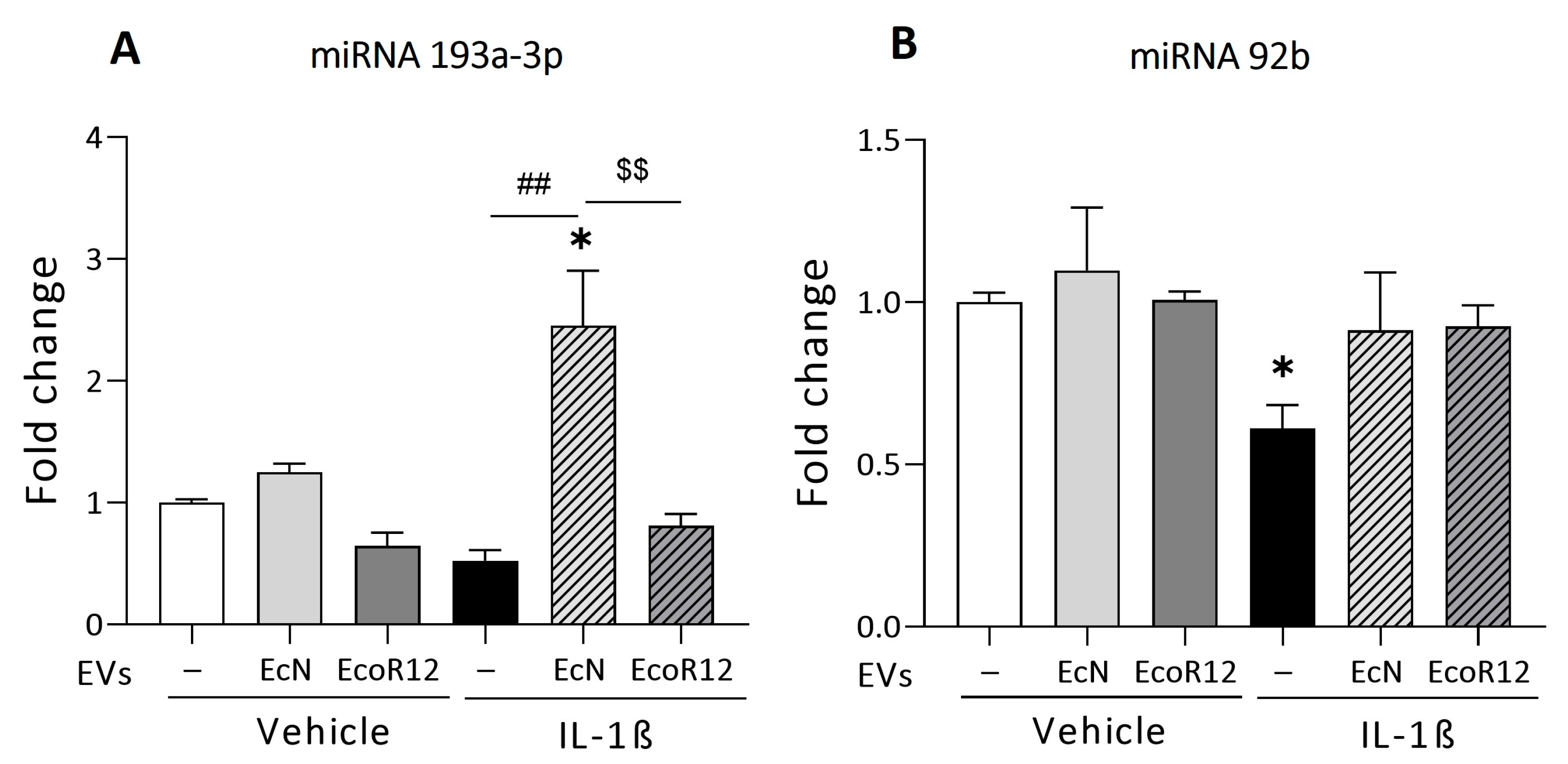
3.2. Regulation of miRNAs That Target PepT1 mRNA by EcN and EcoR12 EVs under Conditions of IL-1β-Induced Inflammation
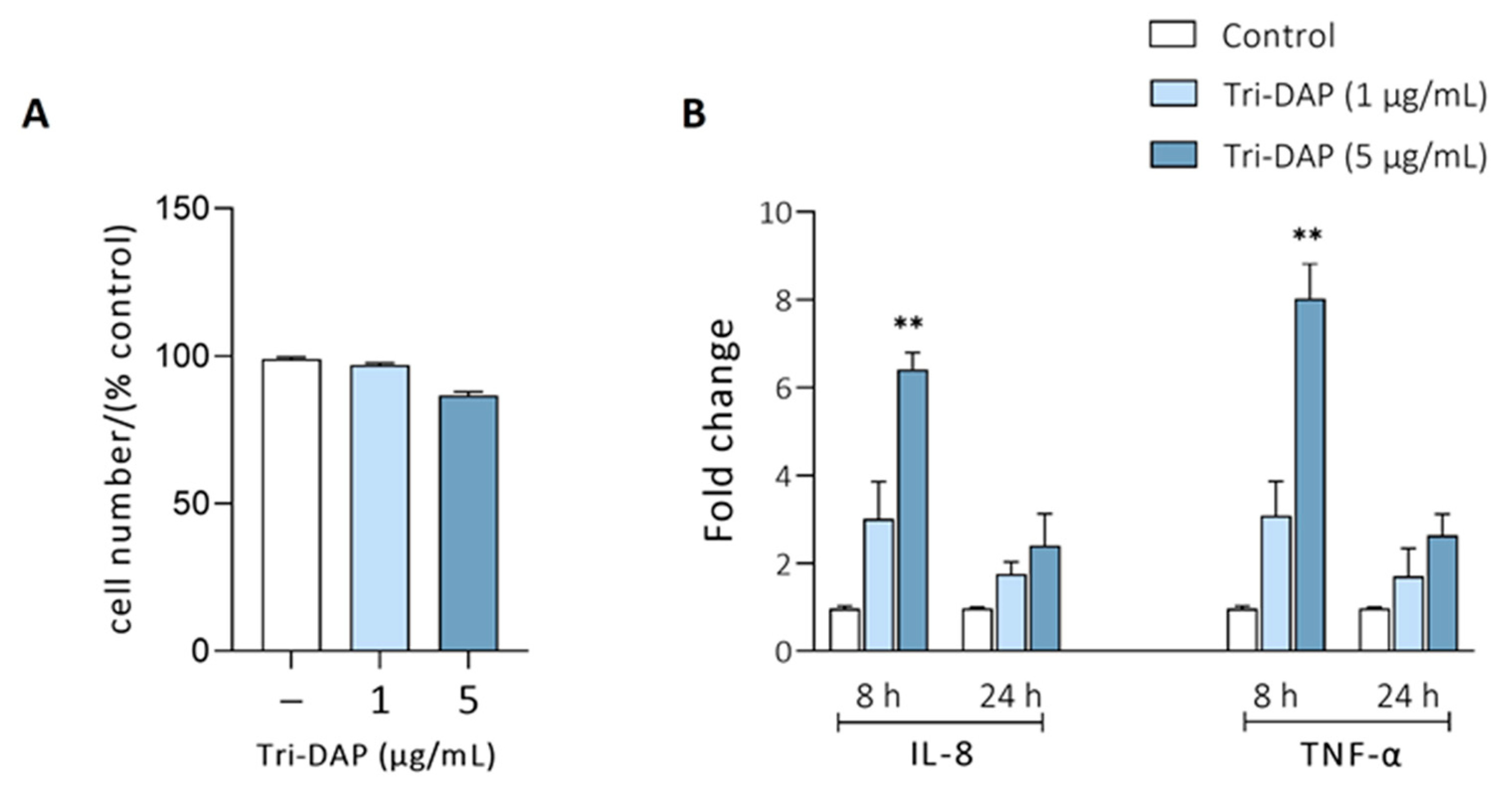
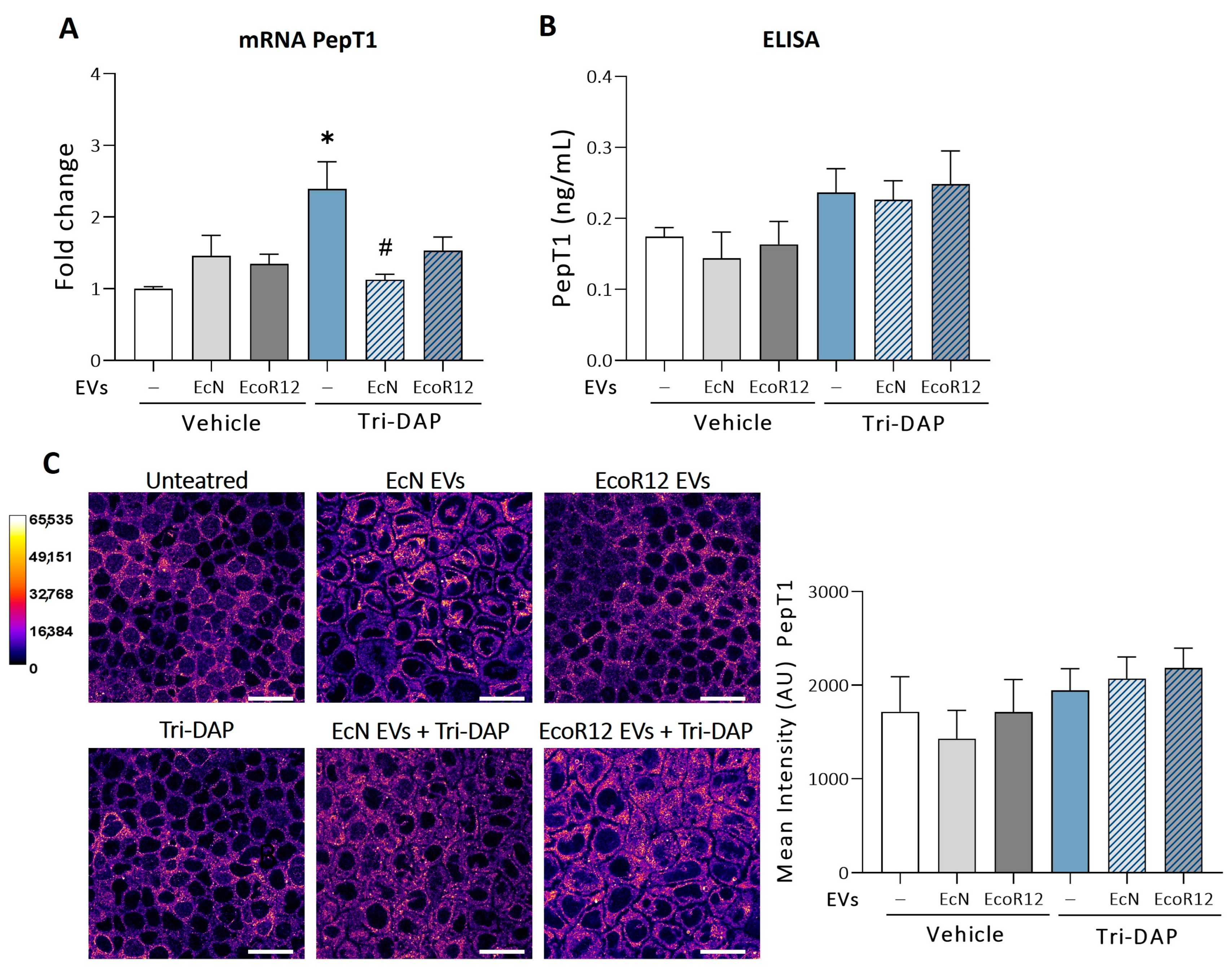
3.3. Effects of EcN and EcoR12 EVs on the Regulation of Tri-DAP Expression by Peptide Substrates
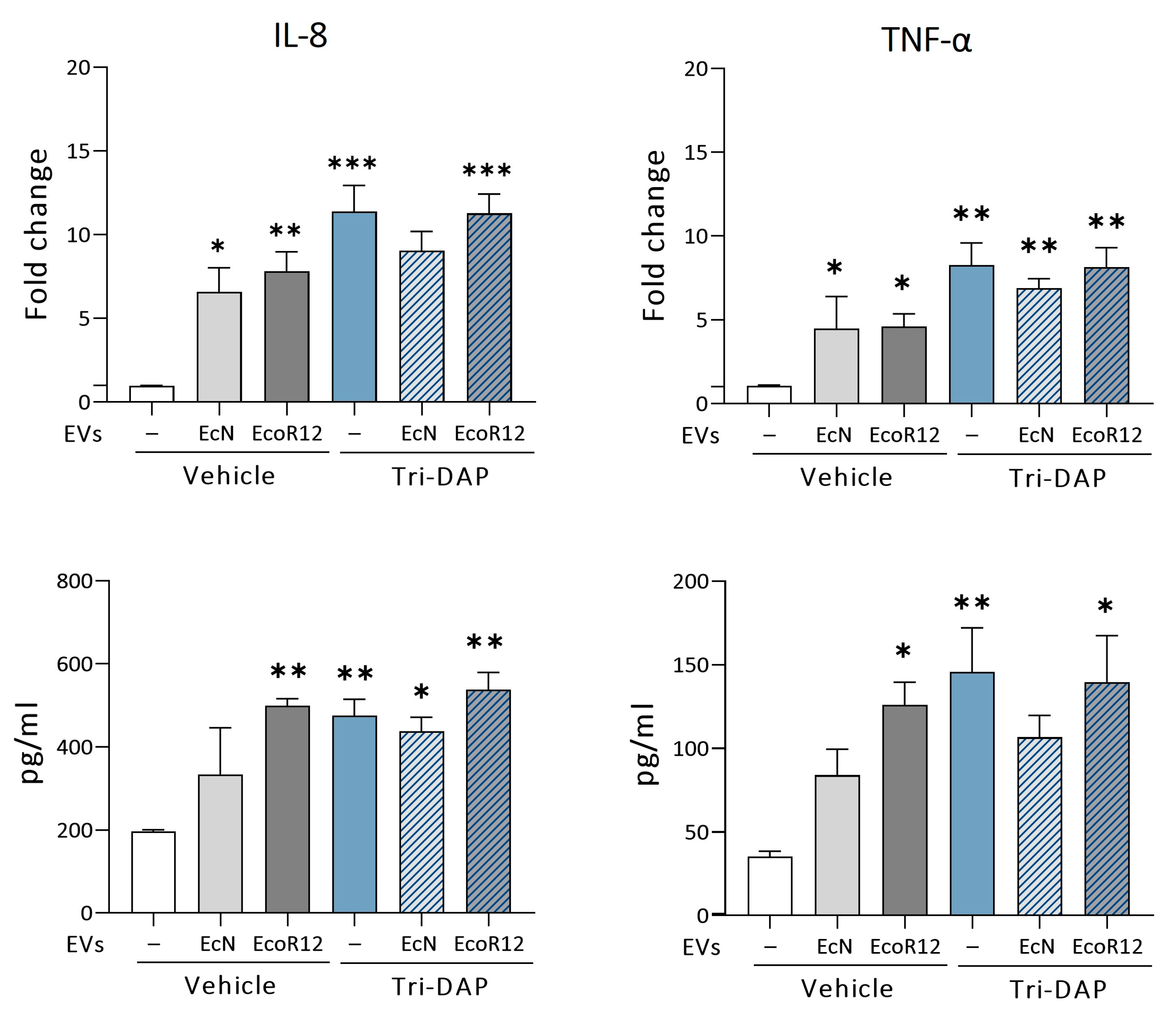
3.4. Modulation of Proinflammatory Cytokines by EcN or EcoR12 EVs in Caco-2 Cells Stimulated with Tri-DAP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Viennois, E.; Pujada, A.; Zen, J.; Merlin, D. Function, regulation, and pathophysiological relevance of the POT superfamily, specifically PepT1 in inflammatory bowel disease. Compr. Physiol. 2018, 8, 731. [Google Scholar] [CrossRef]
- Merlin, D.; Si-Tahar, M.; Sitaraman, S.V.; Eastburn, K.; Williams, I.; Liu, X.; Hediger, M.A.; Madara, J.L. Colonic epithelial hPepT1 expression occurs in inflammatory bowel disease: Transport of bacterial peptides influences expression of MHC class 1 molecules. Gastroenterology 2001, 120, 1666–1679. [Google Scholar] [CrossRef] [PubMed]
- Geillinger, K.E.; Kipp, A.P.; Schink, K.; Röder, P.V.; Spanier, B.; Daniel, H. Nrf2 regulates the expression of the peptide transporter PEPT1 in the human colon carcinoma cell line Caco-2. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 1747–1754. [Google Scholar] [CrossRef]
- Spanier, B.; Rohm, F. Proton coupled oligopeptide transporter 1 (PepT1) function, regulation, and influence on the intestinal homeostasis. Compr. Physiol. 2018, 8, 843–869. [Google Scholar] [CrossRef] [PubMed]
- Ingersoll, S.A.; Ayyadurai, S.; Charania, M.A.; Laroui, H.; Yan, Y.; Merlin, D. The role and pathophysiological relevance of membrane transporter pept1 in intestinal inflammation and inflammatory bowel disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G484–G492. [Google Scholar] [CrossRef]
- Viennois, E.; Ingersoll, S.A.; Ayyadurai, S.; Zhao, Y.; Wang, L.; Zhang, M.; Han, M.K.; Garg, P.; Xiao, B.; Merlin, D. Critical Role of PepT1 in Promoting Colitis-Associated Cancer and Therapeutic Benefits of the Anti-inflammatory PepT1-Mediated Tripeptide KPV in a Murine Model. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 340–357. [Google Scholar] [CrossRef]
- Viennois, E.; Pujada, A.; Sung, J.; Yang, C.; Gewirtz, A.T.; Chassaing, B.; Merlin, D. Impact of PepT1 deletion on microbiota composition and colitis requires multiple generations. NPJ Biofilms Microbiomes 2020, 6, 27. [Google Scholar] [CrossRef]
- Newstead, S. Recent advances in understanding proton coupled peptide transport via the POT family. Curr. Opin. Struct. Biol. 2017, 45, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Viennois, E.; Zhang, M.; Xiao, B.; Han, M.K.; Walter, L.; Garg, P.; Merlin, D. PepT1 Expression Helps Maintain Intestinal Homeostasis by Mediating the Differential Expression of miRNAs along the Crypt-Villus Axis. Sci. Rep. 2016, 6, 27119. [Google Scholar] [CrossRef]
- Wang, C.Y.; Liu, S.; Xie, X.N.; Tan, Z.R. Regulation profile of the intestinal peptide transporter 1 (PepT1). Drug Des. Devel. Ther. 2017, 11, 3511–3517. [Google Scholar] [CrossRef]
- Dalmasso, G.; Nguyen, H.T.; Charrier-Hisamuddin, L.; Yan, Y.; Laroui, H.; Demoulin, B.; Sitaraman, S.V.; Merlin, D. PepT1 mediates transport of the proinflammatory bacterial tripeptide L-Ala-γ-D-Glu-meso-DAP in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G687–G696. [Google Scholar] [CrossRef]
- Laroui, H.; Yan, Y.; Narui, Y.; Ingersoll, S.A.; Ayyadurai, S.; Charania, M.A.; Zhou, F.; Wang, B.; Salaita, K.; Sitaraman, S.V.; et al. L-Ala-γ-D-Glu-meso-diaminopimelic acid (DAP) interacts directly with leucine-rich region domain of nucleotide-binding oligomerization domain 1, increasing phosphorylation activity of receptor-interacting serine/threonine- protein kinase 2 and its interaction with nucleotide-binding oligomerization domain 1. J. Biol. Chem. 2011, 286, 31003–31013. [Google Scholar] [CrossRef] [PubMed]
- Dalmasso, G.; Nguyen, H.T.; Ingersoll, S.A.; Ayyadurai, S.; Laroui, H.; Charania, M.A.; Yan, Y.; Sitaraman, S.V.; Merlin, D. The PepT1-NOD2 signaling pathway aggravates induced colitis in mice. Gastroenterology 2011, 141, 1334–1345. [Google Scholar] [CrossRef]
- Caruso, R.; Warner, N.; Inohara, N.; Núñez, G. NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Wuensch, T.; Ullrich, S.; Schulz, S.; Chamaillard, M.; Schaltenberg, N.; Rath, E.; Goebel, U.; Sartor, R.B.; Prager, M.; Büning, C.; et al. Colonic expression of the peptide transporter PEPT1 is downregulated during intestinal inflammation and is not required for NOD2-dependent immune activation. Inflamm. Bowel. Dis. 2014, 20, 671–684. [Google Scholar] [CrossRef]
- Dai, X.; Chen, X.; Chen, Q.; Shi, L.; Liang, H.; Zhou, Z.; Liu, Q.; Pang, W.; Hou, D.; Wang, C.; et al. MicroRNA-193a-3p reduces intestinal inflammation in response to microbiota via down-regulation of colonic PepT1. J. Biol. Chem. 2015, 290, 99–115. [Google Scholar] [CrossRef]
- Buyse, M.; Tsocas, A.; Walker, F.; Merlin, D.; Bado, A. PepT1-mediated fMLP transport induces intestinal inflammation in vivo. Am. J. Physiol. Cell Physiol. 2002, 283, C1795–C1800. [Google Scholar] [CrossRef]
- Sonnenborn, U.; Schulze, J. The non-pathogenic Escherichia coli strain Nissle 1917-features of a versatile probiotic. Microb. Ecol. Health Dis. 2009, 21, 122–158. [Google Scholar] [CrossRef]
- Jacobi, C.A.; Malfertheiner, P. Escherichia coli nissle 1917 (Mutaflor): New insights into an old probiotic bacterium. Dig. Dis. 2011, 29, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Behnsen, J.; Deriu, E.; Sassone-Corsi, M.; Raffatellu, M. Probiotics: Properties, examples, and specific applications. Cold Spring Harb. Perspect. Biol. 2013, 3, a010074. [Google Scholar] [CrossRef]
- Ma, L.; Tu, H.; Chen, T. Postbiotics in Human Health: A Narrative Review. Nutrients 2023, 15, 291. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef] [PubMed]
- Olovo, C.V.; Wiredu Ocansey, D.K.; Ji, Y.; Huang, X.; Xu, M. Bacterial membrane vesicles in the pathogenesis and treatment of inflammatory bowel disease. Gut Microbes 2024, 16, 2341670. [Google Scholar] [CrossRef]
- González-Lozano, E.; García-García, J.; Gálvez, J.; Hidalgo-García, L.; Rodríguez-Nogales, A.; Rodríguez-Cabezas, M.E.; Sánchez, M. Novel Horizons in Postbiotics: Lactobacillaceae Extracellular Vesicles and Their Applications in Health and Disease. Nutrients 2022, 14, 5296. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Lin, T.L.; Tsai, Y.L.; Wu, T.R.; Lai, W.F.; Lu, C.C.; Lai, H.C. Next generation probiotics in disease amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Garrido, N.; Badia, J.; Baldomà, L. Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J. Extracell Vesicles 2021, 10, e12161. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.S.; Badia, J.; Bosch, M.; Giménez, R.; Baldomà, L. Outer Membrane Vesicles and Soluble Factors Released by Probiotic Escherichia coli Nissle 1917 and Commensal ECOR63 Enhance Barrier Function by Regulating Expression of Tight Junction Proteins in Intestinal Epithelial Cells. Front. Microbiol. 2016, 7, 1981. [Google Scholar] [CrossRef]
- Diaz-Garrido, N.; Badia, J.; Baldomà, L. Modulation of Dendritic Cells by Microbiota Extracellular Vesicles Influences the Cytokine Profile and Exosome Cargo. Nutrients 2022, 14, 344. [Google Scholar] [CrossRef]
- Cañas, M.A.; Fábrega, M.J.; Giménez, R.; Badia, J.; Baldomà, L. Outer membrane vesicles from probiotic and commensal escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front. Microbiol. 2018, 9, 498. [Google Scholar] [CrossRef]
- Fábrega, M.J.; Rodríguez-Nogales, A.; Garrido-Mesa, J.; Algieri, F.; Badía, J.; Giménez, R.; Gálvez, J.; Baldomà, L. Intestinal Anti-inflammatory Effects of Outer Membrane Vesicles from Escherichia coli Nissle 1917 in DSS-Experimental Colitis in Mice. Front. Microbiol. 2017, 8, 1274. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ruiz, S.; Olivo-Martínez, Y.; Cordero, C.; Rodríguez-Lagunas, M.J.; Pérez-Cano, F.J.; Badia, J.; Baldoma, L. Microbiota-Derived Extracellular Vesicles as a Postbiotic Strategy to Alleviate Diarrhea and Enhance Immunity in Rotavirus-Infected Neonatal Rats. Int. J. Mol. Sci. 2024, 25, 1184. [Google Scholar] [CrossRef] [PubMed]
- Olivo-Martínez, Y.; Bosch, M.; Badia, J.; Baldomà, L. Modulation of the Intestinal Barrier Integrity and Repair by Microbiota Extracellular Vesicles through the Differential Regulation of Trefoil Factor 3 in LS174T Goblet Cells. Nutrients 2023, 15, 2437. [Google Scholar] [CrossRef] [PubMed]
- Olivo-Martínez, Y.; Martínez-Ruiz, S.; Cordero-Alday, C.; Bosch, M.; Badia, J.; Baldoma, L. Modulation of Serotonin-Related Genes by Extracellular Vesicles of the Probiotic Escherichia coli Nissle 1917 in the Interleukin-1β-Induced Inflammation Model of Intestinal Epithelial Cells. Int. J. Mol. Sci. 2024, 25, 5338. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Selander, R.K. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 1984, 157, 690–693. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Garrido, N.; Fábrega, M.J.; Vera, R.; Giménez, R.; Badia, J.; Baldomà, L. Membrane vesicles from the probiotic Nissle 1917 and gut resident Escherichia coli strains distinctly modulate human dendritic cells and subsequent T cell responses. J. Funct. Foods 2019, 61, 103495. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.K.; Wang, M.J. Image thresholding by minimizing the measures of fuzziness. Pattern Recognit. 1995, 28, 41–51. [Google Scholar] [CrossRef]
- Dalmasso, G.; Nguyen, H.T.; Yan, Y.; Laroui, H.; Charania, M.A.; Obertone, T.S.; Sitaraman, S.V.; Merlin, D. MicroRNA-92b regulates expression of the oligopeptide transporter PepT1 in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G52–G59. [Google Scholar] [CrossRef] [PubMed]
- Buyse, M.; Charrier, L.; Sitaraman, S.; Gewirtz, A.; Merlin, D. Interferon-γ Increases hPepT1-Mediated Uptake of Di-Tripeptides Including the Bacterial Tripeptide fMLP in Polarized Intestinal Epithelia. Am. J. Pathol. 2003, 163, 1969–1977. [Google Scholar] [CrossRef]
- Vavricka, S.R.; Musch, M.W.; Fujiya, M.; Kles, K.; Chang, L.; Eloranta, J.J.; Kullak-Ublick, G.A.; Drabik, K.; Merlin, D.; Chang, E.B. Tumor necrosis factor-α and interferon-γ increase PepT1 expression and activity in the human colon carcinoma cell line Caco-2/bbe and in mouse intestine. Pflugers Arch. 2006, 452, 71–80. [Google Scholar] [CrossRef]
- Wang, P.; Lu, Y.Q.; Wen, Y.; Yu, D.Y.; Ge, L.; Dong, W.R.; Xiang, L.X.; Shao, J.Z. IL-16 Induces Intestinal Inflammation via PepT1 Upregulation in a Pufferfish Model: New Insights into the Molecular Mechanism of Inflammatory Bowel Disease. J. Immunol. 2013, 191, 1413–1427. [Google Scholar] [CrossRef] [PubMed]
- Berezin, A.E.; Poplyonkin, E.I. Diagnostic and therapeutic value of micro-RNAs in inflammatory bowel disease. Biomed. Res. Ther. 2020, 7, 3622–3632. [Google Scholar] [CrossRef]
- Ayyadurai, S.; Charania, M.A.; Xiao, B.; Viennois, E.; Merlin, D. PepT1 expressed in immune cells has an important role in promoting the immune response during experimentally induced colitis. Lab. Investig. 2013, 93, 888–899. [Google Scholar] [CrossRef]
- Ayyadurai, S.; Charania, M.A.; Xiao, B.; Viennois, E.; Zhang, Y.; Merlin, D. Colonic miRNA expression/secretion, regulated by intestinal epithelial PepT1, plays an important role in cell-to-cell communication during colitis. PLoS ONE 2014, 9, e87614. [Google Scholar] [CrossRef] [PubMed]
- Miyake, M.; Fujishima, M.; Nakai, D. Inhibitory potency of marketed drugs for ulcerative colitis and Crohn’s disease on PEPT1. Biol. Pharm. Bull. 2017, 40, 1572–1575. [Google Scholar] [CrossRef]
- Bruckmueller, H.; Martin, P.; Kähler, M.; Haenisch, S.; Ostrowski, M.; Drozdzik, M.; Siegmund, W.; Cascorbi, I.; Oswald, S. Clinically Relevant Multidrug Transporters Are Regulated by microRNAs along the Human Intestine. Mol. Pharm. 2017, 14, 2245–2253. [Google Scholar] [CrossRef] [PubMed]
- Cañas, M.A.; Giménez, R.; Fábrega, M.J.; Toloza, L.; Baldomà, L.; Badia, J. Outer Membrane Vesicles from the Probiotic Escherichia coli Nissle 1917 and the Commensal ECOR12 Enter Intestinal Epithelial Cells via Clathrin-Dependent Endocytosis and Elicit Differential Effects on DNA Damage. PLoS ONE 2016, 11, e0160374. [Google Scholar] [CrossRef] [PubMed]
- Petrosino, J.F. The microbiome in precision medicine: The way forward. Genome Med. 2018, 10, 12. [Google Scholar] [CrossRef]
- Behrouzi, A.; Nafari, A.H.; Siadat, S.D. The significance of microbiome in personalized medicine. Clin. Transl. Med. 2019, 8, 16. [Google Scholar] [CrossRef]
- Kotla, N.G.; Rochev, Y. IBD disease-modifying therapies: Insights from emerging therapeutics. Trends Mol. Med. 2023, 29, 241–253. [Google Scholar] [CrossRef]
- Long, A.G.; Lundsmith, E.T.; Hamilton, K.E. Inflammation and Colorectal Cancer. Curr. Colorectal. Cancer Rep. 2017, 13, 341–351. [Google Scholar] [CrossRef]
- Pesce, M.; Seguella, L.; Del Re, A.; Lu, J.; Palenca, I.; Corpetti, C.; Rurgo, S.; Sanseverino, W.; Sarnelli, G.; Esposito, G. Next-Generation Probiotics for Inflammatory Bowel Disease. Int. J. Mol. Sci. 2022, 23, 5466. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Esposito, A.; Ercolini, D. Outlook on next-generation probiotics from the human gut. Cell. Mol. Life Sci. 2022, 79, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.; Tong, X.; Wang, R.; Song, X. The clinical effects of probiotics for inflammatory bowel disease: A meta-analysis. Medicine 2018, 97, e13792. [Google Scholar] [CrossRef]
- Martyniak, A.; Medyńska-Przęczek, A.; Wędrychowicz, A.; Skoczeń, S.; Tomasik, P.J. Prebiotics, Probiotics, Synbiotics, Paraprobiotics and Postbiotic Compounds in IBD. Biomolecules 2021, 11, 1903. [Google Scholar] [CrossRef]
- Mosca, A.; Abreu YAbreu, A.T.; Gwee, K.A.; Ianiro, G.; Tack, J.; Nguyen, T.V.; Hill, C. The clinical evidence for postbiotics as microbial therapeutics. Gut Microbes 2022, 14, 2117508. [Google Scholar] [CrossRef]
- Wegh, C.A.M.; Geerlings, S.Y.; Knol, J.; Roeselers, G.; Belzer, C. Postbiotics and Their Potential Applications in Early Life Nutrition and Beyond. Int. J. Mol. Sci. 2019, 20, 4673. [Google Scholar] [CrossRef]
- Afzaal, M.; Saeed, F.; Bibi, M.; Ejaz, A.; Shah, Y.A.; Faisal, Z.; Ateeq, H.; Akram, N.; Asghar, A.; Shah, M.A. Nutritional, pharmaceutical, and functional aspects of rambutan in industrial perspective: An updated review. Food Sci. Nutr. 2023, 11, 3675–3685. [Google Scholar] [CrossRef]
- Yang, J.; Shin, T.-S.; Kim, J.S.; Jee, Y.-K.; Kim, Y.-K. A new horizon of precision medicine: Combination of the microbiome and extracellular vesicles. Exp. Mol. Med. 2022, 54, 466–482. [Google Scholar] [CrossRef]
- Xie, J.; Li, Q.; Nie, S. Bacterial extracellular vesicles: An emerging postbiotic. Trends Food Sci. Technol. 2024, 143, 104275. [Google Scholar] [CrossRef]
- Krzyżek, P.; Marinacci, B.; Vitale, I.; Grande, R. Extracellular Vesicles of Probiotics: Shedding Light on the Biological Activity and Future Applications. Pharmaceutics 2023, 15, 522. [Google Scholar] [CrossRef]
- Shen, Q.; Huang, Z.; Yao, J.; Jin, Y. Extracellular vesicles-mediated interaction within intestinal microenvironment in inflammatory bowel disease. J. Adv. Res. 2022, 37, 221–233. [Google Scholar] [CrossRef]
- Sultan, S.; Mottawea, W.; Yeo, J.; Hammami, R. Gut Microbiota Extracellular Vesicles as Signaling Molecules Mediating Host-Microbiota Communications. Int. J. Mol. Sci. 2021, 22, 13166. [Google Scholar] [CrossRef]
- Kang, C.S.; Ban, M.; Choi, E.J.; Moon, H.G.; Jeon, J.S.; Kim, D.K.; Park, S.K.; Jeon, S.G.; Roh, T.Y.; Myung, S.J.; et al. Extracellular Vesicles Derived from Gut Microbiota, Especially Akkermansia muciniphila, Protect the Progression of Dextran Sulfate Sodium-Induced Colitis. PLoS ONE 2013, 8, e76520. [Google Scholar] [CrossRef]
- Ma, L.; Shen, Q.; Lyu, W.; Lv, L.; Wang, W.; Yu, M.; Yang, H.; Tao, S.; Xiao, Y. Clostridium butyricum and Its Derived Extracellular Vesicles Modulate Gut Homeostasis and Ameliorate Acute Experimental Colitis. Microbiol. Spectr. 2022, 10, e01368-22. [Google Scholar] [CrossRef]
- Molina-Tijeras, J.A.; Gálvez, J.; Rodríguez-Cabezas, M.E. The immunomodulatory properties of extracellular vesicles derived from probiotics: A novel approach for the management of gastrointestinal diseases. Nutrients 2019, 11, 1038. [Google Scholar] [CrossRef]
- Chen, H.Q.; Shen, T.Y.; Zhou, Y.K.; Zhang, M.; Chu, Z.X.; Hang, X.M.; Qin, H.L. Lactobacillus plantarum consumption increases PepT1-mediated amino acid absorption by enhancing protein kinase C activity in spontaneously colitic mice. J. Nutr. 2010, 140, 2201–2206. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Q.; Yang, J.; Zhang, M.; Zhou, Y.K.; Shen, T.Y.; Chu, Z.X.; Zhang, M.; Hang, X.M.; Jiang, Y.Q.; Qin, H.L. Lactobacillus plantarum ameliorates colonic epithelial barrier dysfunction by modulating the apical junctional complex and PepT1 in IL-10 knockout mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G1287–G1297. [Google Scholar] [CrossRef]
- Lin, M.; Zhang, Z.; Gao, M.; Yu, H.; Sheng, H.; Huang, J. MicroRNA-193a-3p suppresses the colorectal cancer cell proliferation and progression through downregulating the PLAU expression. Cancer Manag. Res. 2019, 11, 5353–5363. [Google Scholar] [CrossRef] [PubMed]
- Pande, S.; Yang, X.; Friesel, R. Interleukin-17 receptor D (Sef) is a multi-functional regulator of cell signaling. Cell Commun. Signal. 2021, 19, 6. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olivo-Martínez, Y.; Martínez-Ruiz, S.; Cordero, C.; Badia, J.; Baldoma, L. Extracellular Vesicles of the Probiotic Escherichia coli Nissle 1917 Reduce PepT1 Levels in IL-1β-Treated Caco-2 Cells via Upregulation of miR-193a-3p. Nutrients 2024, 16, 2719. https://doi.org/10.3390/nu16162719
Olivo-Martínez Y, Martínez-Ruiz S, Cordero C, Badia J, Baldoma L. Extracellular Vesicles of the Probiotic Escherichia coli Nissle 1917 Reduce PepT1 Levels in IL-1β-Treated Caco-2 Cells via Upregulation of miR-193a-3p. Nutrients. 2024; 16(16):2719. https://doi.org/10.3390/nu16162719
Chicago/Turabian StyleOlivo-Martínez, Yenifer, Sergio Martínez-Ruiz, Cecilia Cordero, Josefa Badia, and Laura Baldoma. 2024. "Extracellular Vesicles of the Probiotic Escherichia coli Nissle 1917 Reduce PepT1 Levels in IL-1β-Treated Caco-2 Cells via Upregulation of miR-193a-3p" Nutrients 16, no. 16: 2719. https://doi.org/10.3390/nu16162719
APA StyleOlivo-Martínez, Y., Martínez-Ruiz, S., Cordero, C., Badia, J., & Baldoma, L. (2024). Extracellular Vesicles of the Probiotic Escherichia coli Nissle 1917 Reduce PepT1 Levels in IL-1β-Treated Caco-2 Cells via Upregulation of miR-193a-3p. Nutrients, 16(16), 2719. https://doi.org/10.3390/nu16162719






